BD产品图谱
- 格式:pdf
- 大小:13.23 MB
- 文档页数:1

BD Player 介绍2011年9月23日BD特色及功能介绍特及功能介绍BD做为个新兴产品z做为一个新兴产品,那它的优势在哪里?我们怎么样去给客户介绍这样一个新的东西?z从BD产品的名字来看“Blu-ray”,(注意不是Blue-ray,蓝光光盘的英文名称不使用“Blu e-ray”的原因,是“Blue-ray Disc”这个词在欧美的原因是“地区流于通俗、口语化,并具有说明性意义,于是不能构成注册商标申请的许可,因此蓝光于是不能构成注商标申请的许因此蓝光光盘联盟去掉英文字e来完成商标注册。
)那它采用的技术一定是和蓝光有关,做为一种碟它采用的技术定是和蓝光有关做为种碟BD特色及功能介绍特及功能介绍机,它是能够读Blu—ray Disc,它是通过波长机它是能够读Bl ra Disc较短的蓝色激光(波长为405nm)来读取数据或写入数据也因此而得名而传统的红光或写入数据,也因此而得名。
而传统的红光DVD是用红色激光(波长为650nm)来读去或写入数据而CD则是采用780nm的波长,通的波长通常来说,波长越短的激光能够在单位面积上记录或读取的信息越多。
因此,蓝光极大地提高录或读取的信息越多因此蓝光极大地提高了光盘的存储容量,一张蓝光光盘可以做到50G Bytes的容量是现有红光25G Bytes和50G Bytes的容量,是现有红光DVD的5倍多,对光存储产品来说,的特及功能介绍BD的特色及功能介绍蓝光提供了一个跳跃式的发展机会.随着人们对图像质量的要求越来越高,蓝光产品的巨大容量为高清电影1080P,和大容量数据存储到来1080P了可能和方便,很大程度上促进了高清娱乐的发展.蓝光播放器可以完全向下兼容红光DVD.光播完兼光综上,可以得出一个结论,蓝光播放器的一个优点就是:大容量存储和高清播放。
BD如果只有这一个优点还是不够的,除去这个优点,它还是一台多媒体终端,它可以播放的特及功能介绍BD的特色及功能介绍多种高清媒体格式:H.264,VC-1,MPEG-1,MPEG-多种高清媒体格式H264VC1MPEG1MPEG2,MPEG-4, DivX Plus (HD),可支持的封装形式有:.MKV,.TS,.AVI,.MPEG,.MPG,.MP4,.DAT,.ASF., MOV等。

BD-FACSCalibur流式细胞仪培训手册BD FACSCalibur流式细胞仪FACS101 Handbook二、开机、关机标准操作2.1 FACS Calibur 开机1. 开启细胞仪电源。
2. 开启其它周边配备电源,如打印机及MO机。
3. 开启计算机。
4. 确认鞘流液筒有八分满的FACSFlow,确实旋紧(鞘液筒容量为4L)。
5. 将废液倒掉,并在废液筒中参加200ml 家用漂白水(废液筒容量为4L)。
6. 将减压阀方向调在加压〔Pressurize〕位置。
7. 排除液流管路与过滤器中的气泡。
8. 取下样品管,执行PRIME 功能两次。
9. 使用1ml PBS,HIGH RUN 两分钟。
10. 可开始分析样品。
2.2 FACS Calibur 关机关机前必要动作:清洗进样管和外套管,防止进样管堵塞、或有染料残留。
1. 将样品支持架左移,取2 mlFACSClean〔10%Bleach〕上样品,让仪器的真空系统抽取约 1 ml 的液体。
2. 将样品支持架回正,按HI RUN,然后让FACSClean 清洗管路10分钟。
3. 按Standby,取下样品管,执行PRIME功能两次。
4. 取2 ml dH2O,重复上述步骤1-3。
5. 注意最后只留约1 ml dH2O 在试管中。
6. 按STANDBY 五分钟,使风扇冷却雷射后,关闭细胞仪〔必要动作,以保护雷射光源。
〕7. 倒掉废液,并回填200 ml漂白水。
8. 将减压阀放在「VENT 漏气」位置。
将鞘流液筒充填至八分满。
9. 退出软件“File〞→“Quit〞(如有对话选项,选择“Don‘t save〞)。
确认退出计算机中所有BD应用软件,所有数据数据已储存备份。
10. 关闭计算机。
“Special〞→“Shutdown〞。
三、上机分析流程建议首次试机防止进行大量试验,仅需准备以下样品。
〔1〕Negative Control〔不加任何抗体〕。

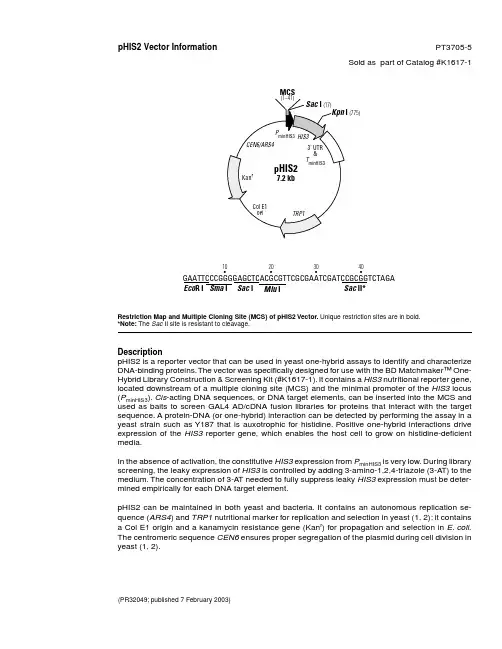
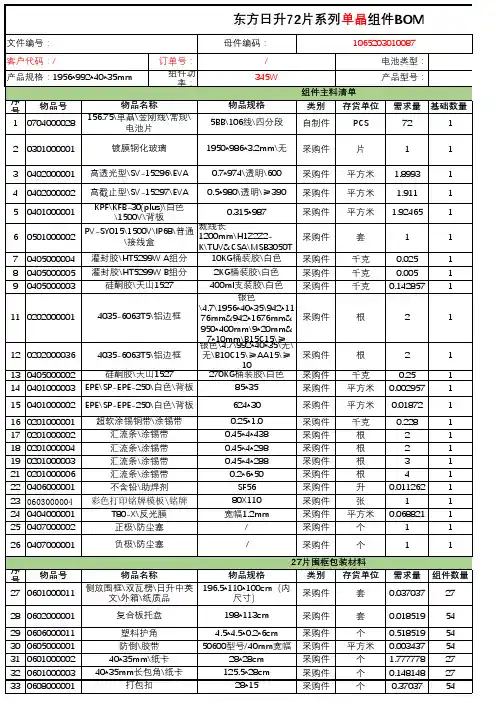
(PR32049; published 7 February 2003)pHIS2 Vector Information PT3705-5Sold as part of Catalog #K1617-1Restriction Map and Multiple Cloning Site (MCS) of pHIS2 Vector. Unique restriction sites are in bold.*Note: The Sac II site is resistant to cleavage.DescriptionpHIS2 is a reporter vector that can be used in yeast one-hybrid assays to identify and characterizeDNA-binding proteins. The vector was specifically designed for use with the BD Matchmaker™ One-Hybrid Library Construction & Screening Kit (#K1617-1). It contains a HIS3 nutritional reporter gene,located downstream of a multiple cloning site (MCS) and the minimal promoter of the HIS3 locus(P minHIS3). Cis -acting DNA sequences, or DNA target elements, can be inserted into the MCS andused as baits to screen GAL4 AD/cDNA fusion libraries for proteins that interact with the targetsequence. A protein-DNA (or one-hybrid) interaction can be detected by performing the assay in ayeast strain such as Y187 that is auxotrophic for histidine. Positive one-hybrid interactions driveexpression of the HIS3 reporter gene, which enables the host cell to grow on histidine-deficientmedia.In the absence of activation, the constitutive HIS3 expression from P minHIS3 is very low. During libraryscreening, the leaky expression of HIS3 is controlled by adding 3-amino-1,2,4-triazole (3-AT) to themedium. The concentration of 3-A T needed to fully suppress leaky HIS3 expression must be deter-mined empirically for each DNA target element.pHIS2 can be maintained in both yeast and bacteria. It contains an autonomous replication se-quence (ARS4) and TRP1 nutritional marker for replication and selection in yeast (1, 2); it containsa Col E1 origin and a kanamycin resistance gene (Kan r ) for propagation and selection in E. coli .The centromeric sequence CEN6 ensures proper segregation of the plasmid during cell division inyeast (1, 2).MCS(775)GAATTCCCGGGGAGCTCACGCGTTCGCGAATCGATCCGCGGTCTAGA10•Sac II*Eco R I Mlu I Sac I Sma I 30•40•20•pHIS2Vector Information BD Biosciences Clontech Protocol # PT3705-52Version # PR32049Notice to PurchaserThis product is intended for research purposes only. It