骨修复Novel therapeutic core–shell hydrogel scaffolds with sequential delivery of cobalt and bone
- 格式:pdf
- 大小:4.60 MB
- 文档页数:14
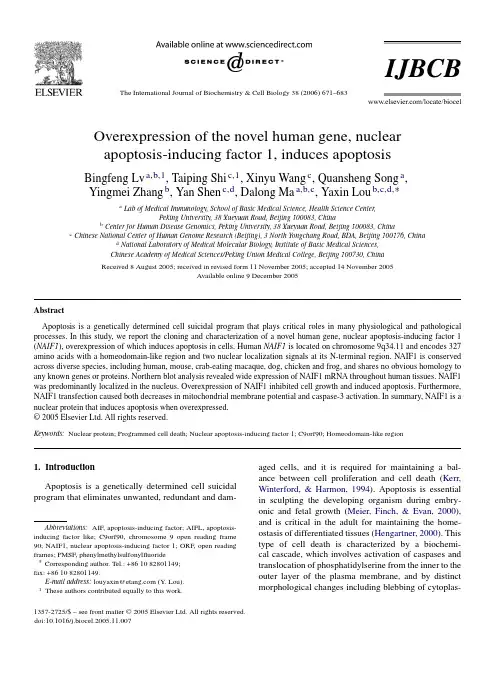
The International Journal of Biochemistry&Cell Biology38(2006)671–683Overexpression of the novel human gene,nuclearapoptosis-inducing factor1,induces apoptosisBingfeng Lv a,b,1,Taiping Shi c,1,Xinyu Wang c,Quansheng Song a,Yingmei Zhang b,Yan Shen c,d,Dalong Ma a,b,c,Yaxin Lou b,c,d,∗a Lab of Medical Immunology,School of Basic Medical Science,Health Science Center,Peking University,38Xueyuan Road,Beijing100083,Chinab Center for Human Disease Genomics,Peking University,38Xueyuan Road,Beijing100083,Chinac Chinese National Center of Human Genome Research(Beijing),3North Yongchang Road,BDA,Beijing100176,Chinad National Laboratory of Medical Molecular Biology,Institute of Basic Medical Sciences,Chinese Academy of Medical Sciences/Peking Union Medical College,Beijing100730,ChinaReceived8August2005;received in revised form11November2005;accepted14November2005Available online9December2005AbstractApoptosis is a genetically determined cell suicidal program that plays critical roles in many physiological and pathological processes.In this study,we report the cloning and characterization of a novel human gene,nuclear apoptosis-inducing factor1 (NAIF1),overexpression of which induces apoptosis in cells.Human NAIF1is located on chromosome9q34.11and encodes327 amino acids with a homeodomain-like region and two nuclear localization signals at its N-terminal region.NAIF1is conserved across diverse species,including human,mouse,crab-eating macaque,dog,chicken and frog,and shares no obvious homology to any known genes or proteins.Northern blot analysis revealed wide expression of NAIF1mRNA throughout human tissues.NAIF1 was predominantly localized in the nucleus.Overexpression of NAIF1inhibited cell growth and induced apoptosis.Furthermore, NAIF1transfection caused both decreases in mitochondrial membrane potential and caspase-3activation.In summary,NAIF1is a nuclear protein that induces apoptosis when overexpressed.©2005Elsevier Ltd.All rights reserved.Keywords:Nuclear protein;Programmed cell death;Nuclear apoptosis-inducing factor1;C9orf90;Homeodomain-like region1.IntroductionApoptosis is a genetically determined cell suicidal program that eliminates unwanted,redundant and dam-Abbreviations:AIF,apoptosis-inducing factor;AIFL,apoptosis-inducing factor like;C9orf90,chromosome9open reading frame 90;NAIF1,nuclear apoptosis-inducing factor1;ORF,open reading frames;PMSF,phenylmethylsulfonylfluoride∗Corresponding author.Tel.:+861082801149;fax:+861082801149.E-mail address:louyaxin@(Y.Lou).1These authors contributed equally to this work.aged cells,and it is required for maintaining a bal-ance between cell proliferation and cell death(Kerr, Winterford,&Harmon,1994).Apoptosis is essential in sculpting the developing organism during embry-onic and fetal growth(Meier,Finch,&Evan,2000), and is critical in the adult for maintaining the home-ostasis of differentiated tissues(Hengartner,2000).This type of cell death is characterized by a biochemi-cal cascade,which involves activation of caspases and translocation of phosphatidylserine from the inner to the outer layer of the plasma membrane,and by distinct morphological changes including blebbing of cytoplas-1357-2725/$–see front matter©2005Elsevier Ltd.All rights reserved. doi:10.1016/j.biocel.2005.11.007672 B.Lv et al./The International Journal of Biochemistry&Cell Biology38(2006)671–683mic membranes,chromatin condensation,margination and fragmentation into apoptotic bodies(Hengartner, 2000;Kerr,Wyllie,&Currie,1972;Thornberry& Lazebnik,1998).A variety of stimuli,including of growth factor deprivation,signaling via certain sur-face receptors,treatment with DNA-damaging agents and inhibitors of macromolecular synthesis,can induce apoptosis via receptor-mediated or mitochondrial path-ways(Ellis,Yuan,&Horvitz,1991;Steller,1995).Dys-regulation of apoptosis has been associated with many serious human diseases,including neoplasia,degener-ative disorders and autoimmune diseases(Nicholson, 2000;Zimmermann,Bonzon,&Green,2001).Thus, studies on apoptosis are of great value for understanding events that occur during development as well as in the adult.By searching the human Refseq and expressed sequence tag(EST)databases,we obtained sequences of hundreds of novel human genes,whose open read-ing frames(ORF)were longer than300base pairs.The ORFs of these genes were cloned into pcDNA3.1-myc-his B vectors and underwent several screening analyses. By observing the morphological changes in transfected cells,we found that a novel human gene,NAIF1(also named C9orf90)caused cells to undergo marked round-ing and eventual detachment from the culture dish.The results of database searching suggested that NAIF1is conserved among human,mouse,crab-eating macaque, dog,chicken and frog,and shared no obvious homology to any known genes or proteins.Northern blot analy-sis revealed ubiquitous expression of NAIF1through-out human tissues and subcellular localization analysis revealed that NAIF1was located in the nucleus.Fur-ther studies indicated that overexpression of NAIF1 inhibited cell growth and survival,while also induc-ing cell shrinkage and chromatin condensation,both of which are hallmarks of apoptosis.Finally,we found that NAIF1caused depolarization of mitochondrial mem-brane potential and caspase-3activation in HeLa cells. Thus,in the present study,we cloned the NAIF1gene and found it to induce apoptosis when overexpressed in cells.2.Materials and methods2.1.MaterialsRPMI1640and DMEM,fetal bovine serum(FBS) and penicillin-streptomycin were purchased from Invit-rogen(USA).MTT[3-(4,5-dimethyltiazol-2-yl)-2,5-diphenyltetrazolium bromide],monoclonal antibod-ies against-actin and FLAG-tag were purchased from Sigma-Aldrich(St.Louis,USA).The cationic lypophilicfluorochrome3,3 -dihexyloxacarbocyanine iodide[DiOC6(3)]was from Molecular Probes(USA). Monoclonal antibodies against caspase-3and PARP were obtained from Transduction Laboratories(Becton Dickinson,USA).2.2.Cell culture and cell linesHeLa cells,a human cervical carcinoma cell line, were maintained in DMEM supplemented with10% heat-inactivated FBS,100U/ml penicillin and100g/ml streptomycin.Human embryonic kidney cells,HEK293, were maintained in RPMI1640medium supplemented as DMEM.Cells were grown at37◦C in a humidified5% CO2atmosphere and used for assays during exponential phase of growth.2.3.Northern blot analysisA470-bp PCR product corresponding to513–982bp of NAIF1coding region was used as a probe and labeled with␣-32P-dCTP using a Random Primers DNA Label-ing System(Invitrogen,USA)as described by the man-ufacturer.Human multiple tissue Northern blots(Clon-tech,USA)were hybridized with the probe and washed according to the procedure provided by the manufacturer. The membranes were exposed to a phosphor-imaging screen overnight and analyzed using the Cyclone TM Stor-age Phosphor System(Packard Instrument Company, USA).2.4.Cloning,plasmids construction and transfectionThe complete coding region of NAIF1was ampli-fied from a human liver cDNA library(Clontech,USA) by PCR using the forward primer P1(5 -GGG GGT GGG TGA GGG GC-3 )and reverse primer P2(5 -ACC CCT GCC CTC ACT GGA TG-3 ).The cDNA encod-ing amino acids71–327of NAIF1,NAIF1(71–327), was amplified using P3(5 -CGA ATT CCA CCA TGT GGT CTG ACC TCA AGA CCG A-3 )and P2as primers.The PCR products were cloned into pGEM-T Easy vector(Promega,USA)and