NeST Long ncRNA 调控IFN-gama
- 格式:pdf
- 大小:1.30 MB
- 文档页数:12

ifn-γ杀伤肿瘤原理
IFN-γ(干扰素γ)是一种由免疫细胞产生的细胞因子,具有
多种免疫调节功能。
它在杀伤肿瘤中起到重要的作用。
IFN-γ杀伤肿瘤的原理主要有以下几个方面:
1. 抑制肿瘤生长:IFN-γ可以通过抑制肿瘤细胞的增殖和促进
肿瘤细胞凋亡,直接抑制肿瘤的生长。
2. 干扰血管生成:IFN-γ可以抑制肿瘤血管生成,使肿瘤缺血、缺氧,从而抑制肿瘤的生长和转移。
3. 增强免疫应答:IFN-γ能够激活和增强免疫细胞的功能,包
括巨噬细胞、NK细胞和T细胞等,使它们对肿瘤细胞的识别
和杀伤能力增强。
4. 调节免疫环境:IFN-γ可以调节免疫环境,提高肿瘤微环境
中的CTLs(细胞毒性T淋巴细胞)浸润和活性,从而增强免
疫细胞对肿瘤的攻击能力。
总的来说,IFN-γ通过多种机制调节免疫和抑制肿瘤生长,发
挥着重要的抗肿瘤作用。
这也是为什么在某些肿瘤治疗中,IFN-γ被用于增强免疫治疗效果的原因。

非编码RNA在肿瘤中的调控作用在癌症的发展过程中,研究人员不断地探索参与调控癌症进程的新颖生物分子,在这过程中,非编码RNA(non-coding RNA,ncRNA)作为一个热门研究领域受到了越来越多的关注。
ncRNA不仅包括一些经典的长非编码RNA,还包括长度短、难以预测的小RNA,例如microRNA(miRNA)和PIWI-interacting RNA(piRNA)等。
近来的研究表明ncRNA在癌症发展过程中可能发挥着重要的调控作用。
ncRNA可以通过多种不同的方式参与调控基因表达,其中最为常见的方式是通过靶向mRNA,从而调节靶向基因的翻译或降解。
例如,miRNA是一种长度约为22 nts的小RNA,多数miRNA可以靶向多种mRNA,从而抑制mRNA的翻译或切断mRNA的降解。
在肿瘤中,研究表明某些miRNA存在着表达异常(over-expression或down-expression)的现象,这种异常的表达与癌症的发展和预后密切相关。
例如在肺癌中,miR-21的表达水平明显升高,而它的靶向基因PDCD4则被抑制,从而增加了肺癌细胞的增殖和侵袭能力。
除了miRNA,长非编码RNA(lncRNA)也参与着调控基因表达。
与miRNA不同的是,大部分lncRNA是具有组织特异性和表达量低的生物分子。
与典型的mRNA不同的是,lncRNA的转录常常伴随着一些不同寻常的规律,例如某些lncRNA可以与DNA或蛋白质形成复合物,在某些情况下也可以与miRNA竞争结合靶向mRNA。
在肿瘤中,研究表明一些lncRNA表达水平发生了改变,它们可能通过获得了新的功能,从而参与着癌症的发展过程。
例如,HOTAIR是一种表达在乳腺癌组织中的lncRNA,它被认为是一种调控各种“表观遗传学”修饰的“开关”,进而影响了许多细胞增殖和转移过程的lncRNA。
另外,ncRNA还可以通过其他的方式参与调控基因表达,例如piRNA可通过与PIWI蛋白的结合形成复合物,从而通过抑制转座子的活性来减少细胞中的DNA损伤。
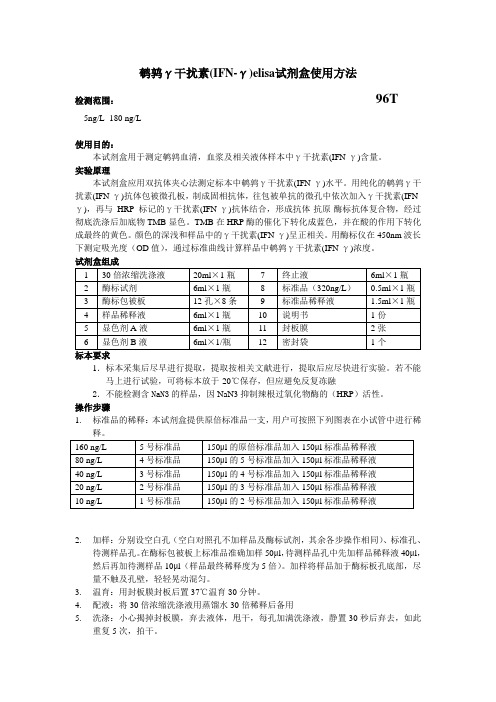
鹌鹑γ干扰素(IFN-γ)elisa试剂盒使用方法检测范围:96T5ng/L -180 ng/L使用目的:本试剂盒用于测定鹌鹑血清,血浆及相关液体样本中γ干扰素(IFN-γ)含量。
实验原理本试剂盒应用双抗体夹心法测定标本中鹌鹑γ干扰素(IFN-γ)水平。
用纯化的鹌鹑γ干扰素(IFN-γ)抗体包被微孔板,制成固相抗体,往包被单抗的微孔中依次加入γ干扰素(IFN-γ),再与HRP标记的γ干扰素(IFN-γ)抗体结合,形成抗体-抗原-酶标抗体复合物,经过彻底洗涤后加底物TMB显色。
TMB在HRP酶的催化下转化成蓝色,并在酸的作用下转化成最终的黄色。
颜色的深浅和样品中的γ干扰素(IFN-γ)呈正相关。
用酶标仪在450nm波长下测定吸光度(OD值),通过标准曲线计算样品中鹌鹑γ干扰素(IFN-γ)浓度。
试剂盒组成1.标本采集后尽早进行提取,提取按相关文献进行,提取后应尽快进行实验。
若不能马上进行试验,可将标本放于-20℃保存,但应避免反复冻融2.不能检测含NaN3的样品,因NaN3抑制辣根过氧化物酶的(HRP)活性。
操作步骤1.标准品的稀释:本试剂盒提供原倍标准品一支,用户可按照下列图表在小试管中进行稀2.加样:分别设空白孔(空白对照孔不加样品及酶标试剂,其余各步操作相同)、标准孔、待测样品孔。
在酶标包被板上标准品准确加样50μl,待测样品孔中先加样品稀释液40μl,然后再加待测样品10μl(样品最终稀释度为5倍)。
加样将样品加于酶标板孔底部,尽量不触及孔壁,轻轻晃动混匀。
3.温育:用封板膜封板后置37℃温育30分钟。
4.配液:将30倍浓缩洗涤液用蒸馏水30倍稀释后备用5.洗涤:小心揭掉封板膜,弃去液体,甩干,每孔加满洗涤液,静置30秒后弃去,如此重复5次,拍干。
6.加酶:每孔加入酶标试剂50μl,空白孔除外。
7.温育:操作同3。
8.洗涤:操作同5。
9.显色:每孔先加入显色剂A50μl,再加入显色剂B50μl,轻轻震荡混匀,37℃避光显色10分钟.10.终止:每孔加终止液50μl,终止反应(此时蓝色立转黄色)。

真核生物3'UTR的转录后水平调控及其与肿瘤的关系康南;王宇;王颖;毛伊雯;燕太强;沈丹华【摘要】真核生物3'非翻译区(3'untranslated regions,3'UTR)在转录后水平调控中起到重要作用,它参与调控mRNA的体内稳定性及降解速率,控制mRNA的利用效率,还决定mRNA的翻译位点及翻译效率,调控mRNA细胞内运输及胞质定位等多种代谢过程.3'UTR既可以与microRNAs或者RNA结合蛋白相互作用来反式调控基因的表达,从而阻止mRNA的翻译或直接降解靶mRNA,同时3'UTR也可以作为独立存在的RNA分子发挥功能,近年来通过对肿瘤全基因组相关研究发现突变发生在3'UTR或与microRNA结合区会破坏细胞内的调控机制,从而影响肿瘤的发生发展,使3'UTR成为目前研究热点,并使其有望成为肿瘤诊断和治疗的新标志物甚至药物靶点.【期刊名称】《医学研究杂志》【年(卷),期】2019(048)002【总页数】4页(P8-11)【关键词】真核生物;3'非翻译区;mRNA代谢;独立分子;突变;肿瘤【作者】康南;王宇;王颖;毛伊雯;燕太强;沈丹华【作者单位】100044 北京大学人民医院病理科;100021 中国医学科学院肿瘤医院分子肿瘤学国家重点实验室;100044 北京大学人民医院病理科;100044 北京大学人民医院病理科;100044 北京大学人民医院病理科;100044 北京大学人民医院病理科【正文语种】中文【中图分类】R34真核生物3′非翻译区(3′untranslated regions,3′UTR)即非翻译区,是指mRNA 分子两端的非编码片段,研究发现,许多mRNA的调控元件存在于5′UTR及3′UTR中,5′UTR主要起始调控mRNA翻译,3′UTR调控mRNA的多种代谢,包括出核、胞质定位、翻译效率及mRNA稳定性等[1]。

茶花鸡2号IFN-α基因克隆及生物信息学分析刘琛;何永江;豆腾飞;杨明华;潘洪彬;赵素梅;李永能;黄英【期刊名称】《中国畜牧杂志》【年(卷),期】2024(60)1【摘要】本研究旨在克隆茶花鸡2号α干扰素基因(IFN-α)CDS区序列并进行生物信息学分析,为后续研究IFN-α基因对茶花鸡2号免疫功能的影响提供参考。
以茶花鸡2号为实验对象设计IFN-α引物,提取总RNA,反转录PCR扩增并克隆其编码区序列,进行生物信息学分析。
结果显示茶花鸡2号IFN-α基因CDS序列全长582 bp,IFN-α蛋白等电点5.05,平均疏水指数-0.514,为酸性亲水蛋白,且存在信号肽和1个跨膜区,主要定位于细胞核。
IFN-α蛋白被4个N糖基化位点、5个O糖基化位点和20个磷酸化位点修饰,主要由α-螺旋与无规卷曲构成。
茶花鸡2号与固始鸡、海兰鸡、惠阳胡须鸡、罗曼鸡和乌骨鸡的氨基酸同源性分别为95.9%、97.9%、97.9%、97.9%和95.9%。
IFN-α蛋白与IRF7、IFNAR1、IFNAR2和IFNK等蛋白存在互作关系。
本研究结果可为进一步探讨IFN-α基因在茶花鸡2号病毒疾病防治中的作用提供理论依据。
【总页数】6页(P217-222)【作者】刘琛;何永江;豆腾飞;杨明华;潘洪彬;赵素梅;李永能;黄英【作者单位】云南农业大学动物科学技术学院/云南省动物营养与饲料重点实验室;云南农业大学组织部【正文语种】中文【中图分类】S831.2【相关文献】1.山茶花翻译控制肿瘤蛋白CjTCTP基因的电子克隆及生物信息学分析2.鸡十二指肠、法氏囊IFN-α基因的克隆和生物信息学分析3.合作猪IFN-β基因编码区克隆及生物信息学分析4.合作猪IFN-ε基因克隆及生物信息学分析5.鸡新城疫Ⅰ系疫苗诱导鸡胚IFN-γ基因转录及cDNA克隆的研究因版权原因,仅展示原文概要,查看原文内容请购买。

非编码RNA对肿瘤的调控机制罗子华;于晓峰;邹健【摘要】非编码RNA(ncRNA)是不参与蛋白质编码的RNA的总称,包括微小RNA(miRNA)、长链非编码RNA(lncRNA)、转运RNA(tRNA)、核糖体RNA (rRNA)等.研究发现,ncRNA在调节组织细胞的发生、分化、增殖、凋亡等方面具有重要的作用.尤其在肿瘤细胞中,ncRNA的表达水平与正常细胞有明显差异,在功能上表现为致癌基因或抑癌基因,提示其在肿瘤的发生发展中可能起重要作用.此文对当前研究较为深入的几种ncRNA调控肿瘤的机制作一综述.【期刊名称】《国际消化病杂志》【年(卷),期】2013(033)002【总页数】4页(P97-100)【关键词】非编码RNA;微小RNA;长链非编码RNA;肿瘤;机制【作者】罗子华;于晓峰;邹健【作者单位】200040上海,复旦大学附属华东医院消化内科【正文语种】中文非编码RNA(ncRNA)是不参与蛋白质编码的RNA的总称,包括微小RNA (miRNA)、长链非编码RNA(lncRNA)、转运 RNA(tRNA)、核糖体RNA (rRNA)等。
人类基因组中编码蛋白质的基因约占2%,绝大部分转录成长链或短链的ncRNA[1],这些曾长期被认为是“垃圾”的转录产物,可能对维持机体的正常生理功能、细胞的正常代谢有重要的作用。
肿瘤的发生过去一直被认为是由于致癌基因或抑癌基因遗传学和(或)表观遗传学的改变所致。
各种调节因子控制这些基因的表达,使细胞正确执行增殖、分化、凋亡等生物学过程。
此类基因的调节失常使得这些生物学过程不受控制,进而导致肿瘤的发生[2]。
最新的研究发现,此类ncRNA在肿瘤的发生、发展、转移、凋亡中起着重要的作用。
如在哺乳动物中,估计miRNA调控超过60%蛋白质编码基因的表达并参与调控大部分的细胞学进程[3]。
研究发现ncRNA在肿瘤细胞中的表达水平与正常组织相比有明显的改变,在功能上表现为致癌基因或抑癌基因,提示其在肿瘤的发生发展中可能起重要的作用。
兰主壁垫垄茎旦查垦兰堕.堡主兰垡垒!!一IFN.Y促进肿瘤免疫逃避及相关的肿瘤基因治疗摘要肿瘤的发生发展包括两个过程:首先是癌变,然后是增殖。
在这两个过程中相应的细胞都不被免疫系统消灭才能最终形成肿瘤。
