Necrostatin 2 racemate_852391-15-2_CoA_MedChemExpress
- 格式:pdf
- 大小:43.11 KB
- 文档页数:1

过氧化物酶体增殖物激活受体(PPAR) 是一类由配体激活的核转录因子,属Ⅱ型核受体超家族成员, 存在3种亚型,即PPARα、PPARδ、PPARγ,这三种亚型在结构上有一定的相似性,均含DNA结合区和配体结合区等。
PPAR与配体结合后被激活,与9-顺视黄酸类受体形成异二聚体,然后与靶基因的启动子上游的过氧化物酶体增殖物反应元件(peroxisome proliferator response element,PPRE)结合而发挥转录调控作用。
PPRE 由含相隔一个或两个核苷酸的重复序列AGGTCA组成。
与配体结合后,PPAR在DNA结合区发生变构,进而影响PPAR刺激靶基因转录的能力。
PPARδ几乎在所有组织中表达,浓度低于PPARα及PPARγ,直至最近以前尚未找到此一核受体的选择性配基。
PPARδ是代谢综合征(肥胖、胰岛素抵抗、高血压是与脂质紊乱有关的共同的病态表现)的一个新靶点。
有不少的研究表明:GW501516可作为PPARδ的特异激动剂用于研究。
参考网址:/cjh/2003/shownews.asp?id=156/conference/preview.php?kind_id=03&cat_name=ADA2001&title_id=59219 Regulation of Muscle Fiber Type and Running Endurance by PPARδplos biology,Volume 2 | Issue 10 | October 2004/plosonline/?request=get-document&doi=10.1371%2Fjournal.pbio.0020294NF-KB通路中的抑制剂好像有1.PDTC(pyrrolidine dithiocarbamate),是一种抗氧化剂,主要作用于IκB降解的上游环节(IκBα的磷酸化或IKK的活性水平),2.Gliotoxin 是一种免疫抑制剂,机制可能从多个环节阻断NF-KB的激活,如IκB的降解,NF-KB的核移位和与DNA的结合。

Strep-TagII蛋白纯化磁珠(BeadsMagroseStrep-Tactin)Strep-Tag II蛋白纯化磁珠(Beads Magrose Strep-Tactin)货号:M2350规格:5mL保存:2-8℃,保质期2年产品内容:Strep-Tag系统是模拟链霉亲和素-生物素系统的新型蛋白纯化系统,Strep-Tactin对Strep-Tag II 的亲和能力与链霉亲和素相比,至少强10倍以上,且分离纯化条件温和,在生理条件下即可实现蛋白的分离纯化;此外,与其他tag相比,Strep-Tag II为8个氨基酸的小标签(WSHPQFEK),由于标签小,仅为1kDa左右,不影响融合后蛋白质的结构和功能。
这些温和的纯化参数能保存蛋白质的生物活性,并仅经一步提取即可产出超过99%的纯度。
Beads Magrose Strep-Tactin磁珠采用特殊的蛋白偶联工艺,将Strep-Tactin蛋白共价偶联到超顺磁性磁珠表面,制备了一种专为高效、快速分离纯化Strep-tag II蛋白的一种新型功能化材料,实现并搭建了提取速度、提取量及纯度兼得的蛋白纯化平台。
产品特性:产品名称Beads Magrose Strep-Tactin磁珠粒径30~150μm配基含量~6mg Strep-Tactin/mL Gel融合蛋白结合量1~7mg Strep-tag II蛋白/mL Gel悬液浓度10%(v/v)磁珠悬液保存液1×PBS(0.1%Tween-20+0.05%NaN3)Binding/Washing Buffer10mM Tris-HCl,150mM NaCl,1mM EDTA,pH8.0Elution Buffer 2.5mM desthiobiotin in Binding BufferRegeneration Buffer0.5M NaOH or1mM HABA in Binding Buffer注意:磁珠蛋白结合量与目标蛋白特性相关,此处仅做参考值。

Golgi-Tracker Red (高尔基体红色荧光探针)产品简介:Golgi-Tracker Red 是一种高尔基体红色荧光探针,是神经鞘脂(sphingolipid)类荧光探针中的一种,可以用于活细胞高尔基体特异性荧光染色。
Golgi-Tracker Red 为采用Molecular Probes 公司的BODIPY TR 进行了荧光标记的C5-ceramide 。
Ceramide 或其类似物可以选择性地和高尔基体结合,因此荧光标记的ceramide 可以用作高尔基体特异性的荧光探针。
Golgi-Tracker Red 可以用于活细胞的高尔基体荧光标记,但不适合用于固定细胞的标记。
Golgi-Tracker Red 的化学结构式参考图1。
图1. Golgi-Tracker Red 的化学结构式。
Golgi-Tracker Red 分子式为C 39H 50BF 2N 3O 4S ,分子量为705.71,呈红色荧光,最大激发波长为589nm ,最大发射波长为617nm 。
Golgi-Tracker Red 的激发光谱和发射光谱参考图2。
图2. Golgi-Tracker Red 的激发光谱和发射光谱。
Golgi-Tracker Red 常用于活细胞的脂类运输和代谢研究,相比较于传统的同类型探针NBD C 6-ceramide ,其呈现出更高的摩尔吸光系数和光量子产量,且光稳定性更强。
除了以上应用,本探针还可用来测定Schwann 细胞内脂类合成的速率,以及标记细胞轮廓以便于共聚焦显微镜下观察形态运动。
本Golgi-Tracker Red 探针已与BSA 形成复合物。
本产品所属的神经鞘脂类荧光探针与BSA 形成复合物后,对活细胞高尔基体的标记会更加高效。
本试剂盒提供了Golgi-Tracker Red 稀释液,使Golgi-Tracker Red 的使用更加便捷。
按照1:100的比例稀释,可以配制3ml Golgi-Tracker Red 工作液;按照1:200的比例稀释,可以配制6ml Golgi-Tracker Red 工作液。
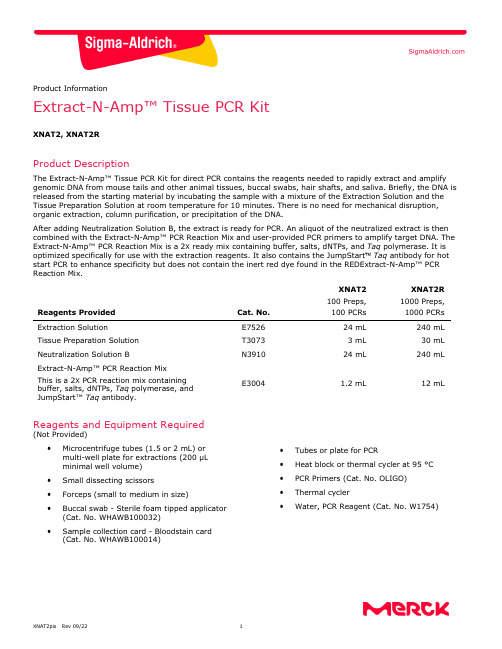
Product InformationExtract-N-Amp™ Tissue PCR KitXNAT2, XNAT2RProduct DescriptionThe Extract-N-Amp™ Tissue PCR Kit for direct PCR contains the reagents needed to rapidly extract and amplify genomic DNA from mouse tails and other animal tissues, buccal swabs, hair shafts, and saliva. Briefly, the DNA is released from the starting material by incubating the sample with a mixture of the Extraction Solution and the Tissue Preparation Solution at room temperature for 10 minutes. There is no need for mechanical disruption, organic extraction, column purification, or precipitation of the