sequenced using ABI PRISM®3100Genetic Analyzer(Applied Biosystems, USA).The inserts were released by cutting with Eco RI and subcloned into the Eco RI site of the mammalian expression vector pcDNA.3.1-myc-His B(Invitrogen, USA).To create the expression vectors carrying GFP at the N-terminal ends of NAIF1or NAIF1(71–327),theB.Lv et al./The International Journal of Biochemistry&Cell Biology38(2006)671–683673ORF of NAIF1and NAIF1(71–327)were subcloned in frame into pEGFP-N1(Clontech,USA).To con-struct a Flag-tagged NAIF1expression vector,the ORF of NAIF1was subcloned in frame into the pFLAG-CMV vector(Sigma-Aldrich,St.Louis,USA),which allowed for expression of NAIF1carrying Flag-tag at its N-terminal in mammalian cells.The resulting vectors,named pEGFP-N1-NAIF1,pEGFP-N1-NAIF1 (71–327)and pFlag-NAIF1were sequenced to confirm correct in-frame fusion between GFP or Flag-tag and NAIF1.The Bax expressing vector was described previ-ously(Wang et al.,2004).DNA transfections were performed using Lipofec-tamine2000(Invitrogen,USA)according to the man-ufacturer’s instruction or by electroporation at a single pulse of120V,20ms,with10g plasmid per106cells in2mm gap cuvettes using an ECM830square wave electroporation system(BTX,USA).2.5.Protein extraction and immunoblottingCells were pelleted by centrifugation,lysed in lysis buffer(20mM Tris–HCl,pH7.4,150mM NaCl,1mM EDTA,1mM EGTA,1mM PMSF)and incubated for 30min on ice.Lysates were centrifuged at18,000×g for10min at4◦C and the supernatant was mea-sured using the BCA protein assay reagent(Pierce, USA).Equal amounts of protein(20g)were separated by sodium dodecyl sulfate/polyacrylamide gel elec-trophoresis(SDS-PAGE)and transferred onto nitrocel-lulose membranes(Amersham Pharmacia,UK).Mem-branes were blocked in Tris-buffered saline containing 0.1%Tween-20(TBS-T)and5%non-fat milk for2h and incubated overnight at4◦C with the appropriate primary antibodies.Blots were washed three times for 10min each with TBS-T and incubated for1h in the dark with the appropriate IRDye TM800-conjugated secondary antibodies(LI-COR Bioscience Inc.,USA)prepared in TBST/5%non-fat milk.The signal was detected using the Odyssey®Imaging System(LI-COR Bioscience Inc.,USA).2.6.MTT assay and colony forming assayFor MTT colorimetric assay,the transfected cells were placed in96-well culture dishes at a density of 2000cells/well in100l medium.At the indicated time points,MTT solution was added and samples were incu-bated for4–6h.The formazan dye was then solubilized and the absorbance was measured at570nm.A colony forming assay was performed as described previously (Wang et al.,2004).Briefly,HeLa and HEK293cells were trypsinized24h post-transfection and a total of 1000cells were seeded in each100mm tissue culture dish.Selection medium with G418(800g/ml)was added.Three weeks later,colonies werefixed and stained with crystal violet.Colonies with a diameter of more than0.5mm were counted.Each group was assayed in triplicate and each experiment was repeated three times.2.7.Detection of phosphatidylserine externalizationTransfected HeLa cells(2×105)were trypsinized, washed twice with PBS,and resuspended in200l bind-ing buffer(10mM HEPES,pH7.4,140mM NaCl,1mM MgCl2,5mM KCl,2.5mM CaCl2).FITC-conjugated Annexin V(Biosea,China)was added to afinal con-centration of0.5g/ml.After incubation for20min at room temperature,propidium iodide was added to afinal concentration of1g/ml and the samples were analyzed on a FACSCaliburflow cytometer(Becton Dickinson, USA)with excitation using a488nm argon ion laser.2.8.Measurement of mitochondrial membrane potentialDiOC6(3)was used to evaluate whether NAIF1-induced apoptosis was associated with the disruption of transmembrane potential in mitochondria.Transfected cells were harvested at the indicated time points and cells were incubated with standard culture medium containing 40M DiOC6(3)for30min.Subsequently,cells were washed with standard culture medium and immediately analyzed byflow cytometric analysis with a FACSCal-iburflow cytometer(Becton Dickinson,USA).2.9.Evaluation of caspase-3activity(DEVDase activity analysis)HeLa cells were transfected with empty vector, NAIF1or Bax expression vectors and harvested in lysis buffer(10mM Tris–HCl,pH7.5,10mM NaHPO4, 130mM NaCl,1%Triton-X100,1mM PMSF).Cell lysates were clarified by centrifugation at18,000×g for20min at4◦C.Cell lysates containing10g pro-tein were incubated at37◦C in buffer containing25mM HEPES,pH7.5,1mM EDTA,100mM NaCl,0.1% CHAPS and10mM dithiothreitol with thefluoro-genic substrate Ac-DEVD-AMC(ALEXIS Biochem-icals,Switzerland).To measure this signal,we used a FLUOStarfluorometer(BMG Labtechnologies,Ger-many)with an excitationfilter of380nm and emission filter of460nm.Results were calculated as a proportion674 B.Lv et al./The International Journal of Biochemistry &Cell Biology 38(2006)671–683of the control over 90min (T90to T0).Samples were prepared in triplicate.2.10.Statistical analysisResults were expressed as means ±standard devia-tions.Student’s t -test was used for statistical comparison.p values of less than 0.01were considered statistically significant.3.Results3.1.Cloning of NAIF1cDNAUsing functional screening of novel human cDNA,we found that a hypothetical gene,chromosome 9open read-ing frame 90(C9orf90),could induce apoptosis.To our knowledge,no functional study has been performed on this hypothetical gene.We named it nuclear apoptosis-inducing factor 1(NAIF1)because it was a nuclear protein that induced apoptosis when overexpressed in cells.The cDNA of NAIF1(Refseq number is NM 197956)is 3198bp long,containing an ORF encoding 327amino acids with an ATG initiation codon whose surrounding sequence (GCCATGG)matches Kozak’s rule (Kozak,1991).EST assembly and Ecgene database searching (http://genome.ewha.ac.kr/ECgene/)revealed that there were at least nine RNA splicing forms of human NAIF1,the length of which ranged from 697to 3406base pair.These nine variants of NAIF1encoded five different proteins,among which the variant encoding 327amino acids was the longest.In this study,NAIF1referred to the 327amino acids variant.We designed primers to amplify the coding region of NAIF1from human cDNAs.The resultant RT-PCR products were in accor-dance with the predicted size (data not shown).The band was purified and cloned into the pGEM-T Easy vector.The insert was sequenced and was identical to the coding region of the hypothetical gene C9orf90.The cDNA sequence of the human NAIF1coding region and predicted amino acid sequence are shown in Fig.1.Alignment of the putative NAIF1cDNA to human genomic sequences revealed that NAIF1is a single copy gene located on chromosome 9q34.11.Database search-ing indicated that NAIF1was conserved among human,mouse,dog,crab-eating macaque,chicken and frog,but shared no obvious homology to any known genes or proteins.The deduced amino-acid sequences of human,mouse,dog,crab-eating macaque and chicken NAIF1proteins were aligned in Fig.2A and thepercentagesFig.1.The cDNA and predicted amino acid sequences of human NAIF1.The predicted glycine-rich region (GRR)is underlined and the predicted NLSs are shown in bold.B.Lv et al./The International Journal of Biochemistry&Cell Biology38(2006)671–683675Fig.2.NAIF1is conserved across diverse species.(A)Amino-acid sequence alignment of NAIF1proteins of different species.Sequence alignment was obtained using the ClustalW algorithm(Slow\Accurate,Identity)of the Lasergene software(DNASTAR Inc.,Madison,WI).The boxed areas indicate matching residues.