不能被免疫系统消灭也正是肿瘤的一个重要特征,这其中包括许多的逃避机制,包括肿瘤细胞表面MHCI类分子的下调,分泌免疫抑制性因子如TNF-B,缺乏T细胞的共刺激信号以及在T细胞遇到肿瘤细胞时发生凋亡等等。
在此研究中,我们首先发现前炎症因子IFN.Y在持续低水平表达时能够促进肿瘤的发生发展,这种作用有一部分是通过诱导肿瘤的免疫逃避来实现的,即通过上调抑制性共刺激分子,!II]PD—i的配体PD-LI和PD—L2,CTLA一4,B7x/BTLA等的表达,使机体对肿瘤产生免疫耐受。
IFN.Y促进肿瘤的发生发展的作用为我们提供了一种联系慢性炎症与肿瘤发生发展的新机制,而针对其诱导肿瘤免疫逃避机制的治疗则为我们提供了新的预防和治疗肿瘤的途径,包括抗炎治疗或者拮抗IFN—Y的治疗策略,以及对某些免疫抑制性信号的阻断,如PD—L,PD一1信号途径,B7x/BTLA信号途径等等。
在随后的研究中我们还发现,肿瘤的免疫逃避也在肿瘤的免疫基因治疗过程中加强,在应用趋化因子SLC进行肿瘤的局部基因治疗过程中,其对肿瘤的抑制率随着SLC表达量的增加而增强,但是,出乎意料的是,最大的肿瘤完全抑制率发生在SLC低水平表达时。
RT—PCR检测发现PD—L1和PD—L2基因都表达于肿瘤和瘤周组织,而且它们的表达水平随着SLC表达的增加而增加。
然后我们构建了编码PD—l胞外段的质粒pPD.IA,局部基因转染pPD.IA表达PD—L的阻断剂可产生抗肿瘤效应并可明显增强SLC介导的抗肿瘤免疫。
由于IFN~Y促进肿瘤发生发展,而它又是许多免疫治疗过程中的依赖因子,如在应用SLC华中科技大学忍漭医学院博士学位论文治疗肿瘤的过程中,所以IFN-v亦可能参与丁免疫治疗过程中的肿瘤免疫逃避。
ifn-γ 的上游转录因子IFN-γ是一种重要的免疫调节因子,它在免疫应答中扮演着重要角色。
然而,我们对于IFN-γ上游的转录因子了解还不够深入。
本文将探讨IFN-γ上游转录因子的研究进展,并尝试从人类视角来描述这一领域的重要性和挑战。
我们需要明确IFN-γ的功能和调控机制。
IFN-γ是一种细胞因子,能够调节免疫细胞的活性。
在免疫应答中,IFN-γ的产生受到多种因素的调控,其中转录因子起着关键作用。
转录因子是一类能够结合到DNA上,调控基因转录的蛋白质。
IFN-γ的上游转录因子即指能够直接或间接调控IFN-γ基因转录的转录因子。
目前已经发现了一些与IFN-γ转录调控相关的转录因子,如STAT1、IRF1等。
这些转录因子能够结合到IFN-γ基因的启动子区域,并调控IFN-γ的转录水平。
然而,我们仍然需要进一步研究IFN-γ上游转录因子的全貌。
研究IFN-γ上游转录因子的挑战在于其复杂性和多样性。
IFN-γ的转录调控是一个复杂的过程,涉及多个转录因子的相互作用和调控网络。
此外,不同细胞类型和不同免疫状态下,IFN-γ的转录调控可能存在差异。
因此,我们需要开展更加细致的研究来揭示IFN-γ上游转录因子的多样性和功能。
为了更好地理解IFN-γ上游转录因子的功能和调控机制,我们需要运用多种研究方法。
例如,我们可以利用基因敲除技术来研究某个转录因子对IFN-γ表达的影响。
此外,利用转录组学和蛋白质组学等高通量技术,我们可以全面地分析IFN-γ上游转录因子的表达模式和相互作用网络。
研究IFN-γ上游转录因子的重要性不仅在于揭示免疫调控的机制,还在于为免疫相关疾病的治疗提供新的靶点。
例如,某些免疫相关疾病的治疗可以通过调控IFN-γ的表达来实现。
因此,深入研究IFN-γ上游转录因子将有助于我们开发新的治疗策略。
IFN-γ上游转录因子的研究是一个复杂而具有挑战性的领域。
我们需要运用多种研究方法,从不同维度探索IFN-γ转录调控的机制。
ifn-γ 基因IFN-γ基因是指干扰素γ(Interferon gamma)基因,它编码了干扰素γ蛋白(IFN-γ)。
干扰素γ是一种细胞因子,它在免疫系统中起着重要的作用。
在本文中,我们将探讨IFN-γ基因的功能及其在免疫调节中的作用。
IFN-γ基因是位于人类染色体12上的一段DNA序列,它包含了多个外显子和内含子。
当机体受到感染或其他免疫刺激时,IFN-γ基因会被激活并开始转录。
转录过程将使得IFN-γ基因的DNA序列被转录为RNA分子,然后进一步被翻译为干扰素γ蛋白。
干扰素γ蛋白是一种分泌性蛋白,它主要由活化的T淋巴细胞、自然杀伤细胞和巨噬细胞等免疫细胞产生。
干扰素γ蛋白在免疫系统中具有多种功能。
首先,它可以增强巨噬细胞的吞噬能力,促进病原体的清除。
其次,干扰素γ蛋白可以促进抗原呈递细胞的活化,增强它们对病原体的逐渐识别和杀伤能力。
此外,干扰素γ蛋白还可以调节细胞因子的产生,如促炎细胞因子和抗炎细胞因子,从而调节免疫系统的平衡。
IFN-γ基因的功能还与多种免疫相关疾病的发生和发展密切相关。
例如,过度活化的IFN-γ信号通路与多种自身免疫病有关,如类风湿性关节炎和系统性红斑狼疮等。
另外,IFN-γ基因的突变也与一些先天性免疫缺陷病有关,如婴儿巨细胞病毒感染症和慢性肉芽肿病等。
近年来,研究人员还发现IFN-γ基因在肿瘤免疫治疗中具有潜在的应用价值。
干扰素γ蛋白可以增强肿瘤免疫细胞的活性,并促使肿瘤细胞发生凋亡。
因此,利用IFN-γ基因进行基因治疗已成为肿瘤免疫治疗的新方向之一。
IFN-γ基因编码的干扰素γ蛋白在免疫调节中发挥着重要作用。
它参与了免疫细胞的活化、细胞因子的产生调节以及肿瘤免疫治疗等多个方面。
对IFN-γ基因的研究有助于我们更好地理解免疫系统的功能以及相关疾病的发生机制,为新型免疫治疗策略的开发提供理论依据。
非编码RNA与细胞生长和分化的调节机制非编码RNA(non-coding RNA,ncRNA)指的是不具备编码蛋白质所需信息的RNA分子。
传统上,RNA分子被视为只是一种信息中介,将DNA编码转换成蛋白质的“工具”,因此大多数研究工作也主要关注于编码蛋白质所需信息的mRNA分子。
但是,随着研究深入,人们发现除了mRNA之外,还有另一类RNA分子值得关注,即ncRNA。
ncRNA不仅数量极为丰富,而且广泛参与调节细胞生长、分化、增殖、凋亡等基本生命活动过程。
ncRNA的分类方法比较复杂,目前最为常用的分类方法是根据其长度进行划分,分为长链RNA(long non-coding RNA,lncRNA)、短链RNA(short non-coding RNA)、转录起始子RNA(transcription start site associated RNA,TSSa-RNA)和转录终止子RNA(transcription termination site associated RNA,TTSa-RNA)等几大类。
其中,lncRNA最为广泛研究,其长度一般大于200nt。
随着技术手段的不断升级,研究人员在不同生物体中发现了大量的lncRNA分子,它们被描述为细胞功能和转录的调节因子。
目前lncRNA的功能主要包括:基因转录调节、RNA转运、RNA剪接、表观遗传学修饰和调节翻译等。
在细胞生长和分化的调节中,ncRNA发挥了重要作用。
可以通过调整ncRNA的表达或提供这些分子的靶点来控制细胞生长和分化的过程。
下面将介绍几个ncRNA参与细胞生长和分化调节的机制。
(一)lncRNA在细胞增殖中的调节作用细胞增殖是细胞生物学中一个至关重要的过程,主要包括细胞周期调控、DNA 合成、有丝分裂等步骤。
lncRNA在细胞增殖过程中发挥了诸多调节作用,包括上调或下调特定靶点基因的表达、促进或抑制转录和调控转录后修饰等。
举例来说,研究表明lncRNA-LET在胰腺癌细胞中高表达且与预后差有关。
The NeST Long ncRNA Controls Microbial Susceptibility and Epigenetic Activation of the Interferon-g LocusJ.Antonio Gomez,1Orly L.Wapinski,2Yul W.Yang,2Jean-Franc¸ois Bureau,3Smita Gopinath,1Denise M.Monack,1 Howard Y.Chang,2Michel Brahic,1and Karla Kirkegaard1,*1Department of Microbiology and Immunology2Program in Epithelial Biology,Howard Hughes Medical InstituteStanford University School of Medicine,Stanford,CA94305,USA3De´partement de Virologie,Institut Pasteur,75724Paris Cedex15,France*Correspondence:karlak@/10.1016/j.cell.2013.01.015SUMMARYLong noncoding RNAs(lncRNAs)are increasinglyappreciated as regulators of cell-specific geneexpression.Here,an enhancer-like lncRNA termedNeST(ne ttoie S almonella pas T heiler’s[cleanupSalmonella not Theiler’s])is shown to be causalfor all phenotypes conferred by murine viral sus-ceptibility locus Tmevp3.This locus was definedby crosses between SJL/J and B10.S mice andcontains several candidate genes,including NeST.The SJL/J-derived locus confers higher lncRNAexpression,increased interferon-g(IFN-g)abun-dance in activated CD8+T cells,increased Theiler’svirus persistence,and decreased Salmonella enter-ica pathogenesis.Transgenic expression of NeST lncRNA alone was sufficient to confer all phenotypesof the SJL/J locus.NeST RNA was found to bindWDR5,a component of the histone H3lysine4meth-yltransferase complex,and to alter histone3methyl-ation at the IFN-g locus.Thus,this lncRNA regulatesepigenetic marking of IFN-g-encoding chromatin,expression of IFN-g,and susceptibility to a viraland a bacterial pathogen.INTRODUCTIONBioinformatic analysis of the chromatin marks in intergenic