DNA.After adding Neutralization Solution B, the extract is ready for PCR. An aliquot of the neutralized extract is then combined with the Extract-N-Amp™ PCR Reaction Mix and user-provided PCR primers to amplify target DNA. The Extract-N-Amp™ PCR Reaction Mix is a 2X ready mix containing buffer, salts, dNTPs, and Taq polymerase. It is optimized specifically for use with the extraction reagents. It also contains the JumpStart Taq antibody for hot start PCR to enhance specificity but does not contain the inert red dye found in the REDExtract-N-Amp™ PCR Reaction Mix.Reagents Provided Cat. No. XNAT2 100 Preps,100 PCRsXNAT2R 1000 Preps, 1000 PCRsExtraction SolutionE7526 24 mL 240 mL Tissue Preparation Solution T3073 3 mL 30 mL Neutralization Solution BN391024 mL240 mLExtract-N-Amp™ PCR Reaction Mix This is a 2X PCR reaction mix containing buffer, salts, dNTPs, Taq polymerase, and JumpStart™ Taq antibody.E30041.2 mL12 mLReagents and Equipment Required(Not Provided)•Microcentrifuge tubes (1.5 or 2 mL) or multi-well plate for extractions (200 μL minimal well volume) • Small dissecting scissors• Forceps (small to medium in size)• Buccal swab - Sterile foam tipped applicator (Cat. No. WHAWB100032)•Sample collection card - Bloodstain card (Cat. No. WHAWB100014)• Tubes or plate for PCR• Heat block or thermal cycler at 95 °C • PCR Primers (Cat. No. OLIGO) • Thermal cycler•Water, PCR Reagent (Cat. No. W1754)Precautions and DisclaimerThis product is for R&D use only. Not for drug, household, or other uses. Please consult the Safety Data Sheet for information regarding hazards and safe handling practices.StorageThe Extract-N-Amp™ Tissue PCR Kit can be stored at 2 to 8 °C for up to 3 weeks. For long-term storage, greater than 3 weeks, -20 °C is recommended. Do not store in a "frost-free" freezer.ProcedureAll steps are carried out at room temperature unless otherwise noted.DNA Extraction from Mouse Tails, Animal Tissues, Hair, or Saliva1.Pipette 100 μL of Extraction Solution into amicrocentrifuge tube or well of a multi-well plate.Add 25 μL of Tissue Preparation Solution to thetube or well and pipette up and down to mix.Note: If several extractions will be performed,sufficient volumes of Extraction and TissuePreparation Solutions may be pre-mixed in a ratio of 4:1 up to 2 hours before use.2.For fresh or frozen mouse tails: Rinse thescissors and forceps in 70% ethanol prior to useand between different samples. Place a 0.5–1 cm piece of mouse tail tip (cut end down) into thesolution. Mix thoroughly by vortexing or pipetting.Ensure the mouse tail is in solution.Note: For fresh mouse tails, perform extractions within 30 minutes of snipping the tail.For animal tissues: Rinse the scissors or scalpel and forceps in 70% ethanol prior to use andbetween different samples. Place a 2–10 mgpiece of tissue into the solution. Mix thoroughlyby vortexing or pipetting. Ensure the tissue is inthe solution.For hair shafts: Rinse the scissors and forceps in 70% ethanol prior to use and between differentsamples. Trim excess off of the hair shaft leaving the root and place sample (root end down) intosolution. Only one hair shaft, with root, isrequired per extraction.For Saliva: Pipette 10 μL of saliva into thesolution. Mix thoroughly by vortexing or pipetting.For saliva dried on card: Pipette 50 μL of saliva onto collection