噬菌体抗体库筛选操作流程tomlinsonij
- 格式:pdf
- 大小:63.74 KB
- 文档页数:14


噬菌体抗体库的构建与初步鉴定目的运用噬菌体展示技术构建抗人β淀粉样肽(Aβ)特异性单链抗体的抗体库并进行初步鉴定.方法从Aβ免疫的BALB/c小鼠脾淋巴细胞中提取总RNA,分离纯化mRNA,经逆转录合成其总cDNA.以PCR方法分别扩增出抗体的重链(VH)和轻链(VL)可变区基因片段,再经重组PCR将两基因片段由编码十五肽(Gly4Ser)3的接头连在一起,克隆到噬菌粒载体pCANTAB5E中,转化至大肠杆菌TG1,经辅助噬菌体M13KO7超感染后回收全部重组噬菌体,构建噬菌体抗体库,SDSPAGE鉴定纯度.结果经琼脂糖凝胶电泳分析,RTPCR方法扩增的抗体VH和VL基因片段大小约350bp和325bp,与预期VH,VL基因片段理论值相符.经重组PCR得到大小约750bp的ScFv基因片段.ScFv基因片段经双酶切后成功定向克隆到噬菌粒载体,回收后构建成库容量为1.1108的噬菌体抗体库,经SDSPAGE鉴定,得到Mr约为30000ku,纯度较好的ScFv抗体.结论成功构建了库容量大的特异性噬菌体抗体库,为抗人Aβ特异性抗体的筛选、生物活性鉴定、临床实验诊断方法建立和治疗应用打下基础.噬菌体展示技术;单链抗体;阿尔茨海默病;β淀粉样肽0引言阿尔茨海默病(alzheimersdisease,AD),又称早老性痴呆症,是一种以记忆丧失、行为失常、人格改变、思维能力下降和认知功能障碍等为特征的中枢神经系统退行性疾病.有研究报道β淀粉样肽(βamyloidpeptide,Aβ)与AD的发病直接相关[1-2].目前国内仅有制备Aβ42多克隆抗体的相关工作,有关建立良好的血液标志物检测方法的研究报道还较少见.本实验通过噬菌体抗体库技术[3-4]构建抗Aβ的单链抗体库,为检测血液或脑脊液中Aβ水平并用于诊断AD[5-6]和抗人Aβ特异性抗体的筛选、生物活性鉴定以及临床治疗应用打下基础.1材料和方法1.1材料雌性BALB/c纯系小鼠20只,6~7wk龄(中山大学实验动物中心);cDNA第一链合成回收试剂盒(美国Fermentas公司);鼠ScFv噬菌体抗体展示系统(瑞典Amershampharmacia公司);Trizol试剂(美国Gibco公司);DNA 多聚酶,QIAquickPCR纯化试剂盒(荷兰Qiagen公司);SfiI内切酶,NotI内切酶(美国NEB公司);T4DNALigase(美国Invitrogen公司);Aβ(美国Americanpeptide公司).1.2方法1.2.1动物免疫将0.5mL的Aβ42(150mg/L)与等量的完全福氏佐剂混合乳化,直到油包水的状态,取5只小鼠用Aβ42进行免疫.第一次基础免疫每只小鼠自后足垫注射0.1mL.基础免疫后2wk再按上述方法用等量的抗原与不完全福氏佐剂乳化后进行加强免疫,每只小鼠腹腔注射0.1mL,共加强免疫2次.45d后引颈法处死免疫小鼠,取其脾脏.1.2.2总RNA抽提与mRNA的纯化用TRIzol试剂抽提小鼠脾细胞的总RNA,并立即纯化mRNA,紫外分光光度计测定其浓度及纯度.1.2.3cDNA第一链的合成取两个灭菌的0.5mL的离心管,在每一管中依次加入mRNA2μL,Oligo(dT)18引物1μL,去RNase水9μL,混匀,置于70℃水浴5min,立即置于冰中,在冰上再加入5倍稀释缓冲液4μL,核糖核酸酶抑制剂1μL,dNTP(10mmol/L)mix2μL,混匀,室温静置5min后再加入MMuLV逆转录酶(2108U/L)1μL,总反应体积为20μL.42℃水浴60min,70℃水浴10min灭活,立即置于冰中5min,完成cDNA第一链的合成.1.2.4PCR扩增抗体的重链和轻链可变区片段取两个灭菌的0.5mL的离心管,在轻链PCR扩增管中分别依次加入cDNA 模板20μL,轻链引物2μL,dNTP(10mmol/L)5μL,10倍稀释缓冲液10μL,DNA聚合酶1μL,ddH2O62μL.重链PCR管cDNA模板20μL,重链引物1∶2μL,重链引物2∶2μL,dNTP(10mmol/L)5μL,10倍稀释缓冲液10μL,DNA聚合酶1μL,ddH2O60μL.95℃热启动5min后,进行如下循环94℃30s,53℃45s,72℃60s,共30个循环,最后一个循环结束后,72℃延伸10min,然后取5μLPCR产物琼脂糖凝胶电泳,同时加DNA分子量标准,用紫外分析仪检测电泳结果,鉴定PCR产物片段大小.1.2.5重链和轻链可变区片段与Linker的连接扩增出的抗体重链(VH)和轻链(VL)的PCR产物按Qiagen公司的QIAquickPCR纯化试剂盒中的说明书进行回收纯化.用琼脂糖凝胶电泳,纯化后的VH和VL与标准浓度的VHmarker分别按1∶1和1∶2稀释后上样电泳进行比较定量,根据条带的亮度来估计基因片段的含量.以相同和接近等摩尔浓度的抗体VH和VL基因混合进行重组PCR反应,按Pharmacia公司的重组噬菌体抗体库系统(RPAS)操作手册进行与Linker的连接PCR反应.然后在PCR产物加上酶切位点后,取5μLPCR产物用15g/L的琼脂糖凝胶电泳,同时加DNA分子量标准,用紫外分析仪检测电泳结果,鉴定PCR反应产物片段的大小.1.2.6连接片段的克隆取一灭菌的0.5mL的离心管,在每一管中依次加入纯化的PCR产物17μL,SfiI1μL,10倍稀释缓冲液2μL,50℃温育2h,酶切产物胶回收纯化.另取一灭菌的0.5mL的离心管,在每一管中依次加入SfiI酶切后的ScFv片段40μL,NotI1μL,BAS0.5μL,10倍稀释缓冲液5μL,ddH2O4.5μL,37℃温育3h,酶切产物胶回收纯化.1.2.7质粒的转化与筛选取4个灭菌0.5mL的离心管,在每一管中依次加入纯化的酶切后ScFv40μL,pCANTAB5E质粒5μL,ATP(10mmol/L)5μL,T4DNA连接酶1μL,5倍稀释缓冲液6μL,ddH2O3μL,24℃温育1h,每个连接产物分别进行回收纯化.1.2.8感受态细菌的制备用划线法将大肠杆菌TG1接种于基本培养基平皿上,于37℃倒置培养过夜,再用无菌牙签挑取一个单菌落至5mL的培养液中,37℃振荡(250r/min)过夜培养.次日取菌液1mL接种至含有100mL培养液的500mL烧瓶中,37℃振荡培养至吸光度(A600nm)达到0.4~0.5时(约需2~3h),4℃,2500r/min离心15min,弃上清液,再加10mL 预冷的TSS重悬菌体,完成制备感受态大肠杆菌,2~3h内进行转化.1.2.9质粒的转化将4种纯化连接产物分别按以下方法进行转化,转化后收集的噬菌体抗体合并到一起组成噬菌体抗体库.在无菌状态下分别取1mL新鲜感受态大肠杆菌TG1至两个预冷的50mL离心管中,置于冰中.其中一个加入50μL纯化的连接产物,另一个加入500pg未经酶切的pUC18质粒,轻轻旋转以混合内容物.冰浴45min.将试管置于42℃水浴2min使细胞热休克,然后立即置于冰中,转化后,用质粒pUC18来检查其转化效率.取100μL转化后至含有900μL的LBG(含20mol/L葡萄糖的LB培养液)的试管中,37℃振荡(250r/min)培养1h,分别取100,10,1μL的培养物在SOBAG琼脂板(含100mg/L氨苄青霉素和2g/L葡萄糖的SOB培养基)上铺盘,同样方法另取100μL未进行转化的感受态大肠杆菌TG1在SOBAG平板上铺盘作为对照组.37℃倒置培养过夜,次日计算菌落数.1.2.10噬菌体抗体库的扩增取900μL转化的大肠杆菌至一无菌的50mL试管中,加入9.1mL含10g/L葡萄糖培养液,37℃振荡(250r/min)培养1h后,再加50μL的20g/L氨苄青霉素和的M13KO7辅助噬菌体,37℃振荡(250r/min)培养1h,1000g离心10min,弃去上清,用10mL的不含葡萄糖的培养液重悬,37℃振荡(250r/min)培养过夜.1000g离心20min,将含有重组噬菌体的上清液转入一无菌的50mL离心管中,0.45μm 过滤后分装保存于2℃~8℃备用,此浓缩后的噬菌体抗体库可用于富集筛选、纯化和鉴定.1.2.11ScFv基因噬菌体抗体库表达抗体的初步鉴定纯化后的单链抗体用SDSPAGE电泳鉴定其纯度,同时设分子量标准电泳,考马斯亮蓝R250染色.2结果2.1总RNA的提取及mRNA的纯化从Aβ免疫后小鼠脾细胞中抽提总RNA,经紫外分光光度计测得总RNA浓度为5.16g/L,纯化后的mRNA的A260nm/A280nm为1.90,浓度为1.28g/L.2.2抗体VL和VH基因的扩增结果显示,VH基因片段约为350bp,VL基因片段约为325bp,与预期VH,VL基因片段理论值大小相符(图1).M1:VHmarker;M2:DNAmarker;1:VHPCR产物;2:VLPCR产物.图1VH和VL基因片段的琼脂糖凝胶电泳分析(略)2.3ScFv基因的连接结果如图2所示,可见约750bpPCR 扩增产物,大小符合预期结果,没有明显的非特异性条带,表明加入VH,VL和Linker引物的浓度准确,并引入了用于在噬菌粒载体pCANTAB5E中进行克隆的限制性内切酶SfiI和NotI 位点.M1:ScFvmarker;M2:DNAmarker;S:ScFvPCR产物.图2ScFv基因片段的琼脂糖凝胶电泳分析(略)2.4抗体ScFv基因的克隆及噬菌体抗体库的构建酶切后抗体ScFv基因片段经琼脂糖凝胶与标准浓度的ScFvmarker比较,ScFv的浓度约20mg/L.在连接反应中,插入的ScFv基因片段与噬菌粒载体pCANTAB5E的量为3∶1效果最理想.ScFv 基因片段通过连接反应成功定向克隆至pCANTAB5E噬菌粒载体的SfiI和NotI位点中.噬菌体载体成功转化至大肠杆菌TG1,经过两次培养,收集所有重组噬菌体,成功构建的噬菌体抗体库,合并计算,噬菌体抗体库的库容量达1.1108.2.5抗体ScFv基因的噬菌体抗体库表达抗体的初步鉴定浓缩提取的ScFv,经蛋白质L琼脂糖亲和纯化得到Mr约为30ku,紫外分光光度计检测结果显示,纯化后单链抗体的表达量约为1.1mg/L,还原性SDSPAGE结果显示纯度较好的ScFv(图3).1:浓缩上清培养液;M:marker;2:纯化ScFv.