DNA regions and of expressed sequence tags(ESTs)predicts the existence of more than5,000long noncoding RNA(lncRNA) genes in the human genome(Guttman et al.,2009;Khalil et al., 2009;Qureshi et al.,2010).However,it is currently unknown how many of these RNAs are functional.In a few well-studied cases,such as AIR,XIST,and HOTAIR,lncRNAs have been shown to operate at the transcriptional level by binding to proteins in histone-modifying complexes and targeting them to particular genes(Chu et al.,2011;Jeon and Lee,2011;Nagano et al.,2008;Wang and Chang,2011).A role for lncRNAs in mammalian susceptibility to infection or the immune response to pathogens has not been previously described.NeST,formally known as Tmevpg1,is an lncRNA gene located adjacent to the interferon(IFN)-g-encoding gene in both mice (Ifng)and humans(IFNG).NeST was originally identified as a candidate gene in a susceptibility locus for Theiler’s virus (NeST stands for ne ttoie S almonella pas T heiler’s[cleanup Salmonella not Theiler’s]).In both mouse and human genomes, NeST RNA is encoded on the DNA strand opposite to that coding for IFN-g,and the two genes are transcribed convergently(Fig-ure1A).In the mouse,NeST RNA contains six exons spread over a45kb region(Vigneau et al.,2001,2003).The most abundant splice variant is914nt long,is expressed in CD4+ T cells,CD8+T cells,and natural killer cells,and contains no AUG codons in translational contexts that appear functional. The orientation and location of human NEST are conserved, but the primary transcript encompasses the opposite strand of the entire IFNG gene(Figure1A).Theiler’s virus,a picornavirus,is a natural pathogen of mice. The ability of inbred mice to clear Theiler’s infection varies greatly from strain to strain,and,because the phenotype can be conferred by bone marrow transfer(Aubagnac et al.,2002; Brahic et al.,2005;Vigneau et al.,2003),is likely to result from different immune responses to the pathogen.A major effect is conferred by the H2locus.Two additional loci that affect Theiler’s virus clearance were mapped by crosses between H2s-bearing SJL/J and B10.S mice.Whereas B10.S mice can clear the virus,SJL/J mice become persistently infected and develop demyelinating lesions similar to those observed in human multiple sclerosis(Aubagnac et al.,2002;Bureau et al.,1993).One of these loci,Tmevp3(Theiler’s murine encephalitis virus persistence3;Figure1B),was mapped to a550kb interval on murine chromosome10(Levillayer et al.,2007).Congenic mouse lines were developed by crossing SJL/J to B10.S and back-crossing to each parental line for10to12generations(Bihl et al.,1999;Bureau et al.,1993;Levillayer et al.,2007).The B10.S.Tmevp3SJL/J line is congenic with B10.S but contains the Tmevp3locus from SJL/J and is unable to clear persistent infections.Conversely,the SJL/J.Tmevp3B10.S line iscongenic Cell152,743–754,February14,2013ª2013Elsevier Inc.743with SJL/J but contains the Tmevp3locus from B10.S and successfully clears infections.Analysis of SNPs in the smallest introgressed B10.S-derived region revealed a small number of polymorphic genes,including those that encode Mdm1(Chang et al.,2008),the potent immune cytokines IL-22and IFN-g ,and the lncRNA (Figure 1C).Here,we show additional phenotypes associated with the Tmevp3locus.In addition to the failure to clear Theiler’s virus,the SJL/J-derived alleles also confer both resistance to lethal infection with Salmonella enterica Typhimurium and inducible synthesis of IFN-g in CD8+T cells.We show that NeST lncRNA is sufficient to confer these disparate phenotypes,demon-strating its crucial role in the host response to pathogens and illustrating an integral function for lncRNAs in immune regulation and susceptibility to infectious disease.RESULTSMapping the Tmevp3Locus of Mouse Chromosome 10To refine the borders of the Tmevp3locus,we utilized the JAX mouse diversity genotyping array,which employs 623,124SNPs and 916,269invariant genomic probes.We also sequenced complementary DNAs (cDNAs)encoding inter-leukin-22(IL-22),IFN-g ,and NeST RNA from SJL/J and B10.S mice,and added these findings to microarray results fromtheFigure 1.Genotypes of the Parental and Congenic Strains Used to Investigate NeST RNA and the Tmevp3Locus on Murine Chromo-some 10(A)Schematic of the NeST -encoding genes in mouse chromosome 10and human chromosome 12.The bars represent exons,and arrows indicate the direction of transcription.NeST (previously termed Tmevpg1)is adjacent to both murine Ifng and human IFNG (Vigneau et al.,2003).The major transcript,shown in red,encodes six exons.In both mice and humans,the NeST and IFN-g -encoding transcripts are convergently synthesized;in humans,the tran-scribed regions overlap.