is not to be used for drug or diagnostic purposes nor is it intended for human use. BD Biosciences Clontech products may not be resold, modified for resale, or used to manufacture commercial products without written approval of BD Biosciences Clontech.BD and BD Logo are trademarks of Becton, Dickinson and Company. ©2003 BDUseTo use pHIS2 in a one-hybrid assay, clone one or more copies of a cis -acting DNA target sequence into the MCS.Then introduce the plasmid into competent yeast cells using the transformation protocol in the BD Matchmaker Library Construction & Screening Kits User Manual (PT3529-1). In contrast to the original BD Matchmaker One-Hybrid System, this reporter vector does not need to be integrated into the yeast genome. Instead, it is maintained as an episome throughout the assay. Inserting your target element may alter the level of background HIS3 expression.Therefore, constructs should be tested for background (leaky) HIS3 expression before you start a one-hybrid analy-sis. Background growth due to leaky HIS3 expression is controlled by adding 3-A T to the selection medium, as described in the User Manual (PT3529-1).Location of Features•Multiple cloning sites: 1–41•HIS3 gene: 152–811•Fragment containing the HIS3 3' UTR & Termination sequence: 812–1446•TRP1 gene: 2855–3529•Fragment containing the Col E1 E. coli origin of replication: 4165–4615•Kanamycin-resistance gene: 5605–4811•CEN6/ARS4 sequences: 6254–5737Propagation in E. coli•Suitable host strains: DH5α and other general purpose strains.•Selectable marker: plasmid confers resistance to kanamycin (50 µg/ml) to E. coli hosts.• E. coli replication origin: Col E1•Copy number: lowPropagation in S. cerevisiae•Suitable host strains: AH109(MAT a ), Y187(MAT α), Y190(MAT a ), SFY526(MAT a ), CG1945(MAT a ), HF7c(MAT a )•Selectable marker: TRP1•S. cerevisiae origin: CEN6/ARS4References1.Sikorski, R. S. & Hieter, P. (1989) Genetics 122:19–27.2. Rose, M. D. & Broach, J. R. (1991) Methods Enzymol. 194:195–230.Note: The attached sequence file has been compiled from information in the sequence databases, published litera-ture, and other sources, together with partial sequences obtained by BD Biosciences Clontech. This vector has not been completely sequenced.。


本产品保密并受到版权法保护2024易观分析2024年3月01电信业人工智能行业应用发展背景电信网络基础设施建设日益完备,但终端用户规模扩大、新兴业务发展对于网络性能要求明显提高来源:工信部,易观分析整理667.2841931996.310831162372544.1575590.2602.7629.515.377.1142.5231.2337.70%2%8%14%21%29%2018201920202021202220232018-2023年移动电话基站规模及变化情况(单位:万个,%)移动电话基站数4G 基站数5G 基站数5G 基站占比17441613143513751337129310841374173723723060356413.4%12.3%10.6%9.4%8.5%7.7%21.8%26.0%26.4%25.2%32.1%19.1%2018201920202021202220232018-2023年电信行业业务收入及变化情况(单位:亿元,%)语音业务收入新兴业务收入语音业务增速新兴业务增速移动互联网接入总流量20182023月户均移动互联网流量711亿GB3015亿GB4.6GB/户月16.9GB/户月蜂窝物联网终端用户数6.7亿户23.3亿户物联网用户占移动终端比重30%电信行业作为我国数字新基建重点领域,已经日益融入社会生产生活的肌理当中,是拉动数字经济增长的重要引擎。
2023年,我国宽带接入端口同比增长6.5%;5G 基站同比增长7.8%,占移动基站总数比例上升至29%。
移动互联网接入流量、物联网用户规模持续稳增,带动2023年电信业务收入同比增长6.2%,按上年价格计算的电信业务总量同比增长16.8%。
从业务结构看,电信行业传统语音业务收入下滑,但数据中心、云计算、大数据、物联网等新兴业务收入保持高速增长,2023年业务收入达到3564亿元,同比增长19.1%。
与此同时,这些新兴业务产生的数据量大幅增加,无论是传输数据规模、实时性、可靠性要求相较传统业务都明显更高,对网络性能的要求也相应提高,需要网络具备更强的处理能力、带宽、延迟控制能力、稳定性和安全性来满足业务需求。
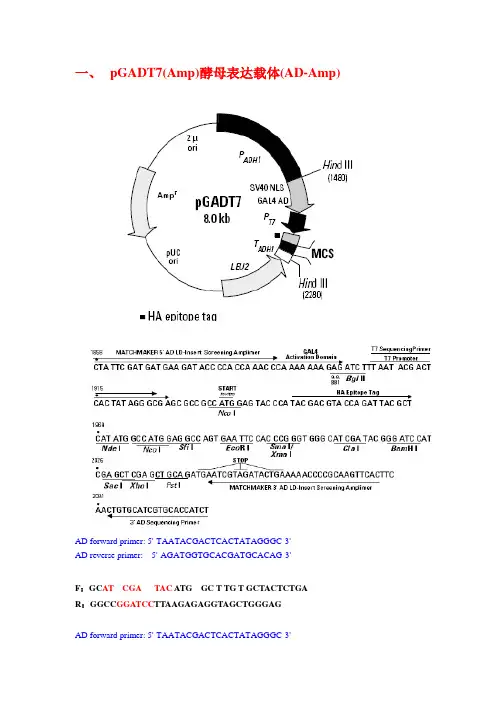
一、pGADT7(Amp)酵母表达载体(AD-Amp)AD forward primer: 5'-TAATACGACTCACTATAGGGC-3'AD reverse primer: 5'-AGATGGTGCACGATGCACAG-3'F:GC AT CGA TAC ATG GC T TG T GCTACTCTGAR:GGCC GGATCC TTAAGAGAGGTAGCTGGGAGAD forward primer: 5'-TAATACGACTCACTATAGGGC-3'AD reverse primer: 5'-AGATGGTGCACGATGCACAG-3'二、pGBKT7(Kan0酵母表达载体(BD-Kan)三、BIFC(Kan)验证互做蛋白表达载体(德国)四、pGR107(Kan)表达载体(PVX-10kb)吴建国MCS:ClaI/SmaI/SalI(pgR107)MCS:ClaI/AscI/NotI/SalI(pgR106) 五、BIFC(Amp)载体(美国)3075(nEYFP-329bp)3076 (cEYFP-218bp)F:GGC GAATTC ATG GCTTGTGCTACTCTGA R:GGC CCTAGG AGAGAGGTAGCTGGGAG TAA TAG TGA六、RNAI(Kan)载体-pFGC5941(王宗华)七、pGEX-4T-1(Amp)原核表达载体八:pET-29a(Kan)原核表达载体九、. pEGAD(Kan)植物表达载体*XbaⅠ cleavage blocked by overlapping dam methylation. Must grow in dam-strain to cut. Note, XhoⅠ is not unique.pTRV2(a) Genome organization of TRV. The TRV-RNA1 open reading frames (ORFs) correspond to 134 and 194 kDa replicase, movement protein (MP) and a 16-kDa cysteine-rich protein. The TRV-RNA2 ORFs corresponds to coat protein (CP), and the 29.4 and 32.8 kDa proteins. gRNA, genomic RNA; sgRNA, subgenomic RNA. Asterisks (*) indicate the readthrough of 134 kDa protein.(b) TRV based VIGS vectors. TRV cDNA clones were placed in betweenthe duplicated CaMV 35S promoter (2×35S) and the nopaline synthase terminator (NOSt) in a T-DNA vector. LB and RB refer to left and right borders of T-DNA. Rz, self-cleaving ribozyme. MCS, multiple cloning sites.十、十一、1103-YFP十二:pBINPLUS十三、pBin438。