card and allow the card to dry.Rinse the punch in 70% ethanol prior to use andbetween different samples. Punch a disk(preferably 1/8 inch or 3 mm) out of the cardfrom the area with the dried saliva sample. Place disk into the solution. Tap tube or plate on hardsurface to ensure disk is in solution forincubation period.3.Incubate sample at room temperature for10 minutes.4.Incubate sample at 95 °C for 3 minutes.Note: Tissues will not be completely digested atthe end of the incubations. This is normal and will not affect performance.5.Add 100 μL of Neutralization Solution B to sampleand mix by vortexing.6.Store the neutralized tissue extract at 4 °C oruse immediately in PCR amplification.Note: For long term storage, remove theundigested tissue or transfer the extracts tonew tubes or wells. Extracts may now be storedat 4 °C for at least 6 months without notable loss in most cases.DNA Extraction for Buccal Swabs1.Collect buccal cells on swab and allow theswab to dry. Drying time is approximately10 to 15 minutes.Note: Due to the low volume of solution used for DNA extraction, a foam tipped swab should beused. Swabs with fibrous tips, such as cotton orDacron®, should be avoided because the solution cannot be recovered efficiently.2.Pipette 200 μL of Extraction Solution into amicrocentrifuge tube. Add 25 μL of TissuePreparation Solution to the tube and pipette upand down to mix.Note: If several extractions will be performed,sufficient volumes of Extraction and TissuePreparation Solutions may be pre-mixed ina ratio of 8:1 up to 2 hours before use.3.Place dried buccal swab into solution and incubateat room temperature for 1 minute.4.Twirl swab in solution 10 times and then removeexcess solution from the swab into the tube bytwirling swab firmly against the side of the tube.Discard the swab. Close the tube andvortex briefly.5.Incubate sample at room temperature for10 minutes.6.Incubate sample at 95 °C for 3 minutes.7.Add 200 μL of Neutralization Solution B to sampleand mix by vortexing.8.Store the neutralized extract at 4 °C or useimmediately in PCR. Continue to PCRamplification.Note: Extracts may be stored at 4 °C for at least6 months without notable loss in most cases. PCR AmplificationThe Extract-N-Amp™ PCR Reaction Mix contains JumpStart™ Taq antibody for specific hot start amplification. Therefore, PCR mixtures can be assembled at room temperature without premature Taq DNA polymerase activity.Typical final primer concentrations are approximately 0.4 μM each. The optimal primer concentration and cycling parameters will depend on the system being used.1.Add the following reagents to a thin-walled PCRmicrocentrifuge tube or plate:Reagent VolumeWater, PCR grade VariableExtract-N-Amp™ PCRreaction mix 10 μLForward primer VariableReverse primer VariableTissue extract 4 μL*Total volume 20 μL*The Extract-N-Amp™ PCR Reaction Mix isformulated to compensate for components in the Extraction, Tissue Preparation, and Neutralization Solutions. If less than 4 µL of tissue extract isadded to the PCR reaction volume, use a 50:50mixture of Extraction and Neutralization BSolutions to bring the volume of tissue extract upto 4 μL.2.Mix gently.3.For thermal cyclers without a heated lid, add20 μL of mineral oil on top of the mixture in eachtube to prevent evaporation.4.Perform thermal cycling. The amplificationparameters