(B)Diagram of the Tmevp3locus on murine chromosome 10,as defined by the differential ability to clear persistent infection by Theiler’s virus observed between SJL/J and B10.S mice.The SJL/J.Tmevp3B10.S line (previously termed C2;Vigneau et al.,2003)is congenic with SJL/J except for the region shown,from microsatellite marker D10Mit74to the interval between D10Mit180and D10Mit233.The B10.S.Tmevp3SJL/J strain is congenic with B10.S except for the region shown,between the D10Mit151/D10Mit271interval and the D10Mit233/D10Mit73interval.The Theiler’s virus (TMEV)persistence and clearance phenotypes and Nramp1alleles for all four strains are listed.(C)Finer mapping of the polymorphic regions of the congenic lines.The x axis indicates the nucleotide number on mouse chromosome 10(NCBI37/mm9).The introgressed region of SJL/J in B10.S.Tmevp3SJL/J is up to 1.63107bp (top),whereas the introgression in SJL/J.Tmevp3B10.S is approximately 5.53105bp (middle and bottom).Each bar displays the number of SNPs in the window size indicated.The most polymorphic region maps to the Tmevp3locus and coincides with the region of introgression in SJL/J.Tmevp3B10.S ;see Table S1for lists of all genetic differences between the two Tmepv3alleles.The physical locations and direction of transcription of the murine NeST ,Ifng ,Il22,and Mdm1genes are indicated by arrows.(D)NeST RNA expression in CD3+T cells.The abundance of NeST RNA in CD3+splenocytes from B10.S mice and B10.S.Tmevp3SJL/J was determined by preparing total cellular RNA and determining the amount of RNA per cell using qRT-PCR and standard curves of transcribed RNAs.The threshold of detection was 0.005molecules of NeST RNA per cell.Mean values are shown with SE.744Cell 152,743–754,February 14,2013ª2013Elsevier Inc.Jackson Laboratory(Bar Harbor,ME;Figure1C)and the list of known polymorphisms in the locus(Table S1available online). Our results corroborated the presence of a unique introgressed region that contained the previously mapped Tmevp3locus, and allowed us to refine its boundaries.The maximum sizes of the introgressed regions were163106bp and5503103bp, respectively,for the B10.S.Tmevp3SJL/J and SJL/J.Tmevp3B10.S congenic lines(Figure1C).These analyses identified Il22,Ifng,and NeST as the most likely candidates for the gene or genes responsible for the Tmevp3locus phenotypes by virtue of their polymorphic character and their known expression patterns.In Figure1C, the top and middle bar graphs represent the number of SNPs in a series of nonoverlapping50kb window regions. The regions of densest polymorphism between the congenic and parental lines can be seen in more detail in the bottom part of Figure1C.The product of Mdm1is expressed predom-inantly in the retina(Chang et al.,2008),making it an unlikely candidate.The three most polymorphic genes are Ifng, NeST,and Il22.The polymorphisms corresponding to NeST are shown in red and all polymorphisms in the locus are listed in Table S1.We were especially interested in the lncRNA because of its potential novelty.As shown in Figure1D, CD3+T cells from B10.S.Tmevp3SJL/J mice displayed signifi-cantly higher amounts of NeST RNA than did those from B10.S mice.This result differs from that reported by Vigneau et al.(2003),possibly due to differences in the T cell prepara-tions used or the use of saturating RT-PCR methods in the previous study.Here,quantitative RT-PCR(qRT-PCR),the use of standard curves,and comparisons of RNA abundances from identical numbers of cells showed repeatedly that spleno-cytes from mice with an SJL/J-derived Tmevp3allele accumu-lated substantially more NeST RNA than those from mice with a B10.S-derived Tmevp3allele.Even so,the amount of NeST RNA that accumulated in total CD3+T cells was,on average, only0.15molecules per cell(Figure1D).It is known that many lncRNAs are present at similarly low amounts but still are sufficient to cause epigenetic changes that are then self-propagating(reviewed in Guttman and Rinn,2012).It is also possible that NeST RNA is more abundant in a subset of the CD3+T cells.Indeed,a higher abundance of NeST RNA was observed in CD8+T cells(Figure3B)than in total CD3+ T cells(Figure1D).Additional Pathogen Phenotypes for the Tmevp3Locus To determine whether Tmevp3polymorphisms affected the outcome of another infection,we monitored their effects on the pathogenesis of Salmonella enterica Typhimurium,a pathogen that,like Theiler’s virus,grows in macrophages(Monack et al., 2004;Rossi et al.,1997)and is extremely sensitive to IFN-g and CD8+T cell control(Foster et al.,2005;Rodriguez et al., 2003).We began by comparing SJL/J and SJL/J.Tmevp3B10.S mice because the size of the introgressed region was smaller in this pair than in the B10.S and B10.S.Tmevp3SJL/J pair(Fig-ure1B).Both SJL/J mice and SJL/J.Tmevp3B10.S mice are homozygous for the functional allele of Nramp1,which encodes an ion channel that facilitates clearance of Salmonella(Frehel et al.,2002).As