pFB-Neo质粒图谱pFB and pFB-Neo Retroviral VectorsINSTRUCTION MANUALCatalog #217563 (pFB Retroviral Vector) and #217561 (pFB-Neo Retroviral Vector) Revision AFor In Vitro Use Only217561-12L IMITED P RODUCT W ARRANTYThis warranty limits our liability to replacement of this product. No other warranties of anykind, express or implied, including without limitation, implied warranties of merchantability orfitness for a particular purpose, are provided by Agilent. Agilent shall have no liability for anydirect, indirect, consequential, or incidental damages arising out of the use, the results of use, orthe inability to use this product.O RDERING I NFORMATION AND T ECHNICAL S ERVICESUnited States and CanadaAgilent TechnologiesStratagene Products Division11011 North Torrey Pines RoadLa Jolla, CA 92037373-6300Telephone(858)424-5444Order Toll Free (800)Technical Services(800) 894-1304Internet techservices@/doc/dba401755b8102d276a20029bd64783e08127d58.htmlWorld Wide Web /doc/dba401755b8102d276a20029bd64783e08127d58.htmlEuropeServices Location Telephone Fax TechnicalAustria 0800 292 499 0800 292 496 0800 292 498Belgium 00800 7000 7000 00800 7001 7001 00800 7400 74000800 15775 0800 15740 0800 15720France 00800 7000 7000 00800 7001 7001 00800 7400 74000800 919 288 0800 919 287 0800 919 289 Germany 00800 7000 7000 00800 7001 7001 00800 7400 74000800 182 **** **** 182 **** **** 182 8234 Netherlands 00800 7000 7000 00800 7001 7001 00800 7400 74000800 023 0446 +31 (0)20 312 5700 0800 023 0448 Switzerland 00800 7000 7000 00800 7001 7001 00800 7400 7400 0800 563 080 0800 563 082 0800 563 081 United Kingdom 00800 7000 7000 00800 7001 7001 00800 7400 74000800 917 3282 0800 917 3283 0800 917 3281 All Other CountriesPlease contact your local distributor. A complete list of distributors is available at/doc/dba401755b8102d276a20029bd64783e08127d58.html .pFB and pFB-Neo Retroviral VectorsC ONTENTSMaterials Provided (1)Storage Conditions (1)Introduction (2)The pFB Vector (3)The pFB-Neo Vector (4)pFB-Luciferase Control Vector (5)Replication-Defective Retroviral Gene Transfer Systems (6)Biosafety Considerations (6)Ligation of Vector and Insert (7)Ligation Guidelines (7)Ligation Protocol (8)Recombination and Host Strains for Subcloning and Propagation (9)Primers (9)Preparation of Media and Reagents (10)References (10)Supplemental References (10)MSDS Information (10)pFB and pFB-Neo Retroviral Vectors M ATERIALS P ROVIDEDMaterial providedQuantityCatalog #217561 Catalog #217563pFB-Neo retroviral vector 10 µg —pFB retroviral vector — 10 µgpFB-Luciferase expression control vector 10 µg 10µgS TORAGE C ONDITIONSAll components:–20°CN OTICE TO P URCHASERUse of the translation enhancer of the pFB-Neo vector is covered by U.S. Patent No. 4,937,190 and is limited to use solely for research purposes. Any other use of the translation enhancer of the pFB-Neo vector requires a license from WARF.B IOSAFETYC ONSIDERATIONSThe host range of a retrovirus is determined not by the vector DNA but by the specific env gene used to construct the packaging cell line. Viruses produced from amphotropic or polytropic packaging lines are capable of infecting human cells. The National Institutes of Health has designated retroviral vectors, such as MMLV, as Biosafety Level 2. Appropriate caution should be used in the production and use of recombinant retrovirus. For more information see Biosafety in Microbiological and Biomedical Laboratories at /doc/dba401755b8102d276a20029bd64783e08127d58.html/od/ors/ds/pubs/ bmbl/contents.htm.Revision A ? Agilent Technologies, Inc. 2008.I NTRODUCTIONThe Stratagene pFB and pFB-Neo plasmid vectors for retroviral genedelivery and expression are derived from the Moloney murine leukemiavirus (MMLV) and can be used to produce high titer viral stocks withMMLV-based packaging cell lines.1,2 Both vectors contain the bacterialorigin of replication and ampicillin-resistance gene from pBR322, anextended MMLV packaging signal (ψ+), and a multiple cloning site (MCS)located between the MMLV 5′ and 3′ long terminal repeat sequences(LTRs), see Figures 1 and 2.The pFB vector is a minimal MMLV-based vector that can accommodatelarger inserts than pFB-Neo. The pFB vector does not contain anyexogenous sequence other than the sequence of the MCS.The pFB-Neo vector differs from the pFB vector only by the presence of acassette consisting of the internal ribosome entry site (IRES) from theencephalomyocarditis virus (EMCV) and the neomycin-resistance gene(neo r). The open reading frame (ORF) for the neo r gene is positioneddownstream of the MCS and follows the IRES. A dicistronic messageencoding the gene of interest and the neo r gene is expressed from the viralpromoter within the 5′ LTR.The pFB VectorFeatureNucleotide position 5′-long terminal repeat (LTR)209–760 transcription initiation (clockwise)616 splice donor818–822 ψ+ extended viral packaging signal760–2046 gag gene (truncated)1236–1723 splice acceptor1751–1753 5′ retro primer binding site2008–2028 multiple cloning site2057–2086 3′ pFB primer binding site2121–2101 3′-long terminal repeat (LTR)2163–2756 pBR322 origin of replication3237–3904 ampicillin resistance (bla ) ORF 4055–4912F IGURE 1 Circular map and features for the pFB retroviral vector. The complete nucleotide sequence and list of restriction sites can be found at /doc/dba401755b8102d276a20029bd64783e08127d58.html .EcoR I Sal I Xho I Not IBamH I pFB Multiple Cloning Site Region (sequence shown 2057?086)The pFB-Neo VectorFeatureNucleotide Position 5′-long terminal repeat (LTR)209–760 transcription initiation (clockwise)616 splice donor818–822 ψ+ extended viral packaging signal760–2046 gag gene (truncated)1236–1723 splice acceptor1751–1753 5′ retro primer binding site2008–2028 multiple cloning site2057–2086 3′ pFB primer binding site2147–2127 internal ribosome entry site (IRES)2093–2586 neomycin resistance ORF2763–3557 3′-long terminal repeat (LTR)3617–4210 pBR322 origin of replication4691–5358 ampicillin resistance (bla ) ORF 5509–6366F IGURE 2 Circular map and features for the pFB-Neo retroviral vector. The complete nucleotide sequence and list of restriction sites can be found at /doc/dba401755b8102d276a20029bd64783e08127d58.html . EcoR I Sal I Xho I Not IBamH I pFB-Neo Multiple Cloning Site Region (sequence shown 2057?086)pFB-L UCIFERASE C ONTROL V ECTORThe pFB-Luc plasmid vector (Figure 3), which contains the luciferase gene,is included with the vectors as an expression control. A retroviral vectorexpressing a reporter gene is useful for optimizing transfection efficiency,confirming viral production, as well as ascertaining whether or not a targetcell line can be infected by a given viral stock.Figure 3 Circular map for the pFB-Luc control vector. The complete nucleotide sequence and list of restriction sites can be found at /doc/dba401755b8102d276a20029bd64783e08127d58.html .R EPLICATION-D EFECTIVE R ETROVIRAL G ENE T RANSFER S YSTEMSReplication-defective retroviral vectors contain all of the cis elementsrequired for transcription of a gene of interest and packaging of thetranscripts into infectious viral particles.2 The retroviral vectors typicallyconsist of an E. coli plasmid backbone containing a pair of viral LTRsbetween which the gene of interest is inserted. In order to generateinfectious virus particles that carry the gene of interest, specializedpackaging cell lines have been generated that contain chromosomallyintegrated expression cassettes for viral proteins Gag, Pol, and Env, all ofwhich are required in trans for production