(B)Sequence comparison of NAIF1proteins.The numbers represent the percentage of sequence identity and divergency determined by the ClustalW method with Slow\Accurate,Identity weight table in the Lasergene software.676 B.Lv et al./The International Journal of Biochemistry &Cell Biology 38(2006)671–683Fig.3.Northern blot analyses.Human multiple tissue Northern blots were hybridized with the ␣-32P-labeled NAIF1probes.The RNA markers are indicated in kilobases.of identity between different species of NAIF1were shown in Fig.2B.PSORT II (http://psort.nibb.ac.jp/)software analysis indicated that there were two NLSs at the N-terminal in the NAIF1protein.Motif analy-sis by Prosite (/prosite/)suggested that there was a glycine-rich region (GRR)from amino acid 92–117of human NAIF1and InterProScan search (/InterProScan/)indicated the 84amino acids of the N-terminal of human NAIF1protein contained a homeodomain-like region,which may bind to DNA (Mannervik,1999).3.2.Northern blot analysesNorthern blot analyses were used to confirm the existence of NAIF1mRNA in human tissues.Human multiple tissue Northern blots were probed with a ␣-32P-labeled probe corresponding to 513–982bp of the human NAIF1coding region.As shown in Fig.3,more than one band was detected in almost all tissues stud-ied,indicating that there were alternative RNA splic-ing forms of human NAIF1in most human tissues,which was in accordance with the result of database searching.3.3.Transfected NAIF1is expressed in human cells The coding region of human NAIF1cDNA was cloned into the pCMV-Flag mammalian expression vec-tor,which expressed Flag-tagged NAIF1protein in human cells.The vectors were transfected into HeLa cells and 24h later,whole cell lysates from transfectedcells were extracted and subjected to Western blot analy-sis.In the lane containing NAIF1-expressing cell lysates,there was one main 45kDa band,which was about 10kDa larger than the predicted size of the NAIF1fusion protein.There were also two weaker bands with molecu-lar masses of 30and 55kDa (Fig.4),indicating that there were different post-translation modifications of recom-binant NAIF1protein.In the lane containing the control vector,there was no obviousband.Fig.4.Transfected NAIF1is expressed in human cells.HeLa cells were transfected with mock or pFlag-NAIF1vectors.Twenty-four hours after transfection,cells were harvested and subjected to Western blot analysis using anti-flag-tag antibody.B.Lv et al./The International Journal of Biochemistry &Cell Biology 38(2006)671–683677Fig.5.Subcellular distribution of human NAIF1.HeLa cells were transfected with expression vectors carrying NAIF1–GFP or GFP using electro-poration.Cells were washed three times with PBS,fixed with 4%paraformaldehyde and incubated with DAPI to stain nuclei.Cells were analyzed with an Olympus IX 71fluorescent microscope and photographs were captured using an Alta U2digital camera (Apogee Instruments Inc.,NY ,USA).3.4.NAIF1protein is localized in the nucleus To determine subcellular localization of NAIF1,we generated a NAIF1–GFP fusion construct carrying GFP at the N-terminal of NAIF1.HeLa cells were tran-siently transfected with NAIF1–GFP fusion constructs or GFP vector.Twenty-four hours after transfection,the transfected cells were stained with DAPI and analyzed by fluorescence microscopy.An intense green fluores-cent signal was exclusively detected inside the nuclei in NAIF1–GFP expressing cells,while the green signal was seen diffusely throughout the cells which expressed only EGFP (Fig.5).Similar subcellular distribution was seen in other cell lines such as HEK293cells (data not shown).These results suggested that NAIF1was a nuclearprotein.Fig.6.Overexpression of NAIF1inhibits cell growth and survival.HeLa (A)and HEK293(B)cells were transiently transfected with the NAIF1expression plasmids and control plasmids.MTT assay (A and B)and colony formation assay (C)were used to determine the effect of overexpression of NAIF1on cell growth and survival.Data are presented as mean ±standard deviation (n =3).*means significantly different from mock-transfected cells.678 B.Lv et al./The International Journal of Biochemistry&Cell Biology38(2006)671–6833.5.Overexpression of NAIF1inhibits cell growth and survivalTo examine the function of NAIF1in cell growth,we analyzed the effect of its overexpression on cell growth using a MTT assay.HeLa and HEK293cells were tran-siently transfected with NAIF1-expressing or control plasmids.Fig.6A and B showed that overexpression of NAIF1significantly inhibited the growth of HeLa and HEK293cells.We also used a colony formation assay to evalu-ate the inhibitory effects of NAIF1on cellgrowth. Fig.7.Overexpression of NAIF1induces apoptosis.HeLa cells were transfected with control(A,D and G),NAIF1(B,E and H)or Bax(C,F and I)expressing vectors and stained with DAPI.Morphological changes of transfected cells(A–F)were observed under a microscope24h(A–C) or48h(D–F)post-transfection.Nuclear changes of transfected cells(G–I)were examined under afluorescent microscope48h post-transfection. Phosphatidylserine externalization of transfected cells was measured by Annexin V binding assay(J)at indicated time points.B.Lv et al./The International Journal of Biochemistry &Cell Biology 38(2006)671–683679HeLa and HEK293cells were transfected with the NAIF1-expressing plasmids containing a neomycin resistance gene.Twenty-four hours after transfection,G418was added and transfected cells were selected for 3weeks.Fig.6C showed that overexpression of NAIF1almost completely inhibited the formation of stable clones as compared with mock-transfected cells.Taken together with data from Fig.6A and B,these results suggested that NAIF1inhibited cell growth and stable colony formation in HeLa and HEK293cells.3.6.Overexpression of NAIF1induces apoptosis We observed morphological changes in transfected cells using fluorescence microscopy.As shown in Fig.7,there were no obvious morphological changes in NAIF1-or mock-transfected HeLa cells 24h post-transfection.However,Bax-transfected cells became round and detached from the cell culture plate within 24h.Forty-eight hours post-transfection,NAIF1-transfected cells underwent marked rounding and eventually detached from the culture dishes,similar to what had been observed in Bax-transfected cells.To confirm whether the NAIF1-transfected cells underwent apoptosis,we examined transfected cells stained with DAPI using fluorescence microscopy.Chromatin con-densation and margination,characteristic of apoptosis,were observed in NAIF1-and Bax-transfected cells 48h post-transfection.An Annexin V binding assay indicated that there were no obvious increases in the portion of Annexin V positive cells compared with mock-transfected cells 24h post-transfection,However,the number of Annexin V positive cells increased significantly in NAIF1-transfected cells,compared with mock-transfected cells 48h post-transfection (Fig.7J).These results indicated that NAIF1-transfected cells underwent an apoptotic process,but that NAIF1-induced apoptosis began much later than apoptosis induced by Bax.This phenomenon was not unique to HeLa cells.We also detected similar changes in HEK293cells (data notshown).Fig.8.Apoptosis of HeLa cells by overexpression of NAIF1is associated with a loss in mitochondrial transmembrane potential.Mitochondrial transmembrane potential ( Ψm )as reflected by DIOC6(3)staining of transfected HeLa cells was determined at indicated time points after trans-fection.Loss of Ψm was visualized as a reduction