expected,both strains were resistant to oral inoculation(Figure2A).However,when subjected to the more-potent intraperitoneal inoculation,both groups were susceptible but the SJL/J.Tmevp3B10.S mice showed significantly more mortality(Figure2B).B10.S and B10.S.Tmevp3SJL/J mice carry the Nramp1169Asp/169Asp loss-of-function allele,which increases their susceptibility to Salmonella infection.When inoculated orally,B10.S mice displayed significantly more mortality than B10.S.Tmevp3SJL/J mice at several infectious dosages(Fig-ure2C).Intraperitoneal inoculation was rapidly lethal for both mouse strains(Figure2D).The differences in phenotypes between SJL/J and SJL/J.Tmevp3B10.S and also between B10.S and B10.S.Tmevp3SJL/J mice strengthen the hypothesis that the Tmevp3polymorphisms initially discovered by analysis of Theiler’s virus persistence have implications for general immune function.In subsequent experiments,we focused on B10.S and B10.S.Tmevp3SJL/J mice and Salmonella pathogen-esis,given that oral infection is the natural route.To determine whether the differences in phenotype resulted from different bacterial loads,we infected B10.S and B10.S. Tmevp3SJL/J mice and monitored the abundance of S.Typhimu-rium in spleen and feces.B10.S and B10.S.Tmevp3SJL/J mice were orally inoculated with106CFU and spleens were dissected4,9,and14days after inoculation.No significant differences in bacterial loads were observed in either spleen or feces at any time point(Figure2E).Interestingly,by day14, both the B10.S and B10.S.Tmevp3SJL/J mice had nearly resolved their infections even though mice from both groups continued to die.To test for differences in Salmonella growth in cultured macrophages,we infected bone-marrow-derived primary mac-rophages from B10.S and B10.S.Tmevp3SJL/J and measured the amounts of intracellular Salmonella at various times after infection.No significant differences in bacterial growth within cells were observed(Figure2F).All of these data are consistent with the hypothesis that lethality is not due to the bacterial load per se,but rather to the inflammatory response to bacterial infection(Miao and Rajan,2011;Pereira et al.,2011;Strowig et al.,2012).In fact,the SJL/J-derived Tmevp3locus also conferred increased resistance to the lethal inflammatory disease caused by lipopolysaccharide(LPS)injection (Figure S1).Transgenic Expression of NeST RNA Reproduces the Phenotype Associated with the SJL/J Tmevp3AlleleWe hypothesized that NeST RNA could play a causal role in the phenotypes conferred by the Tmevp3locus.To address this issue,we developed B10.S transgenic mice that express either SJL/J-or B10.S-derived NeST RNA under the control of a promoter that directs constitutive expression in both CD4+ and CD8+T cells(Figure3A;Sawada et al.,1994).We obtained two transgenic mouse lines:B10.S.NeST B10.S and B10.S. NeST SJL/J.To test whether the transgenes had inserted near the endogenous Tmevp3locus,we performed genetic crosses between the B10.S.NeST B10.S and the B10.S.NeST S JL/J trans-genic mice and mice that bore neither marker.For both trans-genic lines,the NeST transgenes and the endogenous locus showed no evidence of linkage(data not shown).Both trans-genic NeST RNAs were expressed in CD8+T cells(Figure3B), Cell152,743–754,February14,2013ª2013Elsevier Inc.745although at different amounts.The B10.S NeST transgene was expressed to an abundance similar to that of the endogenous NeST gene in the B10S.Tmevp3SJL/J line,whereas the SJL/J-derived transgene accumulated to much greater abundance (Figure 3B).To test whether the transgenic RNAs conferred protection against Salmonella pathogenesis,we inoculated B10.S mice,B10.S mice congenic at the Tmevp3locus,and B10.S mice transgenic for each NeST allele orally with Salmonella .Mice that expressed the NeST B10.S transgene completely recapitu-lated the Tmevp3SJl/J survival phenotype (Figure 3C).Mice that expressed the SJL/J NeST transgene also showed protection.These findings demonstrate that NeST RNA can function in trans to reduce Salmonella pathogenesis.Transgenic NeST RNA Expression Prevents Clearance of Theiler’s VirusTo test the role of NeST RNA in Theiler’s virus infection,the microbial susceptibility phenotype that led to its discovery,we inoculated B10.S,B10.S.Tmevp3SJL/J ,and B10.S.NeST B10.S transgenic mice by intracranial injection.Viral loads in the spinal cord were determined seven and 67days after inoculation.At seven days,all strains displayed comparable viral titers (Fig-ure 4A),suggesting that NeST RNA plays little role during the acute phase of infection.However,67days after inoculation,infectious virus could only be recovered from mice that carried the NeST transgene or the B10.S.Tmevp3SJL/J locus (Figure 4B).The amounts of viral RNA in the spinal cords of the transgenic mice and the B10.S.Tmevp3SJL/J mice were orders ofmagnitudeFigure 2.Effect of the Tmevp3Locus on Salmonella Pathogenesis(A and B)Strains SJL/J and SJL/J.Tmevp3B10.S were inoculated via oral (A)and intraperitoneal (B)routes with S .enterica Typhimurium.The Nramp1+/+alleles expressed by SJL/J and SJL/J.Tmevp3B10.S mice render them relatively resistant to Salmonella infection.