图3纯化ScFv的SDSPAGE(略)3讨论目前已知Aβ蛋白在人类大脑中的沉积和神经纤维缠结的产生是AD的主要组织病理学标志[2],并揭示了细胞内Aβ在大脑内沉积的现象以及Aβ具有毒性的作用机制[7-8].由于临床对AD诊断较为困难,尤其是发病早期,采用血标本进行检测的方法还没有完全建立起来.因此建立新的临床试验诊断方法,尤其是检测血液中的标记物,并且能够区分AD和正常衰老以及其它痴呆的试验方法已成为热点.噬菌体抗体库技术的建立是基因工程抗体研究领域的一次重大突破,使传统的mAb的制备完全改观,为重组抗体的设计、筛选及生产等方面的不断革新及改进提供了高效、可靠的技术和方法,极大地推动了重组抗体在科研、临床诊断及治疗等方面的应用[10-11].此方法简单、快速,而杂交瘤技术从脾细胞融合到获得稳定的单克隆细胞株至少需数月[12].噬菌体抗体库技术可以绕过运用杂交瘤细胞免疫制备抗体,在大肠杆菌中分泌表达,通过发酵大量生产,具有大规模生产、成本低等特点,克服了杂交瘤细胞通过mAb腹水或大规模细胞培养制备抗体的局限性,同时能够保存和传代稳定性,避免了杂交瘤细胞株在传代过程中明显的不稳定性,我们的研究结果显示,直接利用人外周血淋巴细胞构建天然抗体库,筛选出目的噬菌体抗体,使制备全人抗体,尤其是制备自身抗原和其他弱免疫原性抗原的抗体具有独到的优势[13],并可以对抗体基因进行改造.在体外通过基因突变、链更替等技术,进一步提高抗体的亲和力和稳定性.用于制备mAb,并对抗体库进行筛库时,能够得到多克隆重组噬菌体抗体,将该多克隆抗体用于临床试验诊断可提高敏感度,用于治疗可提高疗效,也可以作为分子识别、抗原表位研究的理想工具.综上所述,本实验利用基因工程方法和噬菌体表面展示技术成功构建了免疫小鼠库容量为1.11 08的噬菌体抗体库,为抗人Aβ特异性抗体的筛选、生物活性鉴定、临床实验诊断方法建立以及治疗应用打下基础.。

噬菌体抗体文库淘选1.主要实验仪器表1实验所用主要仪器仪器名称型号/厂家高速冷冻离心机Neofuge15R生物安全柜Heal Force电热恒温水浴锅HHW21.600AⅡ恒温培养箱Heal force(3)5ml5%脱脂牛奶/PBST30℃封闭1h。
(4)5ml PBS洗涤1次。
(5)每管中加入500ul,10^11-10^12pfu的文库噬菌体(或者上一轮的扩增噬菌体),30℃孵育2h。
(6)5ml PBST洗涤4-6次(后几轮可根据富集程度增加洗涤次数)。
(7)每管中加入500ul的Gly-HCl(pH=2.2)洗脱噬菌体,室温振荡孵育6-8min左右。
加入120-130ul的Tris-HCl(pH=9.6)中和溶液至pH=7.0-8.0。
(8)将洗脱后的噬菌体稀释后,侵染对数期的大肠杆菌TG1,铺板测定滴度。
3.2洗脱噬菌体的扩增(1)吸取洗脱后的噬菌体,加入到对数期的大肠杆菌TG1菌液中,37℃静置30min后,220rpm培养30min-1h。
(2)培养基中加入抗生素Amp,37℃,220rpm培养至菌液OD=0.4-0.6左右。
(3)在菌液中加入辅助噬菌体。
37℃静置30min后,220rpm培养45min-1h。
(4)3000-5000rpm离心菌液,弃上清。
用同等体积2YT-Amp-Kan培养基重悬菌体。
30℃,220rpm培养过夜。
(5)次日,8000rpm,4℃,20min离心菌液,将上清转入新的离心管中。
加入1/4体积的(4)用PBS稀释每一轮扩增后的噬菌体,稀释倍数3倍递增。
初始浓度为10^12pfu/ml。
每孔中加入100ul稀释后的扩增噬菌体。
30℃孵育1h,300ul PBST洗涤4-6次。
(5)加入100ul二抗(抗噬菌体M13)稀释液,30℃孵育1h,300ul PBST洗涤4-6次。
(6)加入100ul显色液TMB避光显色3-8min,加入100ul2M HCl终止反应,酶标仪读数(450nm-620nm)。



噬菌体双层平板法操作规程1. 引言噬菌体双层平板法是一种常用的微生物实验方法,用于检测和计数环境中的细菌数量。
该方法利用噬菌体对细菌的寄生作用,形成双层平板,通过观察菌落的形成来进行计数和鉴定。
本文将介绍噬菌体双层平板法的操作规程。
2. 实验所需材料和试剂•LB琼脂培养基•LB液体培养基•目标细菌菌种•噬菌体溶液•加热灭菌器•平板培养皿•称量器具3. 实验步骤3.1 准备工作1.准备好所需材料和试剂,检查其完整性和有效期。
2.将LB琼脂培养基按照规定配制好,并加热至完全溶解。
3.将LB液体培养基按照规定配制好。
3.2 制备平板培养基1.取一定量的配制好的LB琼脂培养基,倒入平板培养皿中,待琼脂凝固。
3.3 细菌接种1.随机选取一支功能良好的细菌培养基,用无菌胶头棉签沾取一小块菌落。
2.在平板培养基的表面连续均匀涂抹菌落,使用一般的“来回”涂抹法。
3.重复上述步骤,将要检测的不同细菌菌种在不同的平板培养基上涂抹。
3.4 噬菌体接种1.取一支已知浓度的噬菌体溶液,用无菌加头棉签沾取一小滴。
2.在相应的细菌涂抹平板上滴加噬菌体溶液,待其完全吸收。
3.5 双层平板培养1.将已接种噬菌体的平板培养皿在室温下倒置放置,并标记。
2.将未接种噬菌体的平板培养皿在室温下正常放置,并标记。
3.6 培养和观察1.将已接种噬菌体的平板培养皿和未接种噬菌体的平板培养皿分别放入37° C恒温培养箱中培养。
2.每过一段时间(如24小时)拿出平板培养皿观察,并记录菌落的数量和形态特征。
4. 结果和分析根据观察到的菌落数量和形态特征,可以进行细菌计数和鉴定。
未接种噬菌体的平板培养皿上菌落数量代表细菌的总数,已接种噬菌体的平板培养皿上菌落数量代表剩余可感染的细菌数量。
通过比较两者的差异,可以估算噬菌体的感染效果。
5. 注意事项1.所有操作必须在无菌条件下进行,避免细菌的污染。
2.使用无菌胶头棉签时,每次接种前必须在无菌火焰中消毒。

ORTHO BioVue
抗体筛选试验操作步骤
d卡一张,平衡至室温;
1、预先取出BioVue抗-IgG卡或抗-IgG,-C
3
将标有条形码的一边面向实验者,标记样本编号;(一卡2人)
2、撕开三孔的锡条(每一个试验),分别滴入50ul BLiss液。
3、用10ul加样枪分别取10ul BioVue筛选红细胞1# /2# /3# 依次加
入三孔中。
4、用40ul加样枪分别取40ul 病人血清依次加入此三孔中。
5、孵育10分钟。
6、离心5分钟,判读结果。
备注:
1、往卡上的反应腔中加样时,注意加样枪应保持45°角,沿管侧壁加入,并
保留空气间隔,切不可垂直加入或用力过猛。
2、严格遵循“先加红细胞,后加血清”的原则。
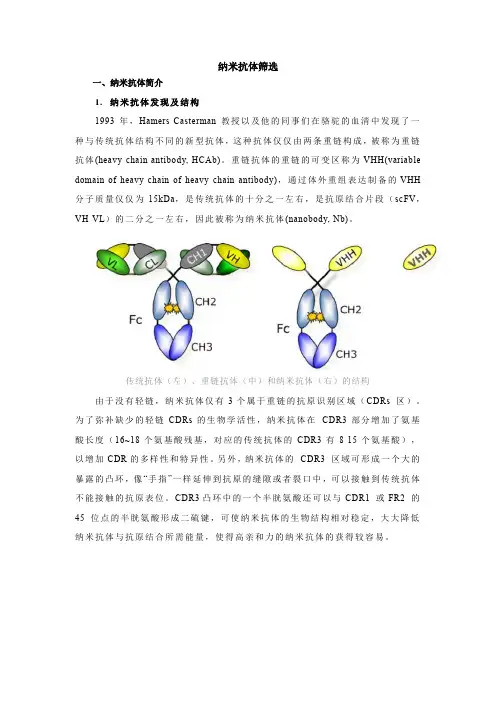
纳米抗体筛选一、纳米抗体简介1.纳米抗体发现及结构1993年,Hamers-Casterman教授以及他的同事们在骆驼的血清中发现了一种与传统抗体结构不同的新型抗体,这种抗体仅仅由两条重链构成,被称为重链抗体(heavy-chain antibody, HCAb)。
重链抗体的重链的可变区称为VHH(variable domain of heavy chain of heavy-chain antibody),通过体外重组表达制备的VHH 分子质量仅仅为15kDa,是传统抗体的十分之一左右,是抗原结合片段(scFV,VH-VL)的二分之一左右,因此被称为纳米抗体(nanobody, Nb)。
传统抗体(左)、重链抗体(中)和纳米抗体(右)的结构由于没有轻链,纳米抗体仅有3个属于重链的抗原识别区域(CDRs 区)。
为了弥补缺少的轻链CDRs的生物学活性,纳米抗体在CDR3部分增加了氨基酸长度(16~18个氨基酸残基,对应的传统抗体的CDR3有8-15个氨基酸),以增加CDR的多样性和特异性。
另外,纳米抗体的CDR3 区域可形成一个大的暴露的凸环,像“手指”一样延伸到抗原的缝隙或者裂口中,可以接触到传统抗体不能接触的抗原表位。
CDR3凸环中的一个半胱氨酸还可以与CDR1 或FR2 的45 位点的半胱氨酸形成二硫键,可使纳米抗体的生物结构相对稳定,大大降低纳米抗体与抗原结合所需能量,使得高亲和力的纳米抗体的获得较容易。
纳米抗体和VH结构示意图2.纳米抗体优势(1)纳米抗体理化特性较稳定(2)纳米抗体的大规模生产较容易实现(3)纳米抗体免疫原性低(4)纳米抗体分子量小,组织渗透能力强,血液清除较快(5)纳米抗体可以识别传统抗体不能识别的位点(6)纳米抗体具备形成多聚体的能力3.纳米抗体筛选纳米抗体筛选和制备纳米抗体的获得现在普遍通过免疫羊驼,从羊驼体内自身的抗体成熟阶段来得到抗体基因,然后通过噬菌体展示筛选技术来从羊驼抗体库中筛选得到高亲和力的抗体序列。

亲和层析法用于噬菌体抗体库的筛选陈瑛炜,罗文新,王明桥,王晋,李利峰,袁权,张军,夏宁邵(福建省医学分子病毒学研究中心,厦门大学细胞生物学与肿瘤细胞工程教育部重点实验室,厦门 361005)摘要:本研究报道一种基于固定化金属亲和层析(IMAC )的噬菌体抗体库液相筛选方法。
将纯化的带有His 标签的抗原与噬菌体抗体库混合,噬菌体抗体与抗原充分结合后再加入亲和介质,使噬菌体抗体抗原复合物通过His 标签与介质结合,然后通过充分洗涤去除非特异性噬菌体抗体,最后将特异性噬菌体抗体洗脱下来,感染TG 1,进行下一轮筛选。
整个筛选过程中抗原与抗体的结合在液相中完成,不仅消除了固相介质对抗原表位的影响,也更有利于噬菌体抗体与抗原的充分作用。
将此方法应用于HEV N E2蛋白特异性人源噬菌体抗体的筛选,抗原竞争EL ISA ,阳性血清阻断,可溶性单链抗体表达检测及测序结果表明,最终获得2个特异性人源抗体。