in the fluorescence signal in the FL1channel.The proportions of cells with reduced DIOC6(3)staining are shown.The results depicted are a representative of three independent experiments.680 B.Lv et al./The International Journal of Biochemistry&Cell Biology38(2006)671–6833.7.Overexpression of NAIF1disrupts mitochondrial transmembrane potentialThe disruption of mitochondrial transmembrane potential is one of the early features of apoptosis.There-fore,we evaluated whether NAIF1-mediated apoptosis was associated with this disruption.DiOC6(3),a cationic lypophilicfluorochrome,is commonly used for quantify-ing the mitochondrial transmembrane potential( Ψm) (Metivier et al.,1998;Petit,O’Connor,Grunwald,& Brown,1990).Transfected cells were pretreated with this dye and were analyzed byflow cytometry.Loss of DiOC6(3)staining indicates disruption of the mitochon-drial inner transmembrane potential( Ψm),which is associated with apoptosis(Susin et al.,1997).In com-parison with mock-transfected cells,NAIF1-transfected cells showed a decrease in their Ψm24and36h post-transfection as indicated by the percentage of cells with DiOC6(3)staining(Fig.8).DiOC6(3)staining was still reduced48h after transfection.Thesefindings suggested that NAIF1-induced apoptosis was associ-ated with a decrease in mitochondrial transmembrane potential.3.8.NAIF1activates caspase-3Activation of effector caspases,such as caspase-3and-7,is responsible for the proteolytic cleavage of a diverse range of structural and regulatory pro-teins in apoptosis(Salvesen&Dixit,1997).To deter-mine whether NAIF1activated caspase-3,we analyzed caspase-3activity in transfected cells using DEVD cleavage assay,a quantitative method used to detect caspase-3-like activity(Rehm et al.,2002).The results showed that DEVD cleavage activity in lysates from NAIF1-overexpressing HeLa cells were at leastfive times higher than mock-transfected cell lysates48h post-transfection(Fig.9A).To further confirm the activa-tion of caspase-3in lysates from NAIF1-overexpressing cells,we detected procaspase-3expression using West-ern blot analysis.We detected decreased procaspase-3 in lysates of NAIF1and Bax overexpressing cells com-pared to mock-transfected cells(Fig.9B).Because pro-teolytic cleavage of specific substrates by activated cas-pases is responsible for cellular dysfunction and struc-tural destruction associated with apoptosis(Thornberry &Lazebnik,1998),we analyzed the cleavage of PARP as a representative substrate in mock-,NAIF1-or Bax-transfected cells.PARP was cleaved in NAIF1-and Bax-transfected cells(Fig.9B).These results strongly suggested that the caspase cascade was activated during NAIF1-inducedapoptosis.Fig.9.NAIF1activates caspase-3.(A)DEVDase activity assay.HeLa cells were transfected with the empty vector or indicated constructs and,at indicated time,DEVDase activity was measured as described in Section2.Data are presented as mean±standard deviation(n=3). *means significantly different from mock-transfected cells.(B)Cellu-lar proteins were extracted from HeLa cells that were transfected with mock,NAIF1or Bax expressing plasmids.Protein levels were detected by western blot analysis with various antibodies as described in Section2.3.9.The N-terminal of human NAIF1is critical for nuclear localization and apoptosis-inducingfunctionComputer analysis indicated that there were two NLSs in the N-terminal of human NAIF1.To test whether this part was important for nuclear localiza-tion of NAIF1,plasmids expressing NAIF1–GFP or NAIF1(71–327)–GFP were transfected into HeLa cells. As shown in Fig.10A,NAIF1–GFP appeared as a nuclear protein,while NAIF1(71–327)–GFP was local-ized mainly in the cytoplasm.To determine whether the N-terminal of NAIF1was responsible for inducing apoptosis,we transfected plasmids expressing NAIF1 or NAIF1(71–327)mutant into HeLa cells.Forty-eight hours later,apoptosis was assessed by staining the nuclei of cells with DAPI.No nuclear morphological alterations were observed in mock-or NAIF1(71–327)-transfected cells.In contrast,NAIF1-transfected cells exhibited chromatin condensation and margination(Fig.10B).B.Lv et al./The International Journal of Biochemistry&Cell Biology38(2006)671–683681Fig.10.The N-terminal of human NAIF1is critical for nuclear localization and apoptosis-inducing function.(A)HeLa cells were transfected with NAIF1–GFP or NAIF1(71–327)–GFP vectors.Twenty-four hour post-transfection,cells werefixed and nuclei were stained with DAPI.(B)HeLa cells were transfected with mock,NAIF1or NAIF1(71–327)expression vectors.Forty-eight hours post-transfection,cells werefixed and nuclei were stained with DAPI.Thus,it appeared that the N-terminal of human NAIF1 was critical for its nuclear localization and apoptosis-inducing function.4.DiscussionIn this study,we cloned and characterized a novel human gene,NAIF1(also named C9orf90),whose over-expression induced apoptosis.NAIF1proteins are con-served in human,mouse,dog,crab-eating macaque, chicken and frog,indicating that it may have important functions in vertebrate animals.NAIF1shows no sig-nificant similarity to any known functional genes in the GenBank puter analysis suggested that the NAIF1protein may be localized in the nucleus.Con-sistent with the computer analysis,our results revealed that NAIF1was expressed mainly in the nucleus.The NAIF1(71–327)mutant was localized mainly in the cytoplasm and could not induce apoptosis,suggesting that amino acids1–70in the NAIF1protein may be an important region not only for apoptosis induction, but also for nuclear localization.It is also possible that NAIF1(71–327)did not induce apoptosis because of its localization in the cytosol,instead of the nucleus.Motif analysis indicated that there was a GRR from amino acid92–117of the NAIF1protein.GRR has been reported to be involved in important roles in cell function,such as RNA-binding,protein–protein interaction,tran-scriptional regulation,processing events,and nucleolar targeting(Gendra,Moreno,Alba,&Pages,2004;Nagao et al.,2000;Orian et al.,1999;Talbot et al.,1997;Yao, Yang,&Seto,2001).Recently,a study described that the GRR(aa152–227)in Sox4served as a novel func-tional domain for promoting apoptotic cell death(Hur et al.,2004).NAIF1may be another example that the GRR serves as a critical domain involved in apoptotic regulation.Further investigations are needed to identify the molecular mechanisms of NAIF1during apoptosis.Several different approaches have confirmed that NAIF1-induced apoptosis when overexpressed.First, NAIF1-transfected cells underwent marked rounding and chromatin condensation and margination(Fig.7). Second,NAIF1transfection increased the Annexin V positive cell population compared with mock-transfected cells(Fig.7).Third,NAIF1caused a depolar-ization in mitochondrial Ψm(Fig.8).Finally,NAIF1-induced caspase-3activation(Fig.9).Interestingly,all of these events appeared much later than in Bax-transfected cells.This delay could be due to the possibility that the apoptosis-inducing ability of NAIF1is simply weaker than that of Bax.Alternatively,the delay could be due to the possibility that NAIF1and Bax might induce apoptosis through different pathways,thus resulting in the induction of apoptosis on different time schedules.。



《中国组织工程研究》 Chinese Journal of Tissue Engineering Research文章编号:2095-4344(2018)18-02813-07 2813www.CRTER .org·研究原著·杜国庆,男,1990年生,山东省青岛市人,汉族,青岛大学口腔医学院在读硕士,主要从事正颌外科与组织工程骨方面的研究。