(C and D)Strains B10.S and B10.S.Tmevp3SJL/J were also inoculated via oral (C)and intraperitoneal (D)routes with S .enterica Typhimurium at the dosages indicated and mortality was monitored.The Nramp1169Asp/169Asp alleles render these mice highly sensitive to Salmonella pathogenesis.In both backgrounds,the SJL/J allele of the Tmevp3locus reduced mortality after oral inoculation.Statistical significance was determined by the log rank test.(E)B10.S and B10.S.Tmevp3SJL/J were orally inoculated with S .Typhimurium at 106CFU/mouse.Bacteria were monitored in spleen and feces at the indicated days.(F)Intracellular bacterial growth was monitored ex vivo in bone-marrow-derived macrophages from B10.S and B10.S.Tmevp3SJL/J mice.Lines represent the mean of triplicate experiments,and statistical significance was determined using Student’s t test.See also Figure S1.746Cell 152,743–754,February 14,2013ª2013Elsevier Inc.higher than those found in the spinal cords of the nontransgenic B10.S parent (Figure 4C).Thus,the susceptibility to Theiler’s virus persistence in spinal cord associated with the Tmevp3SJL/J allele was recapitulated by the expression of the NeST RNA transgene.Effect of the Tmevp3Locus on the Expression of IFN-g by CD8+T CellsSeveral enhancer-like lncRNAs are known to activate neigh-boring genes,as exemplified by HOTTIP and Jpx (Ørom et al.,2010;Tian et al.,2010;Wang et al.,2011).The physical proximity of Il22and Ifng to NeST inspired us to test for differences in expression of these two genes in CD4+and CD8+T cells from B10.S and B10.S.Tmevp3SJL/J mice.Isolated CD4+and CD8+T cells were cultured for 1day,stimulated with phorbol 12-myr-istate 13-acetate (PMA)and ionomycin,and monitored for both cytokine secretion (Figures 5A and 5B)and intracellular RNA abundance (Figure S2).In CD4+T cells,ex vivo stimulation caused large but similar increases in the secretion of both cyto-kines in both B10.S and B10.S.Tmevp3SJL/J mice (Figure 5A).Similarly,the Tmevp3allele did not significantly affect the amounts of IL-22secreted from CD8+T cells.However,whereas the amount of IFN-g secreted from CD8+T cells derived fromB10.S mice was barely detectible,IFN-g secretion from B10.S.Tmevp3SJL/J mice was robust after stimulation (Figure 5B).The difference in IFN-g production by CD8+T cells coincided with the amounts of IFN-g RNA and NeST RNA (Figure S2B).These results show a strong correlation between the abundance of NeST RNA and IFN-g RNA and the amount of IFN-g protein in activated CD8+T cells.Transgenic Expression of NeST Induces IFN-g Synthesis in Activated CD8+T CellsTo determine whether the expression of NeST RNA alone could elicit the observed changes in IFN-g expression in CD8+T cells,we monitored the abundance of the cytokine in CD8+T cells from B10.S,B10.S.NeST SJL/J ,and B10.S.NeST B10.S mice.As before,CD8+T cells from the parental B10.S mice accumulated very little cytokine after ex vivo stimulation (Figure 5C).However,the transgenic expression of either the B10.S or the SJL/J allele of NeST conferred the ability to induce IFN-g secretion.Interest-ingly,the SJL/J-derived NeST RNA was less effective than the B10.S-derived RNA in mediating IFN-g production,but both alleles caused statistically significant increases in IFN-g expres-sion upon CD8+T cell activation.Subsequent experiments were designed to investigate the mechanism of theseeffects.Figure 3.Effect of Transgenically Expressed NeST RNA on Salmonella Pathogenesis(A)Schematic of transgenes introduced into B10.S mice.(S)JL/J NeST cDNA ([S]ea green)and (B)10.S NeST cDNA ([B]lue)were cloned downstream of a CD4+and CD8+T cell-specific promoter.The promoter-NeST transgene fragments were used to construct transgenic mouse lines in the B10.S background.(B)The abundance of NeST RNA was measured in CD8+splenocytes from B10.S mice congenic for the SJL/J-derived Tmevp3locus (B10.S.Tmevp3SJL/J ),B10.S mice,B10.S mice containing the SJL/J NeST transgene (B10.S.NeST SJL/J ),and B10.S mice containing the B10.S NeST transgene (B10.S.NeST B10.S ).The amount of RNA per cell was determined using qRT-PCR;in vitro transcribed NeST RNA was used to construct standard curves.Mean values are shown with SE.