of virus.After the packaging cell line is transfected with the vector DNA, either thesupernatant is collected for transiently produced viral stocks, or stable viralproducer cell lines are selected (provided the vector has an appropriateselectable marker). The supernatant is used to infect dividing target cells.Upon infection, the viral RNA molecule is reverse transcribed by reversetranscriptase (which is present in the virion particle), and the gene ofinterest, flanked by the LTRs, is integrated into the host DNA. Because thevector itself does not express viral proteins, once a target cell is infected, theLTR expression cassette is incapable of another round of virus production.B IOSAFETYC ONSIDERATIONSThe host range of a retrovirus is determined not by the vector DNA but bythe specific env gene used to construct the packaging cell line. Virusesproduced from amphotropic or polytropic packaging lines are capable ofinfecting human cells. The National Institutes of Health has designatedretroviral vectors, such as MMLV, as Biosafety Level 2. Appropriatecaution should be used in the production and use of recombinant retrovirus.For more information see Biosafety in Microbiological and BiomedicalLaboratories at /doc/dba401755b8102d276a20029bd64783e08127d58.html /od/ors/ds/pubs/ bmbl/contents.htm.L IGATION OF V ECTOR AND I NSERTLigation GuidelinesBecause the size of retroviral RNA that can be efficiently packaged islimited to ~11 kb (including the 5′ and 3′ LTRs),1 the size of the insertshould be <8.4 kb for the pFB vector and <7.0 kb for the pFB-Neo vector.Inserts should include both initiation and stop codons. Design the insert tocontain a Kozak sequence. A complete Kozak sequence includesCCACC ATG G, although CC ATG G, or the core ATG, is sufficient.Dephosphorylate the digested plasmid DNA with alkaline phosphatase priorto ligation with the insert DNA. If more than one restriction enzyme is used,the background can be reduced further by gel purification of the digestedplasmid DNA. After gel purification, resuspend the DNA in TE buffer (seePreparation of Media and Reagents) or water.For ligation, the ideal ratio of insert-to-vector DNA is variable; however, areasonable starting point is 2:1 (insert:vector), measured in availablepicomole ends. This ratio is calculated as follows:picomole ends/microgram of DNA2 ?10number of base pairs ?6606=Ligation Protocol1. Prepare the following two experimental and three control ligation reactions by adding the following components in order to separate 0.5-ml microcentrifuge tubes:Component Insertcontrol aDigestcontrol bPhosphatasecontrol cTest I2:1Test IIX:1Prepared vector(10 ng/µl)— 1 µl 1 µl 1 µl 1 µlPrepared insert(10 ng/µl)X µl — — X µl X µl 10 mM rATP (pH 7.0) 0.5 µl 0.5µl 0.5µl 0.5µl 0.5 µl 10× ligase buffer d 0.5µl 0.5µl 0.5µl 0.5µl 0.5 µl T4 DNA ligase (4 U/µl) 0.5 µl — 0.5µl 0.5µl 0.5 µlDouble-distilled water to a final volume of 5 µl X µl 3.0µl 2.5µl X µl X µla Lacks the prepared vector and controls for contamination of the insert by vector DNA.b Lacks the prepared insert and T4 DNA ligase and controls for residual undigested vector.c Lacks the prepared insert and controls for the effectiveness of the alkaline phosphatase treatment; should result in significantly fewer colonies than the test plates.d See Preparation of Media and Reagents.2. Mix the ligation reactions gently.3. Incubate the ligation reactions overnight at 16°C.4. Transform the appropriate competent bacteria with 1–2 µl of ligationreaction (see Recombination and Host Strains for Subcloning and Propagation). Plate transformation reactions on LB–ampicillin agar plates (see Preparation of Media and Reagents).R ECOMBINATION AND H OST S TRAINS FOR S UBCLONING ANDP ROPAGATIONRetroviral vectors are more likely to experience recombination events thanother plasmid vectors because of repeat sequences in the LTR regions. Weoffer RecA–E. coli strains such as XL10-Gold ultracompetent cells or XL1-Blue supercompetent cells which address this issue.P RIMERSThe nucleotide sequence and vector coordinates