通讯作者:孙健,博士,教授,主任医师,硕士生导师,青岛大学附属医院口腔颌面外科,青岛大学口腔数字医学与3D 打印工程实验室,山东省青岛市 266003中图分类号:R318 文献标识码:A稿件接受:2018-05-11Du Guo-qing, Master candidate, Department of Oral and Maxillofacial Surgery, the Affiliated Hospital of QingdaoUniversity, Laboratory of Oral Digital Medicine and 3DPrinting Engineering, Qingdao University, Qingdao266003, Shandong Province,China Corresponding author:Sun Jian, M.D., Professor, Chief physician, Master’ssupervisor, Department ofOral and Maxillofacial Surgery, the Affiliated Hospital of QingdaoUniversity, Laboratory of Oral Digital Medicine and 3DPrinting Engineering,Qingdao University, Qingdao 266003, Shandong Province, China3D 仿生打印组织工程骨修复下颌骨缺损杜国庆,孙 健,李亚莉,陈立强,陈 晨,邓 楠,吴雨桐,李 莉,王志浩(青岛大学附属医院口腔颌面外科,青岛大学口腔数字医学与3D 打印工程实验室,山东省青岛市 266003)DOI:10.3969/j.issn.2095-4344.0868 ORCID: 0000-0002-0182-9203(杜国庆)文章快速阅读:文题释义:3D 仿生打印:3D 仿生打印机可调控打印仓内温度等相关参数来维持仓内细胞及生长因子的活性。

GTR修复牙槽骨缺损的beagle犬动物模型的研究华楠;庄晶【摘要】目的为研究PLGA膜在beagle犬牙槽骨中的生物降解效果,建立一种符合牙槽骨缺损特点的动物模型.方法该研究在2011年12月-2012年2月期间,以3只beagle犬为实验动物,年龄在14~16个月,平均体重(13.0±1.0)kg.拔除其双侧前磨牙.拔除牙后同时放入可吸收膜.在拔牙窝的左侧放置Bio-Guide胶原膜,右侧放置PLGA膜.结果 3只实验犬均无死亡,饮食、活动正常.手术4周后再次进入牙槽骨,发现胶原膜和PLGA膜被完全吸收,生物降解.光学显微镜下显示在放置胶原膜和PLGA膜的拔牙窝上都形成了新的骨小梁.结论该研究采用的建模方法,成功构建了与临床即刻种植骨缺损相似,并可作为研究即刻种植骨界面骨性结合的实验动物模型.【期刊名称】《中外医疗》【年(卷),期】2015(034)026【总页数】2页(P48-49)【关键词】牙槽;骨缺损;引导;组织再生;动物实验模型【作者】华楠;庄晶【作者单位】上海杨思医院口腔科,上海200126;上海杨思医院口腔科,上海200126【正文语种】中文【中图分类】R780.1引导骨再生技术(GBR)是针对骨的引导组织再生(GTR)技术,由GTR延伸而来[1],GTR主要应用于牙齿[2]。
目前,GBR主要运用的是胶原蛋白膜,然而它具有局限性,因此有必要研究合成聚酯纤维的可吸收生物膜。
聚乳酸-羟基乙酸(PLGA)膜是一种可吸收生物膜,具有很好的生物降解性和生物相容性。
而建立动物模型,研究PLGA膜在动物牙槽骨中的生物降解效果,才能进一步阐明PLGA 作为GBR膜在临床使用中的效果。
多位学者先后采用猴、小型猪、比格犬、大耳白兔、鼠等多种实验动物,在其下颌骨制造骨缺损,植入植骨材料,采用组织学的方法评估植骨材料[3]。
但其动物实验模型与种植临床的骨缺损形态各不相同。
为研究PLGA膜在beagle犬牙槽骨中的生物降解效果,建立一种符合牙槽骨缺损特点的动物模型,该研究在2011年12月—2012年2月,采用beagle犬,年龄在14~16个月,按照天然骨缺损动物模型建立的方法制备牙槽骨缺损的实验模型[4],现报道如下。

壳聚糖加强骨组织生物修复机理解析骨组织是人体中最重要的结构之一,具有支撑身体、保护内脏和参与骨骼运动等重要功能。
然而,骨组织容易受到外界损伤和疾病的侵袭,导致骨折、骨质疏松和骨缺损等各种问题。
为了促进骨组织的愈合和修复,许多研究人员致力于寻找新的治疗方法和材料。
壳聚糖作为一种生物可降解的聚合物材料,已被广泛研究,其在骨组织生物修复中具有重要的作用。
壳聚糖是一种天然聚合物,主要存在于海洋生物中,如虾、蟹、贝壳等。
它具有生物相容性、生物降解性和生物可吸收性等优良特性,对人体无毒副作用,因此被广泛应用于医学领域。
壳聚糖加强骨组织生物修复的机理主要涉及以下几个方面:1. 促进骨细胞生长和增殖:研究表明,壳聚糖能够促进骨细胞的增殖和分化,从而提高骨组织的新生和修复。
壳聚糖可以通过调控一系列与骨细胞增殖和分化相关的信号通路来促进骨细胞的再生。
例如,壳聚糖可以通过激活Wnt/β-catenin信号通路和增加TGF-β1的分泌来促进骨细胞增殖和骨基质合成。
2. 促进骨基质沉积:壳聚糖可以促进骨细胞合成和分泌骨基质,从而增加新骨的沉积,并加强骨组织的力学性能。
壳聚糖可以通过刺激骨细胞产生胞外基质蛋白、胶原蛋白和骨基质硫酸葡聚糖等物质,促进骨基质的形成和沉积。
3. 促进血管生成:血管生成是骨组织修复过程中必不可少的一步,壳聚糖通过促进血管内皮细胞的增殖和迁移,以及诱导血管生成因子的表达,可以增加新生血管的密度和分布,提供充足的血液供应和氧气供应,促进骨组织的修复和再生。
4. 抗炎作用:壳聚糖具有抑制炎症反应的作用,可以通过抑制炎性介质的释放和炎症信号通路的调节,减轻骨组织受损区域的炎症反应,促进骨组织的修复和再生。
5. 促进骨组织再生的免疫调节作用:壳聚糖可以调节免疫反应,并促进骨组织再生。
壳聚糖可以通过调节T细胞的分化和功能、调节巨噬细胞的活化和极化等机制,促进骨组织的修复和再生。
总之,壳聚糖作为一种生物可降解的材料,在骨组织生物修复中具有重要的作用。
Novel therapeutic core–shell hydrogel scaffolds with sequential delivery of cobalt and bone morphogenetic protein-2for synergistic boneregenerationRoman A.Perez a ,b ,Joong-Hyun Kim a ,b ,Jennifer O.Buitrago a ,b ,Ivan B.Wall b ,c ,Hae-Won Kim a ,b ,d ,⇑aInstitute of Tissue Regeneration Engineering (ITREN),Dankook University,Cheonan 330-714,South KoreabDepartment of Nanobiomedical Science &BK21PLUS NBM Global Research Center for Regenerative Medicine,Dankook University,Cheonan 330-714,South Korea cDepartment of Biochemical Engineering,University College London,Torrington Place,London WC1E 7JE,UK dDepartment of Biomaterials Science,College of Dentistry,Dankook University,Cheonan 330-714,South Koreaa r t i c l e i n f o Article history:Received 16March 2015Received in revised form 5May 2015Accepted 1June 2015Available online 6June 2015Keywords:Osteogenesis AngiogenesisTherapeutic scaffolds Bone formation Sequential deliverya b s t r a c tEnabling early angiogenesis is a crucial issue in the success of bone tissue engineering.Designing scaffolds with therapeutic potential to stimulate angiogenesis as well as osteogenesis is thus considered a promising strategy.Here,we propose a novel scaffold designed to deliver angiogenic and osteogenic factors in a sequential manner to synergize the bone regeneration event.Hydrogel fibrous scaffolds comprised of a collagen-based core and an alginate-based shell were constructed.Bone morphogenetic protein 2(BMP2)was loaded in the core,while the shell incorporated Co ions,enabled by the alginate crosslinking in CoCl 2/CaCl 2solution.The incorporation of Co ions was tunable by altering the concentra-tion of Co ions in the crosslinking solution.The incorporated Co ions,that are known to play a role in angiogenesis,were released rapidly within a week,while the BMP2,acting as an osteogenic factor,was released in a highly sustainable manner over several weeks to months.The release of Co ions significantly up-regulated the in vitro angiogenic properties of cells,including the expression of angiogenic genes (CD31,VEGF,and HIF-1a ),secretion of VEGF,and the formation of tubule-like networks.However,BMP2did not activate the angiogenic processes.Osteogenesis was also significantly enhanced by the release of Co ions as well as BMP2,characterized by higher expression of osteogenic genes (OPN,ALP,BSP,and OCN),and OCN protein secretion.An in vivo study on the designed scaffolds implanted in rat calvarium defect demonstrated significantly enhanced bone formation,evidenced by new bone volume and bone density,due to the release of BMP2and Co ions.This is the first study using Co ions as an angio-genic element together with the osteogenic factor BMP2within scaffolds,and the results demonstrated the possible synergistic role of Co ions with BMP2in the bone regeneration process,suggesting a novel potential therapeutic scaffold system.Statement of SignificanceThis is the first report that utilizes Co ion as a pro-angiogenic factor in concert with osteogenic factor BMP-2in the fine-tuned core-shell hydrogel fiber scaffolds,and ultimately achieves osteo/angiogenesis of MSCs and bone regeneration through the sequential delivery of both biofactors.This novel approach facilitates a new class of therapeutic scaffolds,aiming at successful bone regeneration with the help of angiogenesis.Ó2015Acta Materialia Inc.Published by Elsevier Ltd.All rights reserved.1.IntroductionRecent advances in bone tissue engineering have focused on developing therapeutic scaffolds with the capacity to load bioactive molecules and deliver them in a controllable and sustain-able manner [1].The in vivo conditions under which damaged tissues are repaired basically require multiple types of bioactive molecules and biological ingredients with their sequential involvement at the correct time and with effective doses [2].Some key elements have long been identified to play key roles in bone repair and regeneration.For instance,bone morphogenetic/10.1016/j.actbio.2015.06.0021742-7061/Ó2015Acta Materialia Inc.Published by Elsevier Ltd.All rights reserved.⇑Corresponding author at:Institute of Tissue Regeneration Engineering (ITREN),Dankook University,Cheonan 330-714,South Korea.Tel.:+82415503081.E-mail address:kimhw@ (H.