英骏载体引物对照表
- 格式:xls
- 大小:101.50 KB
- 文档页数:2



*为必填测序服务客户送样登记表(生产填写)订单号
测序引物备注:1.如用他人引物,请注明他人姓名及引物名称;2.如片断长度大于800,仅填写“通用引物”但没有选择测通,默认两个反应3.自备引物如为插入片断中序列,请备注说明。
需合成测序引物信息(g小写,C/T/A字母大写;长度在17-25之间):
引物名称引物序列
引物名称引物序列
引物名称引物序列
引物名称引物序列
客户签字确认/日期:取样员签字/日期:
测序样品送样要求
友情提醒:1. 所有样品和引物均保存1个月,超过期限恕不提供加测以及返还服务。
2. 投诉期为一个月,如果对测序结果有任何异议,请及时联系;超过期限恕不接受。
3. 一般情况下:菌液和PCR未纯化48小时内完成测序,质粒和PCR已纯化24小时内完成测序。
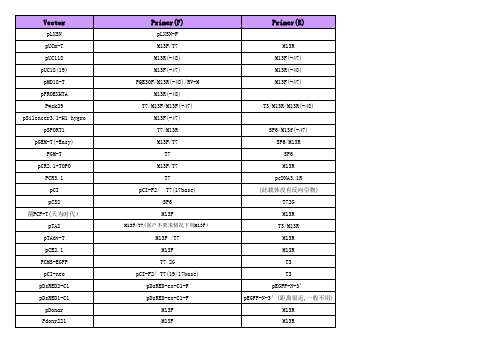
载体名称上游引物(5'primer)下游引物(3'primer)P3*FLAG-CMV CMV-30(825-854)CMV-24(1129-1152)pAc5.1/V5-His(_A,_B,_C) pAc5-5 BGH revpAcG2T pGEX-5 not availablepACT T7EEV T3/EBVrevpACT2GAL4 ADfor pGAL4-ADrvpACT2p17110p12584pACYC184(BamHI-site) PBRforBam PBRrevBampACYCDuet-1(MCSI)ACYCDuetUP1 Primer DuetDOWN1 pACYCDuet-1(MCSII) DuetUp2 T7-TerminatorpAdEasy-1 not available not availablepAdTrack-CMV not available not availablepAS2-1GAL4 Bdfor/P24990pGAL4-BDrv/P31430pB42AD pB42ADF PB42ADRpBABE pBMN_5' pBacPAK8BAC1BAC2pBAD pBAD-F pBAD-RpBAD/HisABC pBAD-5' pBBR1MCS T3/M13rev T7/M13forpBD-GAL4pBD-GAL4-F PBD-GAL4-RpBI12135S NOSpBIND GAL4-Bdfor T3,EBVrevpBK-CMV M13F/T7M13R/T3 pBluescriptIIKS(+/-) T3/M13rev T7/M13for(优先用M13F)pBluescriptIISK(+/-) T3/M13rev T7/M13for pBluescriptKS(+/-) T3/M13rev T7/M13for pBluescriptSK(+/-) T3/M13rev T7/M13forpBR322(BamHI-site) PBRforBam PBRrevBampBR322(EcoRI/HindIII-sites) pBRforEco pBRrevHindpBR325(BamHI-site) PBRforBam PBRrevBampBR329(BamHI-site) PBRforBam PBRrevBampBridge(MCSI) GAL4-Bdfor3'BDpBridge(MCSII) not available not availablepBS-T II T3/M13rev T7/M13forpBS(AMP)T7T3pBSR T3T7terpBudCE4.1(MCSI) CMV-Profor,T7 c-mycrev,EBVrevpBudCE4.1(MCSII)EF-1α Fwd BGHpBudCE4.1/lacZ/CAT CMV-Profor,T7 c-mycrev,EBVrevpBV220pBV220F PBV220RpCAL-c T7/Tac-Profor T7-TerminatorpCAL-n T7/Tac-Profor T7-Terminator pCAMBIA1301(1300)pCAMBIA1301-5'(ECORI)/M13R pCAMBIA1301-3'(HindIII) pCAMBIA1304pCAMBIA1301-5'(ECORI)pCAMBIA1301-3'(HindIII) pCAMBIA2300M13R(-48)M13F(-47)/M13F(-49) pCANTB5E S1S6pCAT3-enhancer RVP3此载体无反向引物pCDFDuet-1(MCSI)ACYCDuetUP1 Primer DuetDOWN1 pCDFDuet-1(MCSII)DuetUP2T7TERpcDNA1.1 CMV-Profor,T7 pCDM8-3pcDNA2.1 M13for/T7 M13rev/Tac-Profor pcDNA3 T7/CMV-Profor BGH rev/SP6pcDNA3.1(+/-) pcDNA3.1F/T7/CMV-Profor BGH revpcDNA3.1/His(_A,_B,_C) T7/CMV-Profor BGH revpcDNA3.1/myc-His(_A,_B,_C) T7/CMV-Profor BGH rev/c-mycrev pcDNA3.1/V5-His(_A,_B,_C) T7/CMV-Profor BGH rev/V5revpcDNA3.1/Hygro(+,-) T7/CMV-Profor BGH rev/V5revpcDNA3.1/Zeo(+.