关键词:噬菌体抗体库;固定化金属亲和层析;液相筛选中图分类号:R373;Q78 文献标识码:A 文章编号:1000-8721(2006)01-0044-06收稿日期:2005-04-27;修回日期:2005-07-12基金项目:福建省科技重大项目(编号:2002F013);国家十五创新药物博士基金(编号:2003AA2Z3539);福建省自然科学基金(编号:C0310005);厦门市科技计划项目(编号:3502Z20055002)。
作者简介:陈瑛炜(1982-),女,硕士,从事基因工程抗体研究。
通讯作者:罗文新,副教授,Tel 186259222184113;Fax :86259222181258;E 2mail :wxluo @jingxian 1xmu 1edu 1cn 噬菌体抗体库技术已经成为目前获得人源抗体的主要途径之一。
从大容量天然人源噬菌体抗体库中,理论上可以获得针对任何抗原的人源单抗。
噬菌体抗体库筛选方法包括纯抗原筛选,非纯抗原筛选,功能性筛选,选择性感染筛选等[1]。
theLibrariesGeneral Introduction toOver the past 10 years Greg Winter’s lab at the MRC Laboratory of Molecular Biology and the MRC Centre for Protein Engineering (Cambridge, UK) has created a number of artificial libraries of antibodies that can be used to derive binders to almost any target molecule using phage display and selection. These binders can be used for all the same applications as conventional monoclonal antibodies (ELISA, Western blotting, FACS, immunohistochemistry etc) but can be isolated in a fraction of the time and without the need for animal immunisation. To date these so called “naïve” or “single pot” phage-antibody libraries have been used successfully in hundreds of molecular biology labs world-wide to derive highly specific antibody reagents to a wide range of different proteins, peptides or small molecule compounds.The latest libraries (Tomlinson I and J) that are being distributed by the MRC HGMP Resource Centre each comprise over 100 million different scFv fragments cloned in an ampicillin resistant phagemid vector and transformed into TG1 E. Coli cells (scFv fragments comprise a single polypeptide with the VH and VL domains attached to one another by a flexible Glycine-Serine linker). By carefully following the protocol provided, large numbers of phagemids can be produced and used to select specific binders to target molecules that are attached to the surface of a tube or biotinylated and captured by streptavidin coated beads (so called “panning”). After each round of panning, the non-binders are washed away and the phagemids bound to the target molecule/s are eluted and amplified by infection into fresh TG1 cells. After producing new phagemids from the previous round of panning, the process can be repeated. Typically two or three rounds of panning are required to ensure that more than half the different scFvs in the selected population bind to the target molecule. The monoclonal scFvs can then be screened for binding (using a simple ELISA based protocol) and then used for further analysis of the target molecule. Since all the functional scFvs in the Tomlinson I and J libraries bind Proteins A and L, either of these secondary reagents can be used for detection, purification or immobilisation. Alternatively, secondary reagents that bind the attached myc or HIS6 tags can be used, although in our experience it is better to use the Protein A or L reagents.Finally, we would like to emphasise that these libraries represent a valuable resource. Whether you are familiar with phage display or not we recommend that you perform test selections and subsequent ELISA screening using the anti-bovine serum albumin and anti-bovine ubiquitin controls provided. Only when these experiments have been successfully carried out should you defrost the libraries and start preparing library phage.Both libraries are based on a single human framework for V H (V3-23/DP-47 and J H4b) and Vκ(O12/O2/DPK9 and Jκ1) with side chain diversity incorporated at positions in the antigen binding site that make contacts to antigen in known structures and are highly diverse in the mature repertoire. The canonical structure (V H: 1-3, Vκ: 2-1-1) encoded by this framework is by far the most common in the human antibody repertoire. The CDR3 of the heavy chain was designed to be as short as possible yet still able to form an antigen binding surface. Both libraries can be selected and affinity matured without knowing the sequences of selected clones. Libraries are in phagemid/scFv format and have been pre-screened for binding to Protein A and Protein L so that the majority of clones in the unselected libraries are functional.Constructed in pIT2 (HIS myc tag). Diversified (DVT) side chains based mainly on those positions which are diverse in the primary repertoire (total of 18 residues - H50, H52, H52a, H53, H55, H56, H58, H95, H96, H97, H98, L50, L53, L91, L92, L93, L94 