for primers suitable forPCR amplification and sequencing of inserts in the pFB and pFB-Neovectors are as follows:Sequence(bp)Primer Coordinates5′ Retro 2008–2028 5′-GGCTGCCGACCCCGGGGGTGG-3′3′ pFB 2121–2101 5′-CGAACCCCAGAGTCCCGCTCA-3′3′ pFB-Neo 2147–2127 5′-GCCAGGTTTCCGGGCCCTCAC-3′P REPARATION OF M EDIA AND R EAGENTSLB Agar (per Liter)10 g of NaCl10 g of tryptone5 g of yeast extract20 g of agarAdd deionized H2O to a final volume of1 literAdjust pH to 7.0 with 5 N NaOHAutoclave(~25 ml/100-mm plate) LB–Ampicillin Agar (per Liter)1 liter of LB agar, autoclavedCool to 55°CAdd 10 ml of 10-mg/ml filter-sterilized ampicillin(~25 ml/100-mm plate)TE Buffer10 mM Tris-HCl (pH 7.5)1 mM EDTA 10× Ligase Buffer500 mM Tris-HCl (pH 7.5)70 mM MgCl210 mM DTTNote rATP is added separately in the ligation reaction.R EFERENCES1. Miller, A. D. (1997). Development and Applications of Retroviral Vectors.In Retroviruses, J. M. Coffin, S. H. Hughes and H. E. Varmus (Eds.), pp. 437-473.Cold Spring Harbor Laboratory Press, Plainview, NY.2. Felts, K., Bauer, J. C. and Vaillancourt, P. (1999) Strategies 12(2):74-77.S UPPLEMENTAL R EFERENCES1. Miller, A. D. (1997) Development and Applications of Retroviral Vectors.In Retroviruses (eds., Coffin, J. M., Hughes, S. H., and Varmus, H. E.) pp. 437–473.Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York.2. Cepko, C., and Pear, W. (1996) In Current Protocols in Molecular Biology,pp. 9.9.1–9.9.16. John Wiley & Sons Inc., New York.MSDS I NFORMATIONThe Material Safety Data Sheet (MSDS) information for Stratagene products is provided on the web at/doc/dba401755b8102d276a20029bd64783e08127d58.html /MSDS/. Simply enter the catalog number to retrieve any associated MSDS’s in a print-ready format. MSDS documents are not included with product shipments.。
Rosetta系列的表达菌株可以提供T7 RNA聚合酶,它能表达PET系列载体上的外源基因;;;pGEX系列载体上的外源基因不需要T7 RNA聚合酶,普通的大肠杆菌经IPTG诱导即可表达Tac启动子是一组由Lac和trp启动子人工构建的杂合启动子,受Lac阻遏蛋白的负调节,它的启动能力比Lac和trp都强;其中Tac 1是由Trp启动子的-35区加上一个合成的46 bp DNA 片段包括Pribnow 盒和Lac操纵基因构成,Tac 12是由Trp的启动子-35区和Lac启动子的-10区,加上Lac操纵子中的操纵基因部分和SD序列融合而成蛋白标签:A myc tag is a polypeptide derived from the gene product that can be added to a protein using technology. It can be used for , then used to separate recombinant, overexpressed protein from wild type protein expressed by the host organism. It can also be used in the isolation of protein complexes with multiple subunits.A myc tag can be used in many different assays that require recognition by an . If there is no antibody against the studied protein, adding a myc-tag allows one to follow the protein with an antibody against the Myc epitope. Examples are cellular localization studies by immunofluorescence or detection by .The peptide sequence of the myc-tag is in 1- and 3-letter codes, respectively:N-EQKLISEEDL-C, N-Glu-Gln-Lys-Leu-Ile-Ser-Glu-Glu-Asp-Leu-C, where N standsfor and C stands for . The tag is approximately 1202 in atomic mass and has 10 amino acids.It can be fused to the and the of a protein. It is advisable not to fuse the tag directly behind the signal peptide of a secretory protein, since it can interfere with translocation into the .A monoclonal antibody against the myc , named 9E10, is available from thenon-commercialpGEX4T1载体基本信息出品公司: GE别名: pGEX-4T-1, pGEX4T1, pGEX 4T 1 质粒类型: 大肠杆菌蛋白表达载体表达水平: 高拷贝启动子: Tac克隆方法: 多克隆位点,限制性内切酶载体大小: 4969 bp5' 测序引物及序列: pGEX5': GGGCTGGCAAGCCACGTTTGGTG 3' 测序引物及序列: pGEX3': CCGGGAGCTGCATGTGTCAGAGG 载体标签: N-GST载体抗性: Ampicillin 氨苄备注: 复制子是pMB1产品目录号: 27-4580-01稳定性: 瞬时表达 Transient组成型: 诱导表达病毒/非病毒: 非病毒pGEX4T1载体质粒图谱和多克隆位点信息原核生物起始点,是DNA链上独特的具有起始DNA复制功能的序列;的复制包括OriC和OriH Thrombin 凝血酶pGEX4T1载体简介pGEX4T1载体序列LOCUS pGEX-4T-1 4969 bp DNA circular SYN DEFINITION pGEX-4T-1ACCESSIONKEYWORDSSOURCEORGANISM other sequences; artificial sequences; vectors. COMMENT This file is created by Vector NTIVNTAUTHORNAME||FEATURES Location/Qualifierssource 1..4969/organism="pGEX-4T-1"/mol_type="other DNA"promoter 184..212/label="tac_promoter"misc_feature 224..246/label="M13_pUC_rev_primer"gene 258..977/label="GST variant"/gene="GST variant"CDS 258..977/label="ORF