-W.Kim).proteins(BMPs)are one of the most efficient bioactive molecules that stimulate osteogenic induction,in orthotopic sites and even in ectopic locations[3].Despite the importance of osteogenic induction,successful bone regeneration requires early stage angio-genesis,and this is particularly important when the defect dimen-sion is large and/or tissue engineered cells are engaged.The secretion of angiogenic factors,recruitment of blood vessel-forming endothelial cells and ultimately the stimulation of vascu-lar network formation are key events taking place at the early stage of bone repair and regeneration[4].Therefore,strategies to improve those initial angiogenic events, accompanied by the stimulation of osteogenesis at a much later stage,have recently been significantly pursued.Noteworthy exem-plar studies include the use of angiogenic factors such as vascular endothelial growth factor(VEGF)and basicfibroblast growth factor (bFGF)at the early phase,while osteogenic molecules,including BMP2,BMP7,and dexamethasone are designed to be delivered at a much later stage[5,6].The success of this strategic concept of‘se-quential drug delivery’is largely dependent on the design of scaf-folds.The incorporation of microparticles within scaffolding matrices and the layered structuring of scaffolds are some key examples.In this sense,PLGA microspheres loaded with BMP2 were embedded in polypropylene scaffolds that rapidly released VEGF($3days)and then facilitated more sustained release of BMP2over50days[6].The same effect was achieved using a dif-ferent approach,whereby sequential delivery of VEGF and BMP2 was possible through the layering of polyelectrolytefilms.In order to achieve this,a degradable tetra layer with a poly(b-amino ester)/polyanion/growth factor/polyanion structure was prepared in sequential layers,enabling sustained release of BMP2over 2weeks,whereas the VEGF within8days[7].Recently,the authors designed a core–shell structuredfibrous scaffold made of a collagen core and an alginate shell using a co-concentric nozzle design[8,9].The core–shellfibrous scaffolds have been demonstrated to be useful platforms for dual drug deliv-ery[8]as well as cell encapsulation[9].The core–shell structure enabled sequential release of two different biomolecules,which were incorporated either in the core or in the shell part,and the release rate was highly dependent on the molecular diffusion rate. Furthermore,for targeting hard tissues,the incorporation of cal-cium phosphate self-setting powders substantially improved the mechanical properties and slowed down the protein release behav-iors,implying usefulness in long-term delivery of therapeutic molecules.Besides their use in drug delivery,the cells encapsu-lated within the core part were shown to survive,proliferate,and properly adopt osteogenic differentiation under osteogenic envi-ronments.This also translated to excellent performance in vivo, suggesting a possible vehicle for cell delivery.In particular,the sys-tem is based on the use of alginate as the shell,which becomes hardened in contact with divalent cations,such as CaCl2,preserv-ing the core–shell structured scaffolds[8].On the basis of the core–shellfibrous scaffolds,in this study we designed a sequential drug delivery system with angiogenic and osteogenic potential,which is ultimately effective for bone regen-eration.We utilized cobalt(Co)ions as the pro-angiogenic factor. The rationale of using Co is that it has been used to induce hypoxia by blocking the degradation of hypoxia inducing factor(HIF)a1 [10].HIF a1is a transcription factor in the oxygen sensing pathway activated in hypoxic environments and is able to alter gene expres-sion and enhance angiogenesis[11].A recent study utilizing Co has demonstrated its considerable effect on angiogenesis[12]. Therefore,we hypothesized that Co incorporated within the core–shellfibrous scaffolds,particularly in the shell part,would be effective in the initial angiogenesis phase.Importantly,the Co ions can be elegantly incorporated within alginate in partial replacement of Ca ions as an alternative cross-linking divalent ionic source[13,14].In this way,we incorporated Co ions within the shell part in the course of alginate hardening in CoCl2/CaCl2 solutions while delivering BMP2in the core part.Fig.1a illustrates schematically the current design of the core–shellfibrous scaffolds to sequentially deliver Co/BMP2.To the best of our knowledge,this co-usage and co-delivery of Co with an osteogenic factor(BMP2)has not yet been reported. While excessive uses of Co may induce cell death,small concentra-tions used in the therapeutic window(reported to be less than 100l M)are expected to have profound positive effects[15,16]. We report this novel design of Co/BMP2sequential delivering scaf-fold,in terms of the process tools,effects on cellular angiogenesis and osteogenesis,and in vivo efficacy in bone regeneration.2.Materials and Methods2.1.Core–shellfibrous scaffold fabricationFor the composition of core and shell materials,we used collagen-and alginate-based hydrogels which also incorporate dif-ferent amounts of self-setting calcium phosphate powder (a-tricalcium phosphate(a-TCP)).The a-TCP powder was obtained by sintering a mixture of calcium hydrogen phosphate(CaHPO4, from Sigma–Aldrich#C7263)and calcium carbonate(CaCO3,from Sigma–Aldrich#239216)at1400°C and subsequent quenching [17,18].The sintered powder was milled in a planetary mill and added with2wt.%hydroxyapatite(HA,from Alco#1.02143)crys-tallites as a seed for the phase-transformation of cement(from a-TCP into HA).The a-TCP powder showed a median particle size of 5.2l m as determined from laser diffraction(Malvern, APA5001SR),and a specific surface area of1.23m2/g as measured by N2adsorption–desorption by the BrunauerÀEmmettÀTeller (BET)method(Quadrasorb SI,Quantachrom Instruments,Ltd., Boynton Beach,FL).Na-alginate(from Sigma–Aldrich#A2158) with an M/G ratio of1.67and a molecular weight of50kDa was dissolved at3%in water.The collagen solution(rat tail type I col-lagen at a concentration of2.05mg/ml,from First Link)was pre-pared by mixing1ml of the collagen solution with100ml of10 Dulbecco’s modified Eagle medium and a proper amount of1N sodium hydroxide which is proper to adjust pH at7.0for subse-quent gelation.A co-concentric nozzle(inner23G and outer17G)was specifi-cally designed and used to produce core–shell structuredfibrous scaffolds.Each solution with a specific composition of10%a-TCP plus alginate or50%a-TCP plus collagen was separately fed into the outer and inner syringe,respectively.Each syringe was attached to an injection pump connected through a microtube. Core–shell structured collagen-alginatefiber(0.3ml)was then injected at a rate of50ml/h through the co-concentric nozzle within a bath containing150mM of divalent ions(Ca2+or Co2+) for1min.The concentration of ions was optimized for the gelation of alginate shell phase while preserving a good cell viability of the fiber scaffolds[8,9].Calcium chloride(CaCl2,from Sigma–Aldrich #223506)and cobalt chloride(CoCl2,from Sigma–Aldrich #232696)were used as the source of Ca and Co ions,respectively. Different crosslinking solutions are summarized in Table1.2.2.Loading and release of Co ion and BMP22.2.1.Co ion loading and releaseThe core–shell scaffolds prepared in the Co ion-containing crosslinking solution incorporate Co ions due to a diffusion of Co through the alginate network.Furthermore,the incorporated Co ions vary depending on the content of Co ions within the crosslink-ing solutions.After preparation of the core–shell scaffolds296R.A.Perez et al./Acta Biomaterialia23(2015)295–308crosslinked in different solutions,the release of Co ions from the scaffolds was analyzed.Each scaffold sample was placed in each well of 24-well plates containing 1.5ml of distilled water.After 1,3,and 6days,the medium was replaced.The supernatant wasanalyzed to measure the amount of Co ions released from the sam-ple using the Co quantification kit (BioVision,Cobalt Colorimetric Assay kit),following the manufacturer’s guidelines.After the col-orimetric reaction,an absorbance was read at 475mm using a spectrophotometer.Data were analyzed in triplicate (n =3).2.2.2.BMP2loading and releaseBMP2protein was obtained as previously described [19].Briefly,the cDNAs of the BMP2were amplified from an adult human cDNA library.According to the GenBank sequence (GeneBank accession number NM002006),a pair of primers,50-G AAGATCTGCCAAACACAAACAGCGG-3and 50-AACTGCAGATCTGTCT TTTCTACCGCTGGACACCCACAACCCTC-30,were designed and used.The PCR products were cloned into pBAD/His A (Invitrogen,Carlsbad,CA)in-frame using the NH 2-terminal 6X His tag.Theillustration showing a current study aim to deliver Co and BMP2cofactors in the core–shell fibrous scaffold system.a designed concentric nozzle into a CoCl 2/CaCl 2solution,become a core–shell fibrous scaffold by the hardening of incorporated.The Co incorporated in the outer shell and the BMP2loaded in the inner core are to be released an early phase of blood vessel formation as well as the osteogenesis by BMP2at a later stage of bone formation.