-) T7BGHrevpcDNA3.3TOPOTA CMV-Profor TKPAreversepcDNA4/HisMax(_A,_B,_C) Xpressfor BGH revpcDNA4/HisMax-TOPO Xpressfor BGH revpcDNA4/HisMax-TOPO/lacZ Xpressfor BGH revpcDNA4/myc-His(_A,_B,_C)T7,CMV-Profor BGH rev,c-mycrev pcDNA4/TO TREfor/CMV-Profor BGH revpcDNA4/V5-His(_A,_B,_C) CMV-Profor,T7 BGH rev,V5revpcDNA4-TET/ON/AMP+CMV-Profor pcDNA5/FRT T7/CMV-Profor BGH revpcDNA6/myc-His(_A,_B,_C) CMV-Profor,T7 BGH rev,c-mycrev pcDNA6/V5-His(_A,_B,_C) CMV-Profor,T7 BGH rev,V5revpCEP4pCEP-F/CMV-Profor EBVrevpCI T7(17BASE) EBVrevpCI-neo T7(17BASE) EBVrev,T3pCMS-EGFP T7 T3pCMS-EGFP T7 EBVrev,T3pCMV-3Tag-4A T3 /PFLAG-CMV-F T7pCMV5pCMV5F PCMV5RpCMV6 CMV-Profor pCMV6-3、HGHrev pCMV6-XL4 CMV-Profor/T7 pCMV6-3、HGHrev pCMV-HA PCMV-F,CMV-Profor,TREfor PCMV-R,EBVrev pCMV-MYC(AMP+)PCMV-F PCMV-R, EBVrev pCMVmyc/nuc CMV-Profor BGH rev,c-mycrevpcmv-scriptEXVector T3 T7pCMV-Sport6 T7/M13for SP6/M13rev pCOLADuet-1(MCSI)ACYCDuetUP1 Primer DuetDOWN1pCOLADuet-1(MCSII)DuetUP2T7TERpCR2.1 T7,M13for M13revpCR2.1-TOPO M13for,T7 M13revpCR3.1 T7 BGH revpCR3.1-Uni T7 BGH revpCR4 T3/M13rev T7/M13forpCR4-Blunt T3/M13rev T7/M13forpCR4-TOPO M13rev,T3 M13for,T7pCR-Blunt M13rev T7,M13forpCR-BluntII M13rev/SP6 T7/M13forpCR-BluntII-TOPO M13rev/SP6 M13for/T7pCRII M13rev/SP6 T7/M13forpCRII-TOPO M13rev/SP6 M13for/T7pCR-ScriptSK(+) T7/M13for T3/M13revpCR-XL-TOPO M13rev T7,M13forpCS2+(MCSI) SP6 EBVrev,T7?pCS2+(MCSII) T3 pSG5-3pDB_GAL4cam T7P pDB_leu PDBLeu-5 PDBLeu-3pDEST22DEST22F DEST22RpDEST15 pGEX-5 T7-TerminatorpDEST20 pGEX-5 pFastBac-3pDEST26 TREfor/CMV-Profor T7/M13forpDisplay T7/CMV-Profor c-mycrevpDNR-LIB(MCS_A) M13for,T7 pSG5-3,M13RpDonar M13F M13RpDONR201PDONR-F PDONR-RpDONR207PDONR-F PDONR-RpDONR223M13F M13RpDrive T7/M13rev SP6/M13forpDsRED1-C1(DsRed1-N-f)DsRed1-C-rpDsRed1-N1 CMV-Profor RFP-Nrev,CMV-R? pDsRed2-N1(DsRed1-N-f)DsRed1-C-rpDsRed2-C1(DsRed1-N-f)DsRed1-C-rpDsRED2-C1(KAN+)pDsRED-ex-C1-F pEGFP-N-3’pDsRed-Express dsRed1_N_primer/CMV-F dsRed1_C_primer/CMV-R pDSRED-monomer-N1(KAN+)dsRed1_N_primer pDSRED-N1(KAN+)pEGFP-N-5’ pDSRED-N-RpEASY-Blunt M13F T7/ M13RpEASY-BluntSimple M13F T7/ M13R(距离插入位点合适,优先安排)pEASY-T1 Simple M13F M13R(距离插入位点合适,优先安排)pEASY-T1M13F/T7M13RpEASY-T3M13F/T7M13R/SP6pECFP-N(C)PECFP-N-5.PECFP-N-3,pEF/myc/cyto/ER/mito/nuc