frame 3"misc_feature complement1019..1041/label="pGEX_3_primer"promoter 1307..1335/label="AmpR_promoter"gene 1377..2237/label="Ampicillin"/gene="Ampicillin"CDS 1377..2237/label="ORF frame 3"rep_origin 2392..3011/label="pBR322_origin"misc_feature 3309..4400/label="lacI"CDS 3441..4400/label="ORF frame 3"promoter 4449..4478/label="lac_promoter"misc_feature 4492..4514/label="M13_pUC_rev_primer"promoter 4513..4531/label="M13_reverse_primer"CDS 4523..81/label="ORF frame 2"misc_feature 4540..4695/label="lacZ_a"promoter complement4543..4559/label="M13_forward20_primer"misc_feature complement4552..4574/label="M13_pUC_fwd_primer"pGEX4T2载体基本信息出品公司: GE别名: pGEX-4T-2, pGEX4T2, pGEX 4T 2 质粒类型: 大肠杆菌蛋白表达载体表达水平: 高拷贝启动子: Tac克隆方法: 多克隆位点,限制性内切酶载体大小: 4970 bp5' 测序引物及序列: pGEX5': GGGCTGGCAAGCCACGTTTGGTG 3' 测序引物及序列: pGEX3': CCGGGAGCTGCATGTGTCAGAGG 载体标签: N-GST载体抗性: Ampicillin 氨苄备注: 复制子是pMB1产品目录号: 27-4581-01稳定性: 瞬时表达 Transient组成型: 诱导表达病毒/非病毒: 非病毒pGEX4T2载体质粒图谱和多克隆位点信息pGEX4T2载体简介pGEX4T2载体序列LOCUS pGEX-4T-2 4970 bp DNA circular SYN DEFINITION pGEX-4T-2ACCESSIONKEYWORDSSOURCEORGANISM other sequences; artificial sequences; vectors. COMMENT This file is created by Vector NTIVNTAUTHORNAME||FEATURES Location/Qualifierssource 1..4970/mol_type="other DNA"promoter 184..212/label="tac_promoter"misc_feature 224..246/label="M13_pUC_rev_primer" gene 258..978/label="GST variant"/gene="GST variant"CDS 258..974/label="ORF frame 3"misc_feature complement1020..1042/label="pGEX_3_primer"promoter 1308..1336/label="AmpR_promoter"gene 1378..2238/label="Ampicillin"/gene="Ampicillin"CDS 1378..2238/label="ORF frame 1"rep_origin 2393..3012/label="pBR322_origin"misc_feature 3310..4401/label="lacI"CDS 3442..4401promoter 4450..4479/label="lac_promoter"misc_feature 4493..4515/label="M13_pUC_rev_primer"promoter 4514..4532/label="M13_reverse_primer"CDS 4524..81/label="ORF frame 3"misc_feature 4541..4696/label="lacZ_a"promoter complement4544..4560/label="M13_forward20_primer" misc_feature complement4553..4575/label="M13_pUC_fwd_primer" pGEX4T3载体质粒图谱和多克隆位点信息pGEX4T3载体简介pGEX4T3载体序列LOCUS pGEX-4T-3 4968 bp DNA circular SYN DEFINITION pGEX-4T-3ACCESSIONKEYWORDSSOURCEORGANISM other sequences; artificial sequences; vectors. COMMENT This file is created by Vector NTIVNTAUTHORNAME||FEATURES Location/Qualifierssource 1..4968/organism="pGEX-4T-3"/mol_type="other DNA"promoter 184..212/label="tac_promoter"misc_feature 224..246/label="M13_pUC_rev_primer" gene 258..976/label="GST variant"/gene="GST variant"CDS 258..980/label="ORF frame 3"misc_feature complement1018..1040/label="pGEX_3_primer"promoter 1306..1334/label="AmpR_promoter"gene 1376..2236/label="Ampicillin"/gene="Ampicillin"CDS 1376..2236/label="ORF frame 2"rep_origin 2391..3010/label="pBR322_origin"misc_feature 3308..4399/label="lacI"CDS 3440..4399/label="ORF frame 2"promoter 4448..4477/label="lac_promoter"misc_feature 4491..4513/label="M13_pUC_rev_primer"promoter 4512..4530/label="M13_reverse_primer"CDS 4522..81/label="ORF frame 1"misc_feature 4539..4694/label="lacZ_a"promoter complement4542..4558/label="M13_forward20_primer"misc_feature complement4551..4573/label="M13_pUC_fwd_primer"pGEX6P1载体基本信息出品公司: GE别名: pGEX-6P-1, pGEX6P1, pGEX 6P 1 质粒类型: 大肠杆菌蛋白表达载体表达水平: 高拷贝启动子: Tac克隆方法: 多克隆位点,限制性内切酶载体大小: 4984 bp5' 测序引物及序列: pGEX5': GGGCTGGCAAGCCACGTTTGGTG 3' 测序引物及序列: pGEX3': CCGGGAGCTGCATGTGTCAGAGG 载体标签: N-GST载体抗性: Ampicillin 氨苄备注: 复制子是pMB1产品目录号: 27-4597-01稳定性: 瞬时表达 Transient组成型: 诱导表达病毒/非病毒: 非病毒pGEX6P1载体质粒图谱和多克隆位点信息pGEX6P1载体简介pGEX6P1载体序列LOCUS pGEX-6P-1 4984 bp DNA circular SYN DEFINITION pGEX-6P-1ACCESSIONKEYWORDSSOURCEORGANISM other sequences; artificial sequences; vectors. COMMENT This file is created by Vector NTIVNTAUTHORNAME||FEATURES Location/Qualifierssource 1..4984/organism="pGEX-6P-1"/mol_type="other DNA"promoter 184..212/label="tac_promoter"misc_feature 224..246/label="M13_pUC_rev_primer"gene 258..992/label="GST"/gene="GST"CDS 258..992/label="ORF frame 3"misc_feature 918..938/label="precision"misc_feature complement1034..1056/label="pGEX_3_primer"promoter 1322..1350/label="AmpR_promoter"gene 