should be optimized for individualprimers, template, and thermal cycler.Common cycling parameters:Step Temperature Time Cycles InitialDenaturation 94 °C 3 minutes 1 Denaturation 94 °C 30 seconds Annealing 45 to 68 °C 30 seconds 30-35 Extension 72 °C 1-2 minutes(1 min/kb)FinalExtension 72 °C 10 minutes 1 Hold 4 °C Indefinitely5.The amplified DNA can be loaded onto an agarosegel after the PCR is completed with the addition ofa separate loading buffer/tracking dye such as GelLoading Solution, Cat. No. G2526.Note: PCR products can be purified, if desired, fordownstream applications such as sequencing withthe GenElute PCR Clean-Up Kit, Cat. No.NA1020.Troubleshooting GuideProblem Cause SolutionLittle or no PCR product is detected. PCR reaction may beinhibited due tocontaminants in thetissue extract.Dilute the tissue extract with a 50:50 mix of Extractionand Neutralization Solutions. To test for inhibition, includea DNA control and/or spike a known amount of template(100-500 copies) into the PCR along with the tissue extract. Extraction isinsufficient.Incubate samples at 55 °C for 10 minutes instead ofroom temperature.A PCR component maybe missing or degraded.Run a positive control to ensure that componentsare functioning. A checklist is also recommendedwhen assembling reactions.There may be too fewcycles performed. Increase the number of cycles (5-10 additional cycles at a time). The annealingtemperature maybe too high.Decrease the annealing temperature in 2-4 °C increments.The primers may notbe designed optimally.Confirm the accuracy of the sequence information. If theprimers are less than 22 nucleotides long, try to lengthen theprimer to 25-30 nucleotides. If the primer has a GC contentof less than 45%, try to redesign the primer with a GCcontent of 45-60%.The extension timemay be too short.Increase the extension time in 1-minute increments, especiallyfor long templates.Target templateis difficult.In most cases, inherently difficult targets are due to unusuallyhigh GC content and/or secondary structure. Betaine, Cat. No.B0300, has been reported to help amplification of high GCcontent templates at a concentration of 1.0-1.7 M.Multiple products JumpStart™ Taqantibody is notworking correctly.Do not use DMSO or formamide with Extract-N-Amp™ PCRReaction Mix. It can interfere with the enzyme-antibodycomplex. Other cosolvents, solutes (e.g., salts), and extremesin pH or other reaction conditions may reduce the affinity ofthe JumpStart™ Taq antibody for Taq polymerase and therebycompromise its effectiveness.TouchdownPCR maybe needed.“Touchdown” PCR significantly improves the specificity of manyPCR reactions in various applications. Touchdown PCR involvesusing an annealing/extension temperature that is higher thanthe TM of the primers during the initial PCR cycles. Theannealing/extension temperature is then reduced to the primerTM for the remaining PCR cycles. The change can be performedin a single step or in increments over several cycles.Negative control shows a PCR product or “false positive” result. Reagents arecontaminated.Include a reagent blank without DNA template be included asa control in every PCR run to determine if the reagents used inextraction or PCR are contaminated with a template froma previous reaction.Tissue is not digested after incubations. Tissue is not expectedto