pEF-5Pcdna3.1R/BGH revpEF1(4,6/myc-His) T7/pEF-5Pcdna3.1R/BGH revpEF1(4,6/V5-His) T7/pEF-5Pcdna3.1R/BGH revpEF1(4;6/His_A;_B;_C) T7/Xpressfor Pcdna3.1R/BGH revpEF5/FRT/V5-D(-TOPO) T7/pEF-5Pcdna3.1R/BGH rev pEGFP M13rev EGFP-NrevpEGFP-1 not available EGFP-Nrev/ PEGFP-N-3′pEGFP-C PEGFP-C-5′PEGFP-C-3′pEGFP-C1 EGFP-Cfor SV40-pArevpEGFP-C2 EGFP-Cfor SV40-pArevpEGFP-C3 EGFP-Cfor SV40-pArevpEGFP-F EGFP-Cfor SV40-pArevpEGFP-N PEGFP-N-5′PEGFP-N-3′pEGFP-N1 CMV-Profor EGFP-NrevpEGFP-N2 CMV-Profor EGFP-NrevpEGFP-N3 CMV-Profor EGFP-NrevpENTR M13F M13RpET-11(-a,-b,-c,-d) T7 T7-TerminatorpET-12(-a,-b,-c) T7 T7-TerminatorpET-14b T7 T7-TerminatorpET-15b T7 T7-TerminatorpET-16b T7 T7-TerminatorpET-17b T7 T7-TerminatorpET-17xb T7 T7-TerminatorpET-19b T7 T7-TerminatorpET-20b(+) T7 T7-TerminatorpET-21(-a,-b,-c,-d)(+) T7 T7-TerminatorpET-22b(+) T7 T7-TerminatorpET-23(-a,-b,-c,-d)(+) T7 T7-TerminatorpET-24(-a,-b,-c,-d)(+) T7 T7-TerminatorpET-25b(+) T7 T7-TerminatorpET-26b(+) T7 T7-TerminatorpET-27b(+) T7 T7-TerminatorpET-28a(-b,-c)(+) T7 T7-TerminatorpET-29a(-b,-c)(+)S.tag/T7 T7-TerminatorpET-3(-a,-b,-c,-d) T7 T7-TerminatorpET-30_Ek/LIC T7 T7-TerminatorpET-30_Xa/LIC T7 T7-TerminatorpET-30a(-b,-c)(+)S.tag/T7 T7-TerminatorpET-31b(+) not available T7-TerminatorpET-32_Ek/LIC TRXfor T7-Terminator pET-32_XA/LIC TRXfor T7-Terminator pET-32a(-b,-c)(+)S.tag/TRXfor T7-Terminator pET-33b(+) T7 T7-Terminator pET-34b(+)S.tag T7-Terminator pET-35b(+)S.tag T7-Terminator pET-36b(+)S.tag T7-Terminator pET-37b(+)S.tag T7-Terminator pET-38b(+) not available T7-Terminator pET-39b(+)S.tag T7-Terminator pET-40b(+)S.tag T7-Terminator pET-41_Ek/LIC GST-Cfor T7-Terminator pET-41a(-b,-c)(+) S.tag/pGEX-5 T7-Terminator pET-42a(-b,-c)(+)S.tag/pGEX-5 T7-Terminator pET-43.1a(-b,-c)(+)S.tag coliDOWNpET-43.1a_EK/LIC S.tag not availablepET-44a(-b,-c)(+)S.tag T7-Terminator pET-44a_EK/LIC S.tag not availablepET-45b(+) T7 T7-Terminator pET-46_Ek/LIC T7 T7-Terminator pET-5(-a,-b,-c) T7 T7-Terminator pET-52b(+) T7 T7-Terminator pET-7 T7 T7-Terminator pET-9(-a,-b,-c,-d) T7 T7-Terminator pETcoco-1 T7 T7-Terminator pETDuet-1(MCSI)pETUP1DuetDOWN1 pETDuet-1(MCSII)DuetUp2T7-Terminator pET-T4 T7 T7-Terminator pEYFP-C1pEGFP-C-5’ pEGFP-C-3' pEYFP-N1pEGFP-N-5’,CMV-Profor pEGFP-N-3’ pFLAG-ATS N26for pTrcHis-rv;C24rev pFLAG-CTC N26for