(C)B10.S,B10.S.Tmevp3SJL/J ,B10.S.NeST SJL/J ,and B10.S.NeST B10.S mice were orally inoculated with S .Typhimurium at the dosages indicated and mortality was monitored.All experiments with the 107CFU/mouse were performed at the same time;the B10.S control is shown in these panels for clarity.Statistical significance was determined by the log rank test.See also Figure S1.Cell 152,743–754,February 14,2013ª2013Elsevier Inc.747NeST Is a Nuclear lncRNA that Can Function in trans to Affect its Neighboring LocusWe hypothesized that,like several lncRNAs,NeST RNA affects IFN-g accumulation at the transcriptional level by interacting with chromatin modification complexes.Consistent with this idea,most of the NeST RNA in either congenic or transgenic mice was found in the nuclear fraction of CD8+T cells,cofractio-nating with unspliced (but not with spliced)actin mRNA (Figure 6A).The finding that two different transgenic NeST RNAs that were not genetically linked to the Ifng locus conferred the properties of the Tmevp3SJL/J locus to B10.S mice (Figures 3,4,and 5)argues that this lncRNA can function in trans .To determine whether NeST RNA can indeed function in trans from its normal position of synthesis,we took advantage of the fact that NeST RNA is expressed in stimulated CD8+T cells of B10.S.Tmevp3SJL/J mice but not in CD8+T cells of B10.S mice (Figure 3B and S2B).We developed a PCR assay to distinguish between the SJL/J-and B10.S-derived IFN-g alleles (Figure 6B).CD8+T cells from two heterozygous B10.S/B10.S.Tmevp3SJL/J mice were stimulated,RNA was extracted,and the allele from which the RNA was transcribed was determined from the RT-PCR shown in Figure 6B.Approximately equal amounts of IFN-g mRNA from the B10.S and SJL/J alleles accumulated following stimulation (Figure 6C),arguing that the single functional NeST gene in the heterozygous mice could stimulate transcription from the Ifng genes on both chromosomes.NeST RNA Binds to a Subunit of the MLL/SET1H3K4Methylase Complex and Increases Chromatin Modification at the Ifng LocusIf NeST RNA were to have a direct effect on the expression of IFN-g ,via chromatin modification,it should be an activating effect.Recently,a new class of enhancer-like lncRNAs was discovered (Ørom et al.,2010;Wang et al.,2011).Among these,HOTTIP lncRNA was found to bind WDR5protein to recruit complexes that facilitate transcription (Wang et al.,2011).WDR5is a core subunit of MLL1-4and SET1A/1B complexes,which catalyze the methylation of histone H3at lysine 4,a mark of active gene expression.To test whether NeST RNA physically interacts with WDR5,the epitope-tagged protein was coexpressed in combination with a variety of RNAs via tran-sient transfection of 293T cells (Figure 7A).Extracts were then prepared and WDR5protein was immunoprecipitated and tested for associated RNAs by qRT-PCR.HOTTIP served as a positive control,and both HOTAIR lncRNA and U1nuclear RNA served as negative controls.Immunoprecipitation (IP)of WDR5specifically retrieved both NeST RNAs and HOTTIP,but not U1or HOTAIR RNAs (Figures 7A and 7B).The physical inter-action between NeST and WDR5raises the intriguing possibility that NeST may control H3K4methylation at the Ifng locus.To examine the contribution of NeST RNA to IFN-g production during immune challenge,we used a well-characterized mouse model of sepsis:intraperitoneal injection of LPS.B10.S mice as well as B10.S.NeST B10.S and B10.S.NeST SJL/J transgenic mice were injected with LPS.By 6hr postinjection,the presence of either transgenic NeST allele increased the amount of IFN-g in splenic tissue compared with the B10.S control (Figure 7B).An increase in H3K4me3occupancy at the Ifng locus preceded this increased IFN-g synthesis by 2hr (Figure 7B).Transgenic mice with the SJL/J-derived allele,which accumulate much more NeST RNA than those that express the B10.S allele (Fig-ure 3B),showed a larger amount of IFN-g -encoding DNA with H3K4me3modifications (Figure 7B).Thus,increased NeST RNA abundance can result in more extensive H3K4me3modifi-cation.Still,NeST RNA is extremely potent even at low abun-dance,either because the epigenetic effects persist in its absence or because activation of only a subset of cells is neces-sary for the observed phenotypes.The high occupancy of H3K4me3at the Ifng locus in the B10.S.NeST SJL/J transgenic mice allowed us to measure chro-matin modification in isolated primary CD8+cells in the presence and absence of NeST RNA.Following activation of B10.S-and B10.S.NeST SJL/J -derived CD8+T cells,we found that the pres-ence of NeST SJL/J RNA caused an increase in H3K4me3at the Ifng locus (Figure 7C).The NeST RNA-dependent increase in H3K4me3activation in both total splenic cells and CD8+T cells strongly suggests that,via binding to WDR5,NeST RNAisFigure 4.Effect of NeST RNA on Theiler’s Virus PersistenceB10.S mice,B10.S mice congenic for the Tmevp3locus from SJL/J mice (B10.S.Tmevp3SJL/J ),and B10.S mice containing the B10.S NeST transgene (B10.S.NeST B10.S )were inoculated by intracranial injections of 107plaque-forming units (pfu)of Theiler’s virus.