(b)from optical microscopy and SEM.Core and shell parts are noted in images.Table 1Crosslinking solutions used for the core–shell formation of collagen-alginate thera-peutic scaffolds,where the concentration of divalent cations of Ca and Co was varied to crosslink alginate as well as to incorporate Co ions.CaCl 2(mM)CoCl 2(mM)015050100140101491150recombinant BMP2protein containing the poly-His tag was expressed and purified using a Ni affinity column under denaturing conditions according to the manufacturer’s protocol(Invitrogen).The BMP2was added directly within the core part of the core–shell scaffolds during the preparation of scaffolds.BMP2solution was mixed with the collagen solution which was then produced into core–shell scaffolds.The crosslinking solution wasfixed at 149mM CaCl2and1mM CoCl2.For the release study of BMP2,each scaffold was placed in each well of24-well plates containing1.5ml of distilled water.After incubation for different durations,the med-ium was replenished,and the supernatant was analyzed by enzyme linked immunosorbent assay(ELISA,Perrotech).2.3.Rat mesenchymal stem cells and cell viability on scaffoldsMesenchymal stem cells(MSCs)derived from rat bone marrow were harvested from the femora and tibiae of adult rats(180–200g)according to the guidelines approved by the Animal Ethics Committee of Dankook University.The harvested product was then centrifuged,and the supernatant was collected and suspended within a cultureflask containing a normal culture medium (a-minimal essential medium(a-MEM)supplemented with10% fetal bovine serum(FBS),100U/ml penicillin,and100mg/ml streptomycin),in a humidified atmosphere of5%CO2in air at 37°C.After incubation for1day,the medium was refreshed and cultured until the cells reached near confluence.After the subcul-ture was maintained in the normal culture condition,cells at2–3 passages were used for further tests.The effects of Co ions incorporated within the scaffolds on the cell viability werefirst assessed.An indirect culture method was used.Cells were seeded at10,000on each well of24-well plates containing1.5ml of normal culture medium.After24h,the scaf-folds were placed on top with the help of Transwell inserts.The scaffolds analyzed were those crosslinked in150mM CaCl2, 149mM CaCl2+1mM CoCl2,and140mM CaCl2+10mM CoCl2. The tissue culture plate without any scaffolds was used as a con-trol.Cell proliferation was measured after1,3,and6days using a cell counting kit-8reagent(CCK-8,Dojindo Molecular Technologies).To perform the CCK-8assay,the medium was removed and replaced with200l l of serum-free medium followed by the addition of20l l of CCK-8solution.The reagent was left to react for2h and the product obtained was then read at an absor-bance of450nm using a spectrophotometer.For the optical microscopy of cell growth behaviors,at each cul-ture period,the cells werefixed with4%paraformaldehyde solu-tion,stained with Alexa Fluor546conjugated Phalloidin (Molecular Probes)and Prolong Gold antifade reagent with DAPI (Molecular Probes),and then visualized underfluorescence signals using a confocal laser scanning microscope(CLSM;LSM510,Zeiss).2.4.In vitro angiogenesis studyThe in vitro performance of the designed scaffolds wasfirst ana-lyzed based on the effects on cellular angiogenesis.The experiment was carried out using an indirect culture method with Transwell inserts,as described above.Four different scaffolds were used for the assays:incorporation of only Co,of only BMP2,of both Co and BMP2,and with no incorporation.The scaffolds that included Co ions were crosslinked within149mM CaCl2+1mM CoCl2, and those that incorporated only BMP2were crosslinked in 150mM CaCl2.Cells cultured without scaffolds were used as a con-trol.Culture periods were up to6days.2.4.1.Expression of angiogenic genesThe expression of genes associated with angiogenesis was assessed by quantitative PCR(qPCR).After3and6days of culture,each sample was washed with PBS twice and the cells were detached by trypsinization.The cell pellet was collected and frozen at-80°C until use.Total RNA was extracted from the cells using the RNeasy Kit(Qiagen)according to the manufacturer’s instructions. The concentration of RNA samples was quantified using a Nanodrop2000spectrophotometer(Thermo Scientific).RNA sam-ples were transcribed into cDNA by combining the RNA with a ran-dom hexamer,followed by denaturation at70°C for5min.After denaturation,the samples were mixed with the RT premix for complete cDNA synthesis.cDNA was then amplified using RT-PCR premix(Bioneer)at94°C for5min,followed by35cycles of94°C for1min denaturation,57°C for1min for annealing,and 72°C for1min for extension.Primers for amplification were CD31: CTCCTAAGAGCAAAGAGCAACTTC(forward),TACACTGGTATTCCAT GTCTCTGG(reverse),VEGF:ACCAGTGACAGAGGCCAATACT(for-ward),GGCCTCCACAGTCAGGTTATAC(reverse),HIF-1a:GCGTACA TAGGGACTGGAGA(forward),AAAGTGGGTAGGAGAAAGGG (reverse)and glyceraldehyde-3-phosphate dehydrogenase (GAPDH):TGAACGGGAAGCTCACTGG(forward),TCCACCACCCTGT TGCTGTA(reverse).The results were normalized with respect to GAPDH.2.4.2.VEGF productionTo determine the ability of the cells to secrete angiogenic fac-tors,the release of VEGF was quantified by ELISA(ELISA, Perrotech).After culturing,the medium was collected every two days and stored at-20°C until further analysis.After6days,an ali-quot of each sample was used to quantify the amount of VEGF pro-duced.The results were normalized to the cell number.2.4.3.Matrigel assay for tubular formationIn vitro tubular formation was examined using a Matrigel(BD Biosciences,BD Matrigel,356231)culture.The Matrigel was coated onto24well plates for1h at37°C for complete gelation,according to the manufacturer’s specifications.80,000cells were then cul-tured on each well plate and Transwell inserts containing different scaffolds were placed on top.The cells were monitored by optical microscopy at3,6,and22h of culture.The images were then ana-lyzed with Metamorph software in order to measure the length of the tubules formed.2.5.In vitro osteogenesis studyIn order to examine the ability of the scaffolds to induce osteo-genic differentiation of rMSCs,a similar experimental design was used as that described above,involving four different scaffold groups and a culture dish control,and the Transwell inserts culture method was used.Culture periods were prolonged up to7and 14days.2.5.1.Expression of osteogenic genesThe expression of genes associated with osteogenesis,including osteocalcin(OCN),alkaline phosphatase(ALP),bone sialoprotein (BSP),and osteopontin(OPN),was assessed by q PCR.After culture for7and14days,the