pTrcHis-rv;C24rev pFLAG1N26for pTrcHis-rv;C24rev pFLAG-CMV(-2)CMV-30;CMV-Profor CMV-24pFLAG-CMV1CMV-30CMV-24pGAD424GAL4_ADfor3'ADpGADGH GAL4_ADfor3'ADpGADT7T7/GAL4_ADfor3'ADpGADT7-Rec T7/GAL4_ADfor3'ADpGADT-T T73,BDpGAD10GAL4_ADfor3'ADpGAPZa-A a-FACTOR3'AOXpGBKT7T73,BDpGBT9MATCHMAKER5'DNA-BD/GAL4BD MATCHMAKER3'DNA-BDpGEM-1 T7 SP6pGEM-3 T7 SP6pGEM-3Zf(+)(-) SP6/M13rev T7/M13for pGEM-4 T7 SP6pGEM-4Z T7/M13for SP6/M13rev pGEM-5Zf(+)(-) SP6/M13rev T7/M13for pGEM-7Zf(+)(-) SP6/M13rev T7/M13for pGEM-9Zf(+)(-) T7/M13for SP6/M13rev pGEM-T T7/M13for SP6/M13rev pGEM-T_Easy T7/M13for SP6/M13rev pGene/V5-His_A(_B,_C) pGENE-5 BGH rev,V5rev pGEX-2T pGEX-5 pGEX-3 pGEX-2TK pGEX-5 pGEX-3 pGEX-3X pGEX-5 pGEX-3 pGEX-4T-1(-2,-3) pGEX-5 pGEX-3 pGEX-5X-1(-2,-3) pGEX-5 pGEX-3 pGEX-6P-1(-2,-3) pGEX-5 pGEX-3pGL2GLP1 GLP2pGL3RVP3GLP2(RVP4)(没有特别说明用GLP2)pGL3-Basic(MCSI) RVP3(PGL3+)? GLP2(PGL3-)?pGL3-Basic(MCSII) RVP4, PLXSN-3'?pGL4 pGlow(-TOPO) T7 pGlow-3pGM-T T7SP6pGWC SP6T7pHook-2 T7/CMV-Profor not availablepIND/V5-His_A(_B,_C) not available BGH rev,V5rev PinpointTMVector PinpointPrimer SP6pIRES(MCSI) T7 pIRES-3pIRES(MCSII) not available T3,EBVrevpIRES2-EGFP CMV-Profor,pIRES2-EGFP.P5’ IRESrev,pIRES2-EGFP.P3’pIRES-hrGFP-2a CMV-Profor,T3 pIRES-hrGFP-3 pIRESneo2 T7/CMV-Profor pIRES-3pIVEX(all) T7/pBRrevBam T7-TerminatorpLEXA pLEXA-F pLEXA-RpLNCX pLNCX-F pLNCX-RpLNCX2CMV-Profor pRevTRE-3pLVTHM(H1启动子下游)H1primer SP6pLXSN pLXSN-F pLXSN-RpMALc MalE primer M13F(-47)pMAL-c2E MalE primer M13F(-47)pMAL-C2x Pmal-c2x-F(P5’)Pmal-C2X-R(P3’ ) pMALd MalE primer M13F(-47)pMALe MalE primer M13F(-47)pMAL-P4X MalE primer M13F(-47)pMAL-p2X MalE primer M13F(-47)pMET A/B/C AUG1F AUG1RpMD18-T M13R(-48)ECORI M13F(-47)(HINDⅢ) pMD19-T M13F47M13R48pMD20-T M13F47M13R48 pMOSBlue T7/M13rev M13forpMSCV-puro pLXSNfw pMSCVrevpMT/V5-His_A(_B,_C) Mtfor BGH rev,V5rev pNTAP T3T7pCTAP pCTAP5'primer T7pOTB7 M13rev/T7 M13for/SP6 pOptiVEC-TOPO TA CMV-F/CMV-Profor EMCVIRES reverse pPC86PPC86-F PPC86-RpPC97PBD-GAL4-F PBD-GAL4-RpPIC3.5K5'AOX3'AOXpPic9k(A+,Z+)5’AOX(Ppic9k-f)/a-factor3’AOX(Ppic9k-R) pPICZa5'AOX/PPICZa-F3'AOX pPROEXHTA M13R(-48)无反向引物pProEX-HTa(b,c)M13R48/ Tac-Profor pBADrv/pTrcHis-3 pPD129.36 M13F20/M13FpQE30or40or60(AMP+)PQE30-F PQE-30RpRC/CMV2/CAT CMV/T7 pRECEIVER CMV-F 此载体无反向引物pRECEIVER-M03pRECEIVER-M03F pRECEIVER-M03R pREP4pREP4 