and L96). After selection, can be matured by incorporating additional diversity based on somatic mutation.Library size (with insert): 1.47 x 108Percentage insert: 96%As above but with NNK side chains.Library size (with insert): 1.37 x 108Percentage insert: 88%1. Check that you have received:a tube of Library I (~500 µl)a tube of Library J (~500 µl)a glycerol stock of a positive control anti-ubiquitin ScFv in bacterial strain TG1 (labeled TG1-antiubi)a glycerol stock of a positive control anti-BSA ScFv in bacterial strain TG1 (labeled 13CG2)a glycerol stock of T-phage resistant E. Coli. TG1 for propagation of phage (labeled TG1Tr)∆(lac-proAB) supE thi hsdD5/F' traD36 proA+B lacI q lacZ∆M15)(K12a glycerol stock of E. Coli. HB2151 for expression of antibody fragmentsara ∆(lac-proAB) thi/F' proA+B lacI q lacZ∆M15)(K12KM133 (~100 µl with 107 pfu/ml)Phage2. Check the library is still frozen and make sure you keep it frozen at -70°C until needed.3. Make stock of KM13 according to Protocol G.4. Run through the protocols using the control clones before you use the library: Streak the controlson TYE plates containing 100 µg/ml ampicillin and 1 % glucose. After overnight growth at 37°C in an incubator pick a single colony from each and grow these overnight (shaking at 37°C) in 5 ml 2xTY1 containing 100 µg/ml ampicillin and 1 % glucose. Make phage for the positive and negative controls separately (use 500 µl of overnight in D1-D10). Use a 1:100 mixture of phage produced from the positive and the negative controls and perform one round of selection (C1-C11) using 100 µg/ml of ubiquitin2 in PBS for coating. Check for enrichment of ubiquitin binders (should be over 50% after one round of selection) by monoclonal phage ELISA (E9-E14).5. Wherever possible use devoted pipettes and disposable plastic ware. The use of polypropylenetubes is recommended as phage may adsorb non-specifically to other plastics.6. For efficient infection of phage, E. coli must be grown at 37°C and be in log phase (OD at 600 nmof 0.4). To prepare this:i. Transfer a bacterial colony from a minimal media plate into 5 ml of 2xTY medium (no antibioticsor glucose). Grow shaking overnight at 37°C.ii. Next day dilute overnight 1:100 into fresh 2xTY medium. Grow shaking at 37°C until OD 600 is0.4 (1.5-2 hrs)7. All centrifugations, except those performed in a micro centrifuge, are performed at 4°C.8.Libraries I and J must be used separately and preferably in parallel. This will ensureselecting the most antigen binding clones.In advanceGather all equipment and reagents (product details are given in the notes at the end of all the protocols). Make sure you have all the necessary media and plates for bacterial growth (the large Bio-Assay plates need to be air-dried in a sterile environment for 2 hrs before use).Plan your time - most of the daily procedures can be performed simultaneously, so read through each protocol carefully before starting.StepsProcedureDay 1 (5 hrs) A1-A6 Grow libraries I and J and make phageB1-B3 Make secondary stock of librariesDay 2 (6 hrs) A7-A12 Grow libraries I and J and make phage (cont.)C1 Coat immunotubes for 1st round of selectionDay 3 (6.5 hrs) C2-C11 1st round of selectionDay 4 (3 hrs) D1-D6 Make phage from 1st round of selectionC1 Coat immunotubes for 2nd round of selectionDay 5 (6.5 hrs) D7-D11 Make phage from 1st round of selection (cont.)C2-C11 2nd round of selectionDay 6 (3 hrs) D1-D6 Make phage from 2nd round of selectionC1 Coat immunotubes for 3nd round of selectionDay 7 (6.5 hrs) D7-D11 Make phage from 2nd round of selection (cont.)C2-C11 3rd round of selectionDay 8 (3 hrs) D1-D6 Make phage from 3rd round of selectionE1 Coat 96 well plate for polyclonal phage ELISADay 9 (6.5 hrs) D7-D11 Make phage from 3rd round of selection (cont.)E2-E8 Polyclonal phage ELISAFurther characterisation of individual clones can be performed by monoclonal phage ELISA (protocol E), monoclonal ELISA using soluble scFv fragments (protocol F), PCR screening (to check for insert, protocol H) and sequencing (protocol I).1. Add the library stock to 200 ml pre-warmed 2xTY containing 100 µg/ml ampicillin and 1 %glucose.2. Grow shaking at 37°C until the OD 600 is 0.4 (1-2 hrs).3. Take 50 ml of this and add 