cells were trypsinized and the cell pellet was collected and frozen at-80°C until used.The total RNA was extracted from differentiated cells using the RNeasy Kit(Qiagen) according to the manufacturer’s instructions.The concentration of RNA samples was quantified using a Nanodrop2000spectropho-tometer(Thermo Scientific).The RNA samples were transcribed into cDNA by combining the RNA with a random hexamer,fol-lowed by denaturation at70°C for5min.After denaturation,the samples were mixed with the RT premix for complete cDNA syn-thesis.cDNA was then amplified using RT-PCR premix(Bioneer) at94°C for5min,followed by35cycles of94°C for1min denat-uration,57°C for1min for annealing and72°C for1min for298R.A.Perez et al./Acta Biomaterialia23(2015)295–308extension.The primers used for amplification were OCN: CATGAAGGCTTTGTCAGACT(forward),CTCTCTCTGCTCACTCTGCT (reverse);ALP:CCTTTGTGGCTCTCTCCAAG(forward),CGATGTCCT TGATGTTGTGC(reverse)BSP:AGAAAGAGCAGCACGGTTGA(for-ward),TCATAGCCATGCCCCTTGTA(reverse);OPN:GAGGAGAAG GCGCATTACAG(forward),AAACGTCTGCTTGTCTGCTG(reverse); and glyceraldehyde-3-phosphate dehydrogenase(GAPDH): TGAACGGGAAGCTCACTGG(forward),TCCACCACCCTGTTGCTGTA (reverse).Results were normalized to GAPDH.2.5.2.OCN immunofluorescence stainingAs the representative osteogenic marker at a relatively later stage,OCN protein expression was examined using immunofluo-rescence staining.After7and14days,the cells grown on each sample werefixed with4%paraformaldehyde and then treated with0.1%Triton X-100.After washing with PBS,the samples were immunostained against OCN antibody to observe osteogenic mar-ker OCN.Alexa Fluor546-conjugated phalloidin(Invitrogen A22283)diluted in PBS was also added to each sample to stain the F-actin.A Prolong Gold antifade reagent with4’,6-diamidi no-2-phenylindole(DAPI;Invitrogen)was used to stain the cells and to produce bluefluorescence signals upon binding to the DNA in the nucleus.Images were visualized under a Zeiss LSM 510microscope.2.6.In vivo bone formation studyThe bone forming ability of the scaffolds was examined in a rat calvarium defect model.Ten week old male Sprague–Dawley rats were used in the experiment.Animals were maintained in a12-h light/dark cycle(lights on from8:00to20:00),in a relative humid-ity(30–70%)and temperature(20–24°C)controlled environment. The rats were allowed a normal diet and water ad libitum.The experimental protocol for all animal care and use in this study was reviewed and approved in accordance with guidelines estab-lished by the Animal Care and Use Committee,Dankook University,Cheonan,Korea.2.6.1.Animal surgery and scaffold implantationThe animals were anaesthetized by intramuscular injection with a mixture of ketamine(80mg/kg)and xylazine(10mg/kg) for surgery.The hair over the skin on the dorsal region of the cra-nium was shaved and the region was aseptically prepared using povidone and70%ethanol for surgery.A linear sagittal midline skin incision was made over the calvarium with a#10surgical blade.A full-thicknessflap of the skin and periosteum was elevated to obtain a sufficient surgicalfield for the drilling.Two 5mm-diameter critical sized circular full-thickness bone defects were created at the center of each parietal bone in the calvarium of each rat.To create the defects without overheating of the bone edges,a trephine bur was used under constant irrigation with nor-mal saline solution,without damaging the underlying sagittal sinus and dura matter.The scaffolds were sterilized prior to use. Each cranial defect was randomlyfilled with0.1ml with the four different groups:with no Co/BMP2,Co only,BMP2only,and Co/BMP2both,having two different defects per animal.In all rats, subcutaneous tissue was closed with absorbable materials,and the skin incisions were sutured with non-absorbable materials.After the implantation,the animals were housed individually in cages and monitored by visual observation during the experimen-tal time for signs of infection,inflammation,and any adverse reac-tion.Six weeks after the operation,the animals were sacrificed to collect the calvariae.The skin was dissected,and the tissues includ-ing the surgical sites and their surrounding tissues were harvested along with the surrounding bone.The specimens werefixed in10%buffered neutralized formalin for24h at room temperature and prepared for micro-CT(l CT)analysis and histology.2.6.2.Micro-computed tomography analysisFollowingfixation,the micro-computed tomography(l CT) images were performed to visualize the samples and to analyze images of new bone formation.The harvested specimens were scanned using a l CT scanner(Skyscan1176;Skyscan,Aartselaar, Belgium)with a camera pixel size of12.56l m;also,a frame aver-aging of3was employed together with afilter of1mm aluminum, a rotation step of0.5°,and a rotation angle of180°.The X-ray tube voltage was65kV and the current was385l A,with an exposure time of279ms.A cylindrical region of interest(ROI)was posi-tioned over the center of the single defect,fully enclosing the new bone within the defect site.The percentages of new bone vol-ume(%),bone surface area(mm2),and bone surface density (1/mm)of newly formed bone within each ROI were measured by assigning a threshold for the total bone content(including tra-becular and cortical bone)through a computer analysis program, CTAn(Skyscan).2.6.3.Histological procedureAfter l CT imaging,the harvested samples were prepared for histological analysis.To achieve this,fixed specimens were decalcified with RapidCal TM solution(BBC Chemical Co., Stanwood,WA,USA),dehydrated through a series of ethanol solu-tions of increasing concentration(from70%to100%),and subsequently embedded in paraffin.Five micrometer thick coronal sections were prepared at the central region of the circular defects and defined as the representative histology.The slides were stained with hematoxylin&eosin(H&E)stain using a routine tech-nique and examined using a light microscope(IX71,Olympus, Tokyo,Japan).2.7.Statistical analysisExperiments were performed in triplicate,unless otherwise specified.Data are expressed as mean±one standard deviation. Statistical analysis was carried out using one way analysis of vari-ance(ANOVA)with a Fisher post hoc test and a significance level was considered at p<0.05.3.Results3.1.Core–shell designed scaffoldsThe images of the core–shellfibrous scaffolds produced are shown in Fig.1b.The optical image shows the transparentfibrous structured scaffolds in the medium and the enlarged image shows the core and shell part of thefiber with an average core diameter of $1000l m and an average shell thickness of$150l m.Thefibrous morphology of the scaffolds was well preserved in the culture medium and easily formed into the shape of a mold.The SEM mor-phologies of the freeze-dried scaffolds exhibited a highly macrop-orous structure and the core–shelled structure can be seen in cross-section.No difference was observed in the morphology of the core–shellfibrous scaffolds depending on the Co/Ca composi-tions used for the alginate crosslinking.The release of therapeutic molecules,including Co in the shell and BMP2in the core,was monitored as shown in Fig.2. Scaffolds were prepared with different crosslinking solutions while varying the concentrations of Co ions from0to150mM which was balanced with Ca ions.The Co and Ca ions will be competitively incorporated into the alginate shell part during the crosslinking, and the incorporated amount of Co could also be assessedR.A.Perez et al./Acta Biomaterialia23(2015)295–308299。