forward primer EBV reverse primer pRK5 SP6 pSG5-3pRSET T7/Xpressfor T7terpRSFDuet-1(MCSI)DuetUP1DuetDOWN1 pRSFDuet-1(MCSII)DuetUP2T7TERpSecTac2 T7/CMV-Profor BGH rev/c-mycrev pSG5 pSG5-5,/T7PSG5-R(SV40) pSHlox1 T7 SP6pSHUTTLE-CMV PSHUTTLE-CMV-F PSHUTTLE-CMV-R pSI T7 EBVrevpSilencer1.0-U6 T7 T3pSilencer2.0-U6 T7 M13forpSilencer2.1-U6neo T7 M13forpSilencer3.1-H1hygro M13F(-47) 3.0REVpSilencer4_1-CMVneo pSilener4.1-5' CMV-Profor pSILENCEV2.0-U6T7 2.0REVpSIneo T7 EBVrev,T3Psit T7 T7TpSK01-T M13F M13RpSK-CMV(KAN+)T7 T3pSOS Psos5'Psos3'pSP64 SP6 M13revpSP64/65SP6 此载体无反向引物pSP65 SP6 M13revpSP70 SP6 T7pSP71 SP6 T7pSP72 SP6 T7pSP73 SP6 T7pSport1SP6/M13F(-47)T7/M13R pSPORT1 SP6/M13for T7/M13revpSPT18 SP6 T7pSTBlue T7/M13rev SP6/M13for pSUPER.Basic T7/M13F M13RpSUPER T7T3pSURE-T M13F M13RpSURE-Tsimple M13F-47M13R-48 pSuperCos pBRforEco T7pT3T7 M13rev/T3 M13for/T7pT7-7 T7 not availablepT7Blue®T7 M13F(-47)pT7T3_18U M13rev/T7 M13for/T3pT7T3D-PAC M13rev/T7 M13for/T3pTA2M13F/T7(优先使用M13F)T3/M13RpT-Adv M13rev T7,M13forpTAg T7/M13rev M13for pTARGETTM pTarget.F(在T7前面)/T7 pTarget.RpTG19-T M13F(-47)M13R(-48) pTARGETTM T7/M13F PTARGET-SEQ pThioHisA,B,andC Trx Forward Trx Reverse / M13for pTO-T7T7 T7TERpTRC99A PTRC99A-F pTrcHis-rvpTrc99a-c PTRC99C-F PTRC99C-RpTRC99a-c PTRC99C-F PTRC99C-R pTrcHisB PTrcHisA-F/Xpressfor PTrcHisA-R pTrcHisA PTrcHisA-F PTrcHisA-RpT-REX-DEST30 TREfor T7pTriEx1 T7/TriExup TriExdownpTriEx1.1 T7/TriExup TriExdownpTriEx1.1_Hygro T7/TriExup not availablepTriEx1.1_Neo T7/TriExup IRESrevpTriplEX1 TriplEx-LD-5 T7,TriplEx-LD-3pTriplEX2 TriplEx-LD-5;pTR5‘ T7,TriplEx-LD-3pTWIN-1T7 T7TERpTXB1 T7 MxeInteinrevpTXB2 T7 MxeInteinrevpTXB3 T7 MxeInteinrevpTYB1 T7 InteinrevpTYB2 T7 InteinrevpTYB3 T7 InteinrevpTYB4 T7 InteinrevpTZ_18U T7/M13rev M13forpUB6/V5-His UB-F BGH rev,V5revpUC13 M13rev/M13rev(-48) M13for/M13for(-49)pUC18 M13rev/M13rev(-48) M13for/M13for(-49)pUC18(19)/118(119)M13F(-47)M13R(-48)/PUC19(HINDⅢ) pUC19 M13rev/M13rev(-48) M13for/M13for(-49)pUC57 M13rev/M13rev(-48) M13for/M13for(-49)pUC8 M13rev/M13rev(-48) M13for/M13for(-49)pUC9 M13rev/M13rev(-48) M13for/M13for(-49)pUM-T M13F(-20)M13R(-20)pUCm-T(AMP+)M13F/T7M13RpUNI/V5-His not available BGH rev,V5revpVAX1T7 T7/CMV-Profor PCDNA3.1R/BGHRpVP22/mycHis VP22-Cfor BGH rev,c-mycrevpVP22/mycHis2 T7/CMV-Profor VP22-NrevpWE15 