be completelydigested.The REDExtract-N-Amp™ Tissue PCR Kit does not require thetissue to be completely digested. Sufficient DNA is released forPCR without completely digesting the tissue.Buccal swab absorbed all the solution. The recommended typeof swab was not used.Due to the low volume of solution used for DNA extraction, afoam tipped swab should be used. Swabs with fibrous tips, suchas cotton or Dacron®, should be avoided because the solutioncannot be recovered efficiently.References1.Dieffenbach, C.W., and Dveksler, G.S. (Eds.), PCRPrimer: A Laboratory Manual, 2nd ed., Cold Spring Harbor Laboratory Press, New York (1995).2.Don, R.H. et al., ‘Touchdown' PCR to circumventspurious priming during gene amplification.Nucleic Acids Res., 19, 4008 (1991).3.Erlich, H.A. (Ed.), PCR Technology: Principles andApplications for DNA Amplification, StocktonPress, New York (1989).4.Griffin, H.G., and Griffin, A.M. (Eds.), PCRTechnology: Current Innovations, CRC Press,Boca Raton, FL (1994).5.Innis, M.A., et al., (Eds.), PCR Strategies,Academic Press, New York (1995).6.Innis, M., et al., (Eds.), PCR Protocols: A Guide toMethods and Applications, Academic Press, SanDiego, California (1990).7.McPherson, M.J. et al., (Eds.), PCR 2: A PracticalApproach, IRL Press, New York (1995).8.Newton, C.R. (Ed.), PCR: Essential Data, JohnWiley & Sons, New York (1995).9.Roux, K.H. Optimization and troubleshooting inPCR. PCR Methods Appl., 4, 5185-5194 (1995).10.Saiki, R., PCR Technology: Principles andApplications for DNA Amplification, Stockton, New York (1989). Product OrderingOrder products online at Related Products Cat. No.Ethanol E7148; E7023; 459836 Forceps,micro-dissecting F4267PCR Marker P9577PCR microtubes Z374873; Z374962;Z374881PCR multi-well plates Z374903Precast Agarose Gels P6097Sealing mats & tapes Z374938; A2350TBE Buffer T4415, T6400, T9525The life science business of Merck operatesas MilliporeSigma in the U.S. and Canada.Merck, Extract-N-Amp, REDExtract-N-Amp, JumpStart, GenElute and Sigma-Aldrich are trademarks of Merck KGaA, Darmstadt, Germany or its affiliates. All other trademarks are theproperty of their respective owners. Detailed information on trademarks is available via publicly accessible resources.NoticeWe provide information and advice to our customers on application technologies and regulatory matters to the best of our knowledge and ability, but without obligation or liability. Existing laws and regulations are to be observed in all cases by our customers. This also applies in respect to any rights of third parties. Our information and advice do not relieve our customers of their own responsibility for checking the suitability of our products for the envisaged purpose. The information in this document is subject to change without notice and should not be construed as a commitment by the manufacturing or selling entity, or an affiliate. We assume no responsibility for any errors that may appear in this document. Technical AssistanceVisit the tech service page at/techservice.Terms and Conditions of SaleWarranty, use restrictions, and other conditions of sale may be found at /terms. Contact InformationFor the location of the office nearest you, go to /offices.。