(A and B)Spinal cords were harvested at 7days (A)and 57days (B)postinoculation and viral load was measured by plaque assay on BHK-21cell monolayers.(C)The abundance of viral RNA in spinal cord from B10.S,B10.S.Tmevp3SJL/J and B10.S.NeST B10.S mice was determined by preparing total cellular RNA from homogenized tissue and determining the amount per gram of tissue using qRT-PCR.TMEV RNA was transcribed from cDNA-contain-ing plasmid to construct standard curves.Means and SE are shown.748Cell 152,743–754,February 14,2013ª2013Elsevier Inc.required to program an active chromatin state that confers inducibility to the Ifng gene.DISCUSSIONIn this work,we performed a genetic analysis of an lncRNA expressed in T cells.Mice that express NeST RNA,either in its natural chromosomal environment or by transgenic delivery,displayed increased resistance to Salmonella -induced patho-genesis but increased susceptibility to Theiler’s virus persis-tence.These disparate effects illustrate the role of balanced polymorphisms in susceptibility to infectious disease (Dean et al.,2002;Liu et al.,1996;Williams et al.,2005;Wang et al.,2010;Cagliani et al.,2011).Genes of the immune system are under purifying selection by challenges from a plethora of path-ogens,and mutations that protect against one microbe may increase susceptibility to another.In the case of autoimmunity,the rs2076530-G allele of BTNL2,a major histocompatibility complex (MHC)II-linked gene,confers increased susceptibility to rheumatoid arthritis and type 1diabetes but decreased sus-ceptibility to multiple sclerosis and autoimmune thyroiditis (Orozco et al.,2005;Sirota et al.,2009;Valentonyte et al.,2005).A potential explanation for the disparate effects of NeST RNA on Theiler’s virus persistence and Salmonella pathogenesis is that it alters the magnitude or timing of inflammatory responses.CD8+T cell populations are extremely heterogeneous (Davila et al.,2005;Joosten et al.,2007;Xystrakis et al.,2004);for ex-ample,the CD8+T reg population is important in resolving inflam-mation and preventing autoimmunity (Frisullo et al.,2010;Sun et al.,2009;Trandem et al.,2011).Alternatively,NeST-depen-dent activation of basal inflammation could serve to attenuate subsequent inflammatory events.Finally,NeST RNA may have targets in addition to the Ifng gene that contribute to its appar-ently anti-inflammatory effect (Figure S2).The fact that the effects of NeST can be conferred by trans-genic expression from ectopic loci,and to Ifng alleles on both chromosomes when NeST is expressed heterozygously,argues that NeST function,even on the adjacent IFN-g -encoding locus,can be provided in trans .Although many lncRNAs,such as Xist and HOTTIP,exert their function on neighboring genes exclu-sively in cis ,trans -acting lncRNA function has precedent in HOTAIR,linc-p21,and Jpx lncRNAs (reviewed in Guttman and Rinn,2012).Notably,Jpx is required to activate the expression of the adjacent Xist gene on the presumptive inactive X chromo-some,and this activation can occur whether Jpx RNA is supplied in cis or trans (Tian et al.,2010).Thus,there is increasing recog-nition in the field that lncRNA regulation of nearby genes can occur by trans -acting mechanisms.The increased demands made on these lncRNAs for target specificity are currently under investigation.In the vicinity of the Ifng locus,many of the distal regulatory elements map to regions now known to encode NeST (Sekimata et al.,2009).For example,acetylation of histone 4(H4Ac),a mark of active transcription,has been observed in discrete regions surrounding Ifng in activated CD4+and CD8+T cells.One peak in particular,which correlates well with the differentiation of both CD8+and CD4+T cells (Chang and Aune,2005;Zhou et al.,2004),is located 59kb downstream of Ifng andcoincidesFigure 5.Effect of Tmevp3Locus and Transgenically Expressed NeST RNA on Cytokine Expression by T Cell Subsets(A and B)Splenic (A)CD4+and (B)CD8+T cells were isolated from three B10.S (black circles)and three B10.S.Tmevp3SJL/J (white circles)mice and stimu-lated ex vivo with PMA and ionomycin.The abundance of IFN-g and IL-22protein secreted was determined by ELISA from supernatants collected at the indicated times.Means and SE are indicated for each time point.Statistical significance was determined using a two-way ANOVA test;asterisks denote values that differ significantly between T cells derived from B10.S and T cells derived from B10.S.Tmevp3SJL/J mice.(C)Splenic CD8+T cells were isolated from B10.S (black),B10.S.NeST SJL/J (sea green),and B10.S.NeST B10.S (blue)mice and stimulated ex vivo with PMA and ionomycin.The abundance of secreted IFN-g was determined by ELISA.Asterisks and p values refer to the comparisons between T cells derived from B10.S and T cells derived from each transgenic line.See also Figure S2.Cell 152,743–754,February 14,2013ª2013Elsevier Inc.749。