pBRrevHind T7pYES2 T7pYes2.RpYES-DEST52 T7/GAL1-Profor V5rev/CYC1-Terminator pYEX_4T_1 Clontech pYEX_4T-3pYEX_4T_2 pGEX-5 pYEX_4T-4pYEX_4T_3 pGEX-5 pYEX_4T-4pYEX_4T_4 pGEX-5 pYEX_4T-4pZeoSV2+/hu-Tyr T7 not availablepZERO-1 M13rev/SP6 T7/M13forpZERO-2 M13rev/SP6 M13for/T7pZome-1-C Zome-F Zome-RpZome-1-N Zome-F Zome-RYCp50(BamHI-site) PBRforBam PBRrevBamYEp13(BamHI-site) PBRforBam PBRrevBamYEp24(BamHI-site) PBRforBam PBRrevBamYIp5(BamHI-site) PBRforBam PBRrevBam RNAi-ReadypSIREN-RetroQ-U6ZsGreen U6 菌PCDB T7BGH天为时代TA载体T7T3m13(+)T7?T3?pMSCV-neo pMSCV-5'pMSCV-3'。
3个水稻RNAi载体的构建与应用柯建浩;廖耀平;王骆艺;穆虹【摘要】利用RNAi(RNA interference)干扰技术,构建了启动子和干涉区域不同的3种水稻干涉载体,通过农杆菌介导转化水稻粳稻品种中花11.表型和分子分析表明,3种载体都能有效降解靶基因OsUGP2,暗示pRNAi-ubi-UGP2载体能同时沉默OsUGP2家族基因,pRNAi-ubi-UGP2PRO载体能特异沉默OsUGP2,pRNAi-IP-UGP2载体对OsUGP2基因的沉默不具特异性,但其沉默具有人为可调控性.【期刊名称】《广东农业科学》【年(卷),期】2011(038)009【总页数】4页(P1-3,封2)【关键词】水稻;RNAi;载体构建;OsUGP2【作者】柯建浩;廖耀平;王骆艺;穆虹【作者单位】广东省农科院水稻研究所,广东广州510640;华南农业大学生命科学学院,广东广州510640;广东省农科院水稻研究所,广东广州510640;广东省农科院水稻研究所,广东广州510640;华南农业大学生命科学学院,广东广州510640【正文语种】中文【中图分类】S511;Q785水稻是世界上最重要的粮食作物之一,全球一半以上的人口以稻米为主食。
水稻是我国最重要的粮食作物,总面积、总产量及单位面积产量均居全国粮食作物首位。
同时,水稻也是单子叶植物研究的模式植物,其基因组在禾谷类作物中是最小的,约由4.3亿个碱基对组成[1-2]。
目前,水稻全基因组测序已基本完成,对其基因进行功能分析已成为世界研究热点。
对基因功能进行分析可采取正向遗传学和反向遗传学两种策略[3-4]。
其中,RNAi 作为反向遗传学策略的一种方法,已成为揭示水稻基因功能的一件重要利器。
植物中常用的 RNAi技术包括转录后基因沉默技术(posttranscriptional gene silencing,PTGS)和转录基因沉默技术(transcriptional gene silencing,TGS)[5]。
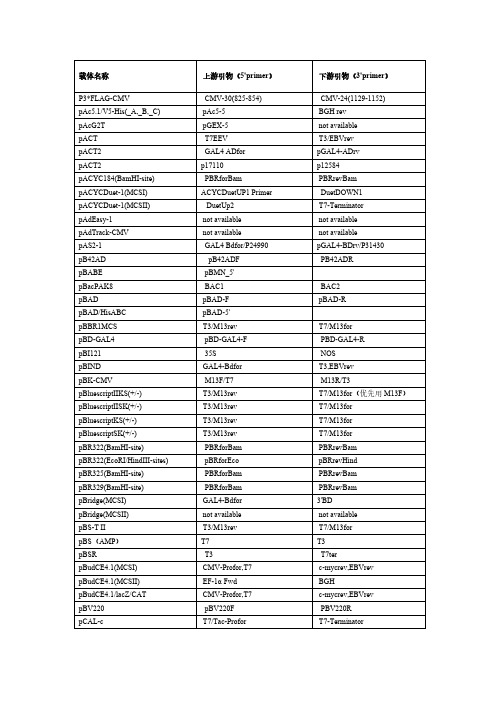
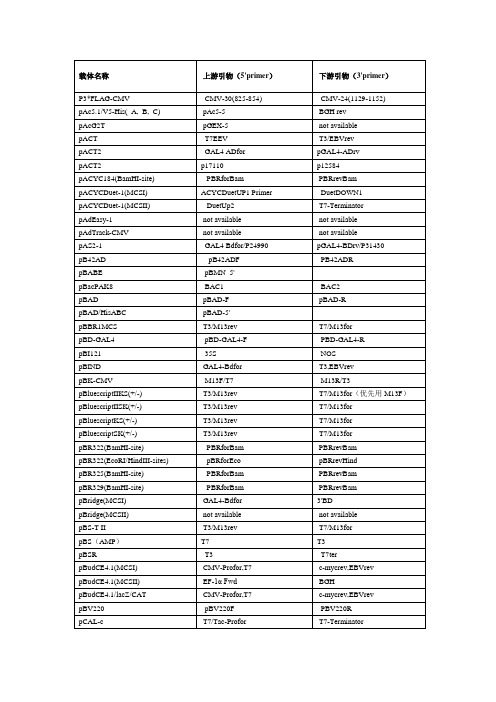
常用载体测序引物列表常用载体测序引物列表载体名称正向引物反向引物pACT T7 T3pACT2 GAL4 AD(5’AD) 3'AD/pACT2-R PACYCDUET-1(MCSII) DuetUP2 T7TERpAD/pl-DEST CMV-FpAD-Track pAD-Track-F 无pAS2-1 5’BD M13-26/48 PB42AD PB42ADF PB42ADR PBAD PBAD-F PBAD-R pBABE pBABE5’ pBABE3’ PBACPAK8 BAC1 BAC2 pBK-CMV T7/M13-20 T3/M13-26 PBS(SK/KS)/M13- M13F/T7T3/M13R pBV220 PBV220F PBV220R pBI121需根据酶切位点确认需根据酶切位点确认 pCAMBIA 1301(1300) 需根据酶切位点确认需根据酶切位点确认pCAMBIA 2300 需根据酶切位点确认需根据酶切位点确认PCANTB5E S1/M13R S6pCAT3-enhancer pGL3+pcDNA3.0 pEGFP-N5/T7 SP6/BGHpcDNA3.1 pEGFP-N5/T7 BGHpcDNA4 T7/pEGFP-N5 BGHpcDNA6 T7/pEGFP-N5 BGHpcDNAII T7SP6 pCE2.1 M13FM13R pCEP4 pCMV5R/PCEP-F pEGFP-N5/EBV-R pCF-T M13FM13R pCIT7(17Base ) pCMV5R pCI-neo T7(17Base ) T3pCMS-EGFP T7T3 pCMV-3Tag-4A T3 /pFlag-CMV-F T7pCMV5 pCMV5F pCMV5R pCMV5-Flag pCMV5FpCMV5R pCMV-MYC/NUC pCMV5FpCMV5R PCMV-HA pCMV5FpCMV5R