2x1011 KM13 helper phage3. (Use the remaining 150 ml to make asecondary bacterial stock of the library by following protocol B).4. Incubate without shaking in a 37°C water bath for 30 min.5. Spin at 3,000 g for 10 min (3,600 rpm in Centra 8 or equivalent). Resuspend in 100 ml of 2xTYcontaining 100 µg/ml ampicillin, 50 µg/ml kanamycin and 0.1% glucose.6. Incubate shaking at 30°C overnight.7. Spin the overnight culture at 3,300 g for 30 min (4,000 rpm in Centra 8 or equivalent).8. Add 20 ml PEG/NaCl (20 % Polyethylene glycol 6000, 2.5 M NaCl)to 80 ml supernatant. Mix welland leave for 1 hr on ice.9. Spin 3,300 g for 30 min (4,000 rpm in Centra 8 or equivalent). Pour away PEG/NaCl. Respinbriefly and aspirate any remaining dregs of PEG/NaCl.10 Resuspend the pellet in 4 ml PBS and spin at 11,600 g for 10 min in a micro centrifuge to removeany remaining bacterial debris.11. Store the phage at 4°C for short term storage or in PBS with 15 % glycerol for longer termstorage at -70°C.12. To titre the phage stock dilute 1µl phage in 100µl PBS, 1µl of this in 100µl PBS and so on untilthere are 6 dilutions in total. Add 900µl of TG1 at an OD 600 of 0.4 to each tube and incubate at 37°C in a waterbath for 30 mins. Spot 10 µl of each dilution on a TYE5 plate containing 100 µg/ml ampicillin and 1 % glucose and grow overnight at 37°C. Phage stock should be 1012-1013/ml, enough for at least 10 selections.1. Grow the remaining 150 ml from A3 for a further 2 hr shaking at 37°C.2. Spin down the cells at 10,800 g for 10 min. Resuspend in 10 ml of 2xTY containing 15 % glycerol.3. Store this secondary stock in 20x 500 µl aliquots at -70°C. Use one aliquot for each phagepreparation according to protocol A - this will only be necessary if you wish to do more than 10 selections.(Alternatively, phage can be selected using biotinylated antigen in solution or affinity chromatography. For details see Winter et al. (1994) Annu. Rev. Immunol.12, 433)immunotube6 overnight with 4 ml of the required antigen. The efficiency of coating can1. Coatdepend on the antigen concentration, the buffer and the temperature. Usually 10-100 µg/mlantigen in PBS is used.2. Next day wash tube 3 times with PBS (pour into the tube and then pour it immediately out again).3. Fill tube to brim with 2 % MPBS (2 % Marvel milk powder7 in PBS). Incubate at rt. Standing onthe bench for 2 hr to block.4. Wash tube 3 times with PBS.1012 to 1013 phage from A11 in 4 ml of 2 % MPBS. Incubate for 60 min at rt. rotating using 5. Addan under-and-over turntable and then stand for a further 60 min at rt. Throw away supernatant.6. Wash tubes 10 (round 1)-20 (rounds 2 and 3) times with PBS containing 0.1 % Tween 20.7. Shake out the excess PBS and elute phage by adding 500 µl of trypsin-PBS (50 µl of 10mg/mltrypsin stock solution8 added to 450 µl PBS) and rotating for 10 min at rt using an under-and-over turntable.8. Take 1.75 ml of TG1 at an OD 600 of 0.4 and add 250 µl of the eluted phage (the remaining 250µl should be stored at 4°C). Incubate for 30 min at 37°C in a water bath without shaking.µl, 10 µl of a 1:102 dilution and 10 µl of a 1:104 dilution on TYE plates containing 1009. Spot10µg/ml ampicillin and 1 % glucose and grow overnight at 37°C to titre the phage.10. In round 1 if using a complex antigen (eg cells, cell lysates etc): take the remaining TG1 cultureand spin at 11,600 g in a micro centrifuge for 5 min. Resuspend the pelleted bacteria in 1 ml of 2xTY and plate on a large square Bio-Assay dish9 containing TYE, 100 µg/ml ampicillin and 1 % glucose.In round 1 if using a single hapten, carbohydrate or protein antigen and in subsequent rounds for all antigens: take the remaining TG1 culture and spin at 11,600 g in a micro centrifuge for 5 min.Resuspend the pelleted bacteria in 50 µl of 2xTY and plate on a regular TYE plate containing 100 µg/ml ampicillin and 1 % glucose.11. Grow plates at 37°C overnight.The first round of selection is the most important. Any errors made at this point will be amplified in subsequent rounds of selection. After each round you should get back at least 100 infective phage. If you get less than this it is probable that a mistake has occurred. If so, try repeating the infection with a freshly grown TG1 culture (from a new overnight) at an