1392..2252/label="Ampicillin"/gene="Ampicillin"CDS 1392..2252/label="ORF frame 3"rep_origin 2407..3026/label="pBR322_origin"misc_feature 3324..4415/label="lacI"CDS 3456..4415/label="ORF frame 3"promoter 4464..4493/label="lac_promoter"misc_feature 4507..4529/label="M13_pUC_rev_primer"promoter 4528..4546/label="M13_reverse_primer"CDS 4538..81/label="ORF frame 2"misc_feature 4555..4710/label="lacZ_a"promoter complement4558..4574/label="M13_forward20_primer" misc_feature complement4567..4589/label="M13_pUC_fwd_primer"pET28a载体基本信息出品公司: EMD Biosciences Novagen别名: pET28a, pet 28a, pET-28a+ 质粒类型: 大肠杆菌蛋白表达表达水平: 高克隆方法: 多克隆位点,限制性内切酶载体大小: 5369 bp5' 测序引物及序列: T7: 5'-TAATACGACTCACTATAGGG-3' 3' 测序引物序列: T7t: 5'-GCTAGTTATTGCTCAGCGG-3' 载体标签: N-His, N-Thrombin, N-T7, C-His载体抗性: 卡那备注: N端含有Thrombin蛋白酶切位点; pET28a, b, c 的差异仅仅存在于多克隆位点处;产品目录号: 69864-3稳定性: 瞬时表达 Transient组成型: 组成型 Constitutive 病毒/非病毒: 非病毒pET28a载体质粒图谱和多克隆位点信息pET28a载体简介pET-28a-c+载体带有一个N端的His/Thrombin/T7蛋白标签,同时含有一个可以选择的C端His标签;pET28a载体的单一的多克隆位点见上面的环状质粒图谱;注意:载体序列是以pBR322质粒的编码规矩进行编码的,所以T7蛋白表达区在质粒图谱上面是反向的;T7 RNA聚合酶启动的克隆和表达区域在质粒图谱中也被标注了出来;质粒的F1复制子是被定向的,所以在T7噬菌体聚合酶的作用下,包含有蛋白编码序列的病毒粒子能够产生,并启动蛋白表达,同时蛋白表达将被T7终止子序列Cat. No. 69337-3的作用下终止蛋白翻译;pET30a载体基本信息出品公司: EMD Biosciences Novagen别名: pET30a, pet 30a质粒类型: 大肠杆菌蛋白表达表达水平: 高克隆方法: 多克隆位点,限制性内切酶载体大小: 5422 bp5' 测序引物及序列: T7: 5'-TAATACGACTCACTATAGGG-3'3' 测序引物序列: T7t: 5'-GCTAGTTATTGCTCAGCGG-3' 载体标签: N-His, N-S, N-Thrombin, N-EK, C-His载体抗性: 卡那备注: pET30a, b, c的差异只存在于多克隆位点,其他位点不存在;产品目录号: 69909-3稳定性: 瞬时表达组成型: 组成型病毒/非病毒: 非病毒pET30a载体质粒图谱和多克隆位点信息pET30a载体简介pET-30a+载体携带有一个N端的His标签/Thrombin酶切位点/ S标签/ EK蛋白酶切标签以及一个可选择的C端His标签;载体的多克隆位点见上面的图谱;需要注意的是图谱中的核苷酸编码顺序是以pBR322载体的复制方向为正方向,所以T7表达区域在图谱中是处于反向的位置;T7 RNA聚合酶启动的克隆和表达区域在质粒图谱中也被标注了出来;质粒的F1复制子是被定向的,所以在T7噬菌体聚合酶的作用下,包含有蛋白编码序列的病毒粒子能够产生,并启动蛋白表达,同时蛋白表达将被T7终止子序列Cat. No. 69337-3的作用下终止蛋白翻译;pet32a载体基本信息出品公司: EMD BiosciencesNovagen别名: pET32a, pet 32a质粒类型: 大肠杆菌蛋白表达表达水平: 高克隆方法: 多克隆位点,限制性内切酶载体大小: 5900bp5' 测序引物: T7: 5'-TAATACGACTCACTATAGGG-3' 3' 测序引物: T7t: 5'-GCTAGTTATTGCTCAGCGG-3' 载体标签: thioredoxin N端; His 中间和C端载体抗性: 氨苄备注:pET32a载体用于表达可溶性,具有活性功能的蛋白;pET-32a+载体具有N端Thrombin凝血酶酶切位点,N 端肠激酶酶切位点;pET32啊,pET32b,pET32c载体的差异主要是在于多克隆位点,其他地方完全一致;产品货号: 69015-3稳定性: 瞬时表达组成型: 组成型病毒/非病毒: 非病毒pet32a载体质粒图谱和多克隆位点信息pet32a载体简介pET32系列载体是设计用来克隆构建和高水平融合表达表达带有109个氨基酸Trx标签的蛋白质序列;将基因插入到载体的多克隆位点,能够获得融合表达蛋白质;同时融合蛋白包含有His标签和S标签,这些可以用于蛋白表达检测和纯化过程;载体的多克隆位点见上面的图谱;需要注意的是图谱中的核苷酸编码顺序是以pBR322载体的复制方向为正方向,所以T7表达区域在图谱中是处于反向的位置;T7 RNA聚合酶启动的克隆和表达区域在质粒图谱中也被标注了出来;质粒的F1复制子是被定向的,所以在T7噬菌体聚合酶的作用下,包含有蛋白编码序列的病毒粒子能够产生,并启动蛋白表达,同时蛋白表达将被T7终止子序列Cat. No. 69337-3的作用下终止蛋白翻译;pET23a载体基本信息出品公司: EMD BiosciencesNovagen别名: pET23a, pet 23a质粒类型: 大肠杆菌蛋白表达表达水平: 高克隆方法: 多克隆位点,限制性内切酶载体大小: 3666bp5' 测序引物: T75' 测序引物序列: 5'-TAATACGACTCACTATAGGG-3'载体标签: N-T7, C-His载体抗性: Ampicillin克隆菌株: DH5a表达菌株: BL21系列备注: Same as pET21abcd+ but no lac; a,b,c,d vary by MCS.产品目录号: 69745-3稳定性: 稳表达组成型: 组成型病毒/非病毒: 非病毒pET23a载体质粒图谱和多克隆位点信息pET23a载体简介The pET-23a-d+ vectors carry an N-terminal T7•Tag® sequence plus an optional C-terminal His•Tag® sequence. These vectors differ from pET-21a-d+ by the “plain” T7 promoter instead of the T7lac promoter and by the absence of the lacI gene. Unique sites are shown on the circle map. Note that the sequence is numbered bythe pBR322 convention, so the T7 expression region is reversed on the circular map. The cloning/expression region of the coding strand transcribed by T7 RNA polymerase is shown below. The f1 origin is oriented so that infection with helper phage will produce virions containing single-stranded DNA that corresponds to the coding strand. Therefore, single-stranded sequencing should be performed using the T7 terminator primer Cat. No. 69337-3.pGEXKG载体基本信息出品公司: ATCC别名: pGEX-KG, pGEXKG, pGEX KG质粒类型: 大肠杆菌蛋白表达载体表达水平: 高拷贝启动子: Tac克隆方法: 多克隆位点,限制性内切酶, BamHI,SmaI, EcoRI, XbaI, NcoI, SalI, XhoI, SacI, HindIII载体大小: 5006 bp5' 测序引物及序列: pGEX5': GGGCTGGCAAGCCACGTTTGGTG 3' 测序引物及序列: pGEX3': CCGGGAGCTGCATGTGTCAGAGG 载体标签: N-GST载体抗性: Ampicillin 氨苄备注: 复制子是pMB1, 来源于pGEX-2T 产品目录号: 77103稳定性: 瞬时表达 Transient组成型: 诱导表达病毒/非病毒: 非病毒pGEXKG载体质粒图谱和多克隆位点信息pMAL-C2X 表达载体使用直链淀粉树脂amylose resin分离融合蛋白。