pCMV-SportSP6/M13RpCMV-Tag T3T7 pCMV-TNT T7/SP6pCMV5R pCR2.1-TOPO M13F/T7M13R pCR3.1 T7BGH pCS2 SP6T7PDNR-LIB M13F M13RpDonar M13F M13R pDONR201 pDONR-F pDONR-R pDONR221 M13F M13RpDrive T7 SP6 pDRIveR T7 SP6pDsRED1-C1 pDsRED-C1-F pECFP-C-3 pDsRed1-N1 pEGFP-N-5 RFP-NrevpDsRed-Monomer-N1 pEGFP-N-5 pDsRed1-NpDsRED2-C1 pDsRED-ex-C1-F pEGFP-N-3’pEGFP-N-5 pDsRed1-N pDSRED2-N1pDSRED-N1 pEGFP-N-5’ PDSRED-N-R pEASY-BLUNT M13F/T7 M13RTerpEASY-E1 T7 T7pEASY-T1 M13F/T7 M13RpEASY-T3 M13F/T7 M13R/SP6 pECFP-C pEGFP-C-5’ pECFP-C-3' pECFP-N1 pEGFP-N-5’ pEGFP-N-3’pEF/myc/ER pEF-F pCDNA3.1R pEGFP-C pEGFP-C-5’ pEGFP-C-3' pEGFP-N pEGFP-N-5’ pEGFP-N-3’ pENTR/D-TOPO M13F M13RTerpET-*(His) T7 T7pET22(a,b,c) T7 T7TerTerpET28,30,24,49 T7 T7TerpET32(a,b,c) T7/S.tag T7pET-42a (-b, -c)(+) Stag T7 TerpET50 T7/s.tag T7TERpET44 T7/s.tag T7Ter pEYFP-N1 pEGFP-N-5’ pEGFP-N-3’ PETBlue M13-20 PETBlueDOWN PETDUET-1(MCSI) PBRrevBam DuetDOMN1 PETDUET-1(MCSII) DuetUP2 T7TERpEYFP-C1 pEGFP-C-5’ pEGFP-C-3' pEYFP-N1 pEGFP-N-5’ pEGFP-N-3’ pFastBac pFastBac-F pFastBac-R pFastBac HT_A,B,C pFastBac-PH pECFP-C-3 pFastBac1 pFastBac-PH pECFP-C-3 pFastBac Dual (MCSI) pFastBac-PH pECFP-C-3 pFLAG-CMV CMV30/pFLAG-CMV-F CMV-24/pFLAG-CMV-Rp3xFlag-CMV-7.1 CMV-F/CMV24 CMV30pGAD424 5’AD 3'ADpGADGH 5’AD 3'ADpGADT7-Rec 5’AD/T7 3'ADpGAPZa-A/B/C a-FACTOR 3'AOXforward 3'AOXpGAPZ-A/B/C pGAPpGBKT7 T7 3'BDpGEM-T(-Easy) M13F/T7 SP6/M13RpGEX-(*)T pGEX-5’ pGEX-3’pGL2 pCMV5R PGL3-Pgl3-BASIC pGL3+ PGL3-pGL3-Enhancer pGL3+ PGL3-PinPoint TM PinPoint Primer SP6pIRES PIRES-F/T7 T3/PIRES-RpIRES2-EGFP pIRES2-EGFP.P5’pIRES2-EGFP.P3’ pLenti6/v5-Dest pEGFP-N-5’PLEXA PLEXA-F PLEXA-R PLNCX PLNCX-F PLNCX-R pLXSN pLXSN-F pLXSN-RPjc1-tac T7pJet.1.21.Blunt T7primer M13F(-47) pMAL-c2E MalEP3/PMAL-C2X-R pMAL-C2x P5/PMAL-C2X-Fprimer M13F(-47) pMAL-p2X MalEPMD18-T M13R(-48) M13F(-47) PMD19-T M13F(-47) M13R(-48) pMIR-report M13FPPC86 PPC86-F PPC86-R3’AOXpPIC9K 5’AOX/a-factor3'AOX pPICZa 5'AOX/PPICZa-FpPROEXHTA M13R(-48)pQE30or40 pQE30+ pQE30- pRECEIVER CMV-FpRSET T7 T7TER PRSFDUET-1(MCSI) DuetUP1 DuetDOWN1 PRSFDUET-1(MCSII) DuetUP2 T7TERpShuttle-CMV pShuttle-CMV-F pShuttle-CMV-R PSILENCE-1.0-U6 T7 T3pSilencer3.1-H1 hygro M13F(-47) 3.0revpSILENCEV2.0-U6 T7 2.0revpSK01-T M13F M13RpSK-CMV T7 T3Psos H1PSP64/65 SP6pSP72 T7 SP6T7/M13RpSport1 SP6/M13F(-47)pSTBlue-1 T7 SP6pSUPER