OD 600 of 0.4 using the remaining 250 µl of eluted phage from C7. If this still yields less than 100 phage, repeat the selection starting at C1.1. After overnight growth add 7 ml of 2xTY 15 % glycerol to the large square Bio-Assay dish or 2mlsto the regular plates and loosen the cells with a glass spreader, mixing them thoroughly. After inoculating 50 µl of the scraped bacteria to 50 ml of 2xTY containing 100 µg/ml ampicillin and 1 % glucose, store 1 ml of the remaining bacteria at -70°C in 15% glycerol.2. Grow shaking at 37°C until the OD 600 is 0.4 (1-2 hrs).3. Take 10 ml of this culture and add 5x1010 helper phage.4 Incubate without shaking in a 37°C water bath for 30 min.5. Spin at 3,000 g for 10 min. Resuspend in 50 ml of 2xTY containing 100 µg/ml ampicillin, 50 µg/mlkanamycin and 0.1% glucose.6. Incubate shaking at 30°C overnight.7. Spin the overnight culture at 3,300 g for 15 min.8. Add 10 ml PEG/NaCl (20 % Polyethylene glycol 6000, 2.5 M NaCl)to 40 ml supernatant. Mix welland leave for 1 hr on ice.9. Spin 3,300 g for 30 min. Pour away PEG/NaCl. Respin briefly and aspirate any remaining dregsof PEG/NaCl.10. Resuspend the pellet in 2 ml PBS and spin at 11,600 g for 10 min in a micro centrifuge to removethe remaining bacterial debris.11. Use 1 ml of this phage for the next round of selection (protocols C and D) and store 1 ml at 4°C.12. Repeat selection for another 2 rounds.Populations of phage produced at each round of selection can be screened for binding by ELISA to identify "polyclonal" phage antibodies. Phage from single colonies can then be screened by ELISA to identify "monoclonal" phage antibodies. Alternatively, after a polyclonal phage ELISA you could proceed directly to making monoclonal soluble antibody fragments, see protocol F. In general, we have found that 2% Marvel in PBS is best for blocking during phage ELISA whereas 3% BSA in PBS is best for blocking during scFv ELISA.1. Coat a 96 well flexible assay plate10 overnight with 100 µl per well of antigen in the same bufferand at the same concentration as used for selection.2. Wash wells 3 times with PBS. Plates can be immersed in a shallow bath containing PBS but youshould check that all wells fill with wash solution (if they do not you may create false positivesduring later washes). Discard liquid by flipping plate over and then shaking it. Add 200 µl per well of 2 % MPBS (2 % Marvel in PBS) or 3% BSA-PBS (3% bovine serum albumin in PBS) to block and incubate for 2 hr at rt.3. Wash wells 3 times with PBS. Add 10 µl PEG precipitated phage from the end of each round ofselection in 100 µl 2 % MPBS (or 3 % BSA-PBS).4. Incubate for 1 hr at rt. Discard phage solution and wash 3 times with PBS-0.1 % Tween 20.5. Add 1 in 5000 dilution of HRP-anti-M1311 in 2 % MPBS (or 3 % BSA-PBS). Incubate for 1 hr atrt., wash 3 times with PBS-0.1 % Tween 20.µl of substrate solution (100 µg/ml TMB12 in 100 mM sodium acetate, pH 6.0. with10 µl 1006. Addof 30 % hydrogen peroxide added to 50 ml of this solution directly before use) to each well and leave at rt. for 2-15 min. A blue colour should develop.7. Stop the reaction by adding 50 µl 1 M sulphuric acid. The blue colour should turn yellow.9. Inoculate individual colonies from the titration plates from each round of selection (see C11) into100 µl 2xTY containing 100 µg/ml ampicillin and 1 % glucose in 96 cell-well plates13. Growshaking (250 rpm) overnight at 37°C.10. Use a 96 well transfer device14 to transfer a small inoculum (about 2 µl) from this plate to asecond 96 cell-well plate containing 200 µl of 2xTY with 100 µg/ml ampicillin and 1 % glucose per well. Grow shaking (250 rpm) at 37°C for 2 hr. (Make glycerol stocks of the original 96-well plate, by adding glycerol to a final concentration of 15 %, and then storing the plates at -70°C).11. After 2 hr incubation (of the second plate) add 25 µl 2xTY containing 100 µg/ml ampicillin, 1 %glucose and 109 helper phage.12. Shake (250 rpm) at 37°C for 1 hr. Spin 1,800 g for 10 min, then aspirate off the supernatant.13. Resuspend pellet in 200 µl 2xTY containing 100 µg/ml ampicillin and 50 µg/ml kanamycin. Growshaking (250 rpm) overnight at 30°C.14. Spin at 1,800 g for 10 min and use 50 µl of the supernatant in phage ELISA as detailed above.Individual