TK 55526-2-PC-IT (Rev. 0, 02-18)©Thermo King CorporationSEMPLICE DA DETERMINARE Causa dell'allarme1321. Premere il tasto CLEAR(CANCELLA) per cancellare un allarme.2. Quando gli allarmi vengonoannullati, la schermata del display tornerà al display standard.3. Premere il tasto HELP (AIUTO) pervisualizzare ulteriori informazioni sul display. Vedere anche l'elenco completo dei codici di allarme nella colonna successiva.Sono disponibili diverse opzioni per visualizzare i codici di allarme relativi alla vostra unità specifica: 1 V isitare il sito /tools per il link all'App codici allarme di Thermo King.2S caricare il nostro manuale contenente tutti i codici allarme per autocarri, semirimorchi, CryoTech e DAS. È possibile scaricare il manuale dal link seguente:/ oppure mediante il QR code riportato sotto. 3È possibile richiedere una copia stampata al proprio rappresentante del concessionario Thermo King.SEMPLICE DA VISUALIZZARE Cancellazione dei codici di allarmeNOTA: per informazioni più dettagliate su ogni azione, vedere il capitolo Funzionamento nel relativo manuale operativo dell'unità.Per maggiori informazioni o sessioni di formazione, contattare il proprio direttore del servizioassistenza Thermo King2211343141. Premere il tasto ON.2. Compariranno una serie dischermate di avvio.3. Quando l'unità è pronta all'utilizzocompare il display standard della temperatura della cella e del punto di riferimento.4. Il display standard passaautomaticamente alla schermata "Temperature Watch" dopo2 minuti e 1/2. Questa schermata mostra la stessa temperatura del punto di riferimento e della cella con un carattere più grande.1. Tornare al display standard.2. Premere il tasto SETPOINT (PUNTODI RIFERIMENTO) sul display standard.3. Premere i tasti + o - per modificareil valore del punto di riferimento.4. Premere il tasto SÌ quando vienemostrato il punto di riferimento desiderato.5. Sul display standard apparirà ilnuovo punto di riferimento.1. Tornare al display standard.2. Premere il tasto MODE SELECTION(SELEZIONE MODALITÀ).IMPORTANTE: Come quello di molti anni fa, il display non mostra il test nella parte alta per indicare "Cycle-Sentry" o "Continuo".3. Se l'unità è in Cycle-Sentry, l'iconaCycle-Sentry comparirà nell'angolo superiore destro del display come mostrato.4. Se l'unità opera in modalitàContinua, l'icona Cycle-Sentry non viene visualizzata.5. Premere nuovamente il tastoMODE SELECTION (SELEZIONA MODALITÀ) per far nuovamente funzionare l’unità nella modalità precedente.ManometriSensori1345345211. Tornare al display standard.2. Premere il tasto DEFROST(SBRINAMENTO).3. Compariranno una serie dischermate per lo sbrinamento.4. Verrà visualizzato il display dellosbrinamento. L'indicatore a barra mostrerà il tempo rimanente per il completamento del ciclo disbrinamento. Una volta completato il ciclo di sbrinamento, vienevisualizzato nuovamente il display standard.121. Tornare al display standard.2. Premere il tasto GAUGES(MANOMETRI).3. Premere i tasti BACK(PRECEDENTE) o NEXT (SUCCESSIVO) per scorrere i seguenti manometri: Temperatura del liquido diraffreddamento, Livello del liquido di raffreddamento, Olio motore, Pressione, Ampere, Tensione della batteria, Giri/min. del motore, Pressione di mandata, Pressione di aspirazione, Posizione ETV, I/O. Se non viene premuto alcun tasto1. Tornare al display standard.2. Premere il tasto SENSORS(SENSORI).3. Premere i tasti BACK(PRECEDENTE) o NEXT(SUCCESSIVO) per scorrere le varie schermate