T7 T3pT7Blue(R) T7 M13F(-47)pTA2 M13F/T7 T3/M13R/T7 M13RpT-Adv M13FpTARGET TM pTarget-F/T7 arget-R PTHIOHISA,B,C TRX-FORWARD TRX-REVERSEpTO-T7 T7 T7TERPBV220-/PTRC99C-R pTRC99a-c PQE30+/PTRC99C-F pTriPLEx2 PT5/5'TriplEx2 T7pTz57r/t M13+ M13-pTWIN-1 T7 T7TERpUC18(19)/118(119) M13F(-47) M13R(-48)M13F(-47)/M13F pUC57 M13R(-48)/M13RpUCm-T M13F/T7 M13RpVAX1 T7 BGHT3/M13R/M13R(-48) pWSK29 T7/M13F/M13F(-47) pXT7 T7 SP6pYES2 T7 pYes2.R。
利用In-Fusion技术构建存活素-增强型绿色荧光蛋白融合基因重组慢病毒表达载体【摘要】目的:本研究以构建存活素增强型绿色荧光蛋白(survivin &efp)融合基因慢病毒表达载体(pcfusurvivin)为例来探讨infusion克隆技术在常规载体构建中的应用价值。
方法:根据infusion技术原理,克隆引物设计时,在survivin 同源序列的两侧分别引入经ae i线性化的载体pcfu两端各15个碱基,将以此引物扩增的聚合酶链式反应(pcr)产物与线性化pcfu用infusion交换酶在室温下作用30min,使survivin特异性扩增产物两端的序列与线性化载体两端的序列发生同源交换,取2μl交换液进行转化,挑取阳性克隆,进行酶切和测序鉴定。
将鉴定正确的阳性克隆瞬时转染293t 细胞,观察survivin &efp融合蛋白在293t细胞中的表达。
结果:每2μl克隆交换液获得大约103个克隆数,阳性率达90%以上,瞬时转染pcfusurvivin可获得survivin &efp融合蛋白在293t细胞中的表达。
结论:该技术是一种非连接酶依赖性克隆技术,使基因克隆步骤简化并大大节省了实验时间和经费。
关键词】基因;克隆细胞;基因,病毒value of infusion clonin techhique on routine vector construction lin chao ui fan lin chen lian londepartment of cardioloy,union hospital,fujianmedical university,fuzhou,fujian,350001,abstract:objective:to introduce a simple method for the clonin of pcr products.methods:the infusion clonin technique was described by constructin a rebinant lentivirus vector (pcfu) with survivin &efp fusion ene as a sample.the survivin cdna was amplified with survivin ene specific primers with 15 bp extensions homoloous to the pcfu ends.by the action of the infusion enzyme at room temperature for 30 minutes,the sinlestranded pcr frament and vector ends were fused due to the 15 bp homoloy.finally,clones derived from transformation were chosen randomly and identified.results:about 103 positive clones for inserts were obtained after transformation with 2 μl of exchanin products,and the ratio of the positive colonies was more than 90%.after 24 h the pcfusurvivin was transfected into 293t eukaryotic cells,the expression of the survivin &efp fusion ene can be confirmed with fluorescence microscope.conclusion:the liation independent property makes the infusion pcr clonin technique rapid,reliable and hiher costeffective,avoidin the need for multiple sub clonin steps.key words:enes;clone cells;enes,viral传统的聚合酶链式反应(pcr)产物克隆技术包括补平末端克隆、ta克隆以及连接酶依赖性克隆等。