colonies picked from the various rounds of selection (as plated on the dilution series) can be induced in TG-1 to produce soluble scFv (F2-F6). This will ensure the expression of all selected clones including those in which the scFvs contain TAG stop codons (TG-1 as able to suppress termination and introduce a glutamate residue at these positions). Unfortunately, since the TAG stop codon between the scFv and the gIII gene is also suppressed this leads to co-expression of the scFv-pIII fusion which tends to lower the overall levels of scFv expression, even in clones where there are no TAG stop codons in the scFv itself. To circumvent this problem, the selected phage can be used to infect HB2151 (a non-suppresor strain) which is then induced to give soluble expression of antibody fragments (scFv genes that do not contain TAG stop codons will now yield higher levels of soluble scFv than in TG-1, but those that contain TAG stop codons will not produce any soluble scFv) (F1-F6). The expressed scFvs can then be used in ELISA. Detection of bound scFv can be performed using either Protein A-HRP15 or Protein L-HRP16 conjugates.1. From each selection take 10 µl of eluted phage and infect 200 µl exponentially growing HB2151bacteria (OD 600 of 0.4) for 30 min at 37°C in a water bath. Plate 50 µl, 50 µl of a 1:102 dilution,50 µl of a 1:104 dilution and 50 µl of a 1:106 dilution on TYE plates containing 100 µg/mlampicillin and 1 % glucose and grow overnight at 37°C.2. Pick individual colonies into 100 µl 2xTY 100 µg/ml ampicillin and 1 % glucose in 96 cell-wellplates and grow shaking (250 rpm) overnight at 37°C.e a 96 well transfer device14 to transfer a small inocula (about 2 µl) from this plate to a second96 cell-well plate containing 200 µl 2xTY containing 100 µg/ml ampicillin and 0.1 % glucose perwell. Grow shaking (250 rpm) at 37°C until the OD 600 is approximately 0.9 (about 3 hr). (A stock can be made of the first plate, by adding glycerol to a final concentration of 15 % and storing at -70°C).4. Once OD 0.9 is reached (wells look quite cloudy) add 25 µl 2xTY containing 100 µg/ml ampicillinand 9 mM IPTG (isopropyl β-D-thiogalactoside, final concentration 1 mM IPTG). Continueshaking (250 rpm) at 30°C overnight.5. Coat a 96 well flexible assay plate overnight with 100 µl per well of antigen in the same buffer andat the same concentration as used for selection.6. Spin the plate from step F4 at 1,800 g for 10 min and use 50 µl of the supernatant (take care notto transfer any bacteria) for ELISA in 3% BSA-PBS (final concentration) (protocol E) except usinga 1:5000 dilution of Protein A-HRP15 or Protein L-HRP16 to detect binding in step E5.µl TG1 at an OD 600 of 0.4 with 10 µl of 100-fold serial dilutions of KM13 helper 2001. Infectphage3 (in order to get well separated plaques) in a 37°C water bath (without shaking) for 30 min.Add to 3 ml molten H-top agar (42°C) and pour onto warm TYE plates (no antibiotics). Allow to set and incubate overnight at 37°C.2. Pick a small plaque into 5 ml of fresh TG1 at an OD 600 of 0.4. Grow for about 2 hr shaking at37°C.3. Add to 500 ml 2xTY in a 2 litre flask and grow shaking at 37°C for 1 hr. Add kanamycin to a finalconcentration of 50 µg/ml (no glucose). Grow overnight shaking at 30°C.4. Spin overnight culture at 10,800 g for 15 min. Add 100 ml PEG/NaCl (20 % polyethylene glycol6000, 2.5 M NaCl) to 400 ml supernatent and leave for 1 hr on ice.5. Spin 10,800 g for 30 min. Pour away PEG/NaCl.6. Resuspend the pellet in 8 ml PBS and add 2 ml PEG/NaCl. Mix well and leave for 20 minutes onice.7. Spin 3,300 g for 30 min. Respin briefly and aspirate any remaining dregs of PEG/NaCl.8. Resuspend pellet in 5 ml PBS and spin at 11,600 g for 10 min in a micro centrifuge to remove anyremaining bacterial debris.9. Store the helper phage at 4°C for short term storage or in PBS with 15 % glycerol for longer termstorage at -70°C.10. To titre the helper phage take 45µl phage and add 5µl trypsin stock solution. Incubate for 30 minsat 37 °C. Dilute 1µl of trypsin treated phage in 1ml PBS and make five 100 fold serial dilutions of this in 1ml aliquots of PBS. Add 50µl of the six dilutions to six separate tube containing 1ml of TG1 at an OD 600 of 0.4. Mix, add 3 ml molten H-Top and pour evenly onto TYE plates.Perform the same dilution series using 1µl of non-trypsin treated phage. The titre of the trypsin treated phage should be 105-108 lower than for the non-trypsin treated phage. If not, pickanother plaque and repeat helper phage preparation.Once the libraries have been selected you may wish to check individual clones for the presence of full length VH and Vκ insert. All PCRs are at annealing temperature of 55°C. 1 min extension for V H or Vκon their own, 2 min extension for V H and Vκ together.For V H only use LMB3: CAG GAA ACA GCT ATG AClink seq new: CGA CCC GCC ACC GCC GCT Gwith insert = 527 bpwithout insert = 227 bpFor Vκ only use DPK9 FR1 seq: CAT CTG TAG GAG ACA GAG TCpHEN seq: CTA TGC GGC CCC ATT CAwith insert = 368 bpwithout insert = no bandFor V H and Vκ use LMB3: CAG GAA ACA GCT ATG ACpHEN seq: CTA TGC GGC CCC ATT CAwith insert = 935 bpwithout insert = 329 bpFor sequencing of selected clones the following primers are recommended.For V H use link seq new CGA CCC GCC ACC GCC GCT GFor Vκ use pHEN seq CTA TGC GGC CCC ATT CAXhoI NotIRBSCAGGAAACAGCTATGACCATGATTACGCCAAGCTTGCATGCAAATTCTATTTCAAGGAGACAGTCATA ATG AAA TAC CTA ----------------> M K Y L LMB3SfiI __NcoI__TTG CCT ACG GCA GCC GCT GGA TTG TTA TTA CTC GCG GCC CAG CCG GCC ATG GCC GAG GTG TTT L P T A A A G L L L L A A Q P A M A E V F XhoI linkerGAC TAC TGG GGC CAG GGA ACC CTG GTC ACC GTC TCG AGC GGT GGA GGC GGT TCA GGC GGA GGTD Y W G Q G T L V T V S S G G G G S G G GSalI NotI HIS-tag GGC AGC GGC GGT GGC GGG TCG ACG GAC ATC CAG ATG ACC CAG GCG GCC GCA CAT CAT CAT CACG S G G G G S T D I Q M T Q A A A H H H H<------------------------link seq newmyc-tagCAT CAC GGG GCC GCA GAA CAA AAA CTC ATC TCA GAA GAG GAT CTG AAT GGG GCC GCA TAG ACTH H G A A E Q K L I S E E D L N G A A * T<---------------------pHEN seq1. 2xTY is 16g Tryptone, 10g Yeast Extract and 5g NaCl in 1 litre.2. Bovine ubiquitin (5 mg) is available from Fluka, Chemika-BioChemika, Industriestrasse 25, CH-9470 Buchs, Switzerland, Tel +41 81 755 2511, Fax +41 81 756 5449.3. KM13 is the protease cleavable helper phage described i n Kristensen and Winter, Folding andDesign3, 321-328 (1998).4. PBS is5.84 g NaCl, 4.72 g Na2HPO4 and 2.64 g NaH2PO4.2H20, pH 7.2, in 1 litre.5. TYE is 15g Bacto-Agar, 8g NaCl, 10g Tryptone, 5g Yeast Extract in 1 litre.6. Nunc Maxisorp immuno test tubes (Cat. No. 4-44202) are available from Gibco BRL, LifeTechnologies Ltd., P. O. Box 35, Trident House, Washington Road, Paisley, PA3 4EF, Scotland, U.K, Tel +44 141 814 6100, Fax +44 141 887 1167.7. 'Marvel' is dried skimmed milk powder.8. Trypsin (T-1426 Type XIII from Bovine Pancreas - Sigma Chemical Company Ltd., Fancy Rd.,Dorset, BH17 7NH, U.K, Tel +44 1202 733114; Fax +44 1202 715460) made up in 50mM Tris-HCl pH7.4, 1mM CaCl2 and stored at -20°C9. Nunc Bio-Assay dish is available from Gibco BRL (see note 6).10. Falcon MicroTest III flexible 96 well flat bottomed assay plates are available from BectonDickinson Labware, Becton Dickinson and Co., 2 Bridgewater Lane, Lincoln Park, NJ 07035,USA.11. HRP-anti-M13 is available from Amersham International plc, Amersham Place, Little Chalfont,Buckinghamshire, HP7 9NA, UK. Tel: +44 01494 544000; Fax: +44 01494 542929.12. TMB is 3,3',5,5'-tetramethylbenzidine and is available from Sigma (see Note 8). A 10 mg/ml stocksolution can be made by dissolving the TMB in DMSO.13. Corning 'Cell Wells' flat-bottomed multiple well tissue culture treated plates are available fromCorning Glass Works, Corning N.Y. 14831. USA.14. The 96 well transfer device is a piece of wood the size of a microtitre plate with a handle on oneside and 96 metal pins (each 7 cm long with a concave end) on the other. This can be sterilised between bacterial transfer by immersion in a bath of ethanol and then by flaming (hold well away from your body and any flamable objects). If you haven't got one of these (or something similar) you will have to make one yourself or alternatively use a multichannel pipette for bacterialtransfer.15. Horse Radish Peroxidase conjugated Protein A is available from Amersham International plc (seenote 11)14. Horse Radish Peroxidase conjugated Protein L is available from Actigen Ltd, 5 Signet Court,Swanns Road, Cambridge, CB5 8LA, UK. Tel: +44 01223 319101; Fax: +44 01233 316443.。