dei sensori:Controllo della temperatura dell'aria di ritorno, Display della temperatura dell'aria di ritorno, Controllo della temperatura dell'aria di mandata, Display della temperatura dell'aria di mandata, Differenziale di temperatura, Temperatura dellaserpentina dell'evaporatore, Temperatura ambiente dell'aria, Temperaturasostitutiva 1, Sensori 1-6 di temperatura registratore dati opzionale e Sensore di temperatura del pannello. Se non viene premuto alcun tasto entro 30 secondi, il display tornerà alla visualizzazione standard.4. Premere il tasto LOCK (BLOCCATO) per soffermarsi sulla schermata di un sensoreper 15 minuti. Premere nuovamente il tasto per sbloccare la schermata.5. Premere il tasto EXIT (ESCI) per tornare al display standard.entro 30 secondi, il display tornerà alla visualizzazione standard.4. Premere il tasto LOCK (BLOCCATO) per soffermarsi sulla schermata di unmanometro per 15 minuti. Premere nuovamente il tasto per sbloccare la schermata.5. Premere il tasto EXIT (ESCI) per tornare al display standard.NOTA: è necessario selezionare il tasto YES (S Ì) entro 10 secondi dalla selezione del nuovo punto di riferimento, altrimenti la modifica sarà annullata.5721Contaore23NOTA: Per informazioni più dettagliate, vedere il capitolo Funzionamento nel relativo manuale operativo dell’unità.1. Cancellare tutti i codici di allarme.2. Tornare al display standard.3. Premere il tasto MENU.4.Premere il tasto NEXT(SUCCESSIVO) fino a visualizzare il menu Pretrip (Verifica prima della partenza).5. Premere il tasto SELECT(SELEZIONA) per avviare una verifica prima della partenza.6. Se l'unità non è in funzione, verràavviata una verifica completa. Se l'unità è in funzione in modalità diesel o elettrica, verrà eseguita una verifica prima della partenza con motore in funzione.7. Al termine di tutte le verifiche,i risultati vengono indicati come PASS (SUPERATO), CHECK (CONTROLLARE) o FAIL (FALLITO). Se i risultati sono CHECK (CONTROLLARE) o FAIL (FALLITO), vengono generati dei codici di allarme per consentire ai tecnici di risalire all'origine del problema.1. Tornare alla schermata del displaystandard.2. Premere il tasto MENU.3. Scorrere il Menu principalepremendo ripetutamente i tasti NEXT (SUCCESSIVO) e BACK(PRECEDENTE) finché compare la schermata del menu principale dei contaore.4. Premere il tasto SELECT(SELEZIONA) per accedere al menu contaore.5. Premere i tasti NEXT (SUCCESSIVO)e BACK (PRECEDENTE) pervisualizzare i display del contaore.。

CS-2100i/2000i根据SYSMEX在血栓与止血领域多年积累的经验,为全面满足各类实验室的需求率先开发CS-2100i/2000i。
四种方法学:凝固法/发色底物法/免疫比浊法/聚集法unique!多波长高精度光学检测:5种波长检测提供可靠数据新型智能监测:特有的HIL check 功能排除溶血/黄疸/脂血干扰更多新检测项目的选择:FXIII,vWF:Rco等更可靠的实验室质量保证:SNCS实时在线质控全面满足血栓与止血检测需求!CA-7000以全球最快的分析速度提供全面和高精准的止凝血项目检测结果,仪器集中了凝固法,发色底物法和免疫法于一体,设计高度人性化和智能化,操作简便,成为大规模实验室的首选。
● 多参数测试,500测试/小时高速分析能力● 操作简便,灵活对待各种需求● 优秀的试剂管理系统(SRS)● 安全、实用的系统设计● 大容量的数据管理能力,完整的质控系统CA-1500汇集了当今血栓/止血分析仪最新的各种先进功能于一身,是市场上少见的性能/价格比极高的一台仪器,是大型教学医院,综合医院实验室的首选。
它具有快速处理能力,最快180测试/小时,集多种检测功能于一身:凝固法、发色底物法、免疫法。
具有全能随机组合能力,两种方法测定纤维蛋白质,适合常规大量和急诊使用。
● 拥有高速处理能力、随机测试功能和自动再检查功能● 三种分析方式,包括多规则监视的广泛质控文件和平行线生物分析功能● 卓越的性能可以灵活适应实验室的多样化需求CA-500系列CA-500系列包含了六款机型,设计新颖、符合经济原则,是各中小型实验室开展血栓/止血实验的最佳选择,也是半自动升级到全自动的理想机型。
小型台式仪实用可靠,具备三种检测系统即凝固法、发色底物法、免疫法的自由组合用户可根据需要选择相应机型。
CA-50设计上完全沿用了全自动CA系列的检测原理,锁定人为误差因素的设计确保它有别于其他半自动血凝仪,达到全自动仪器的准确性与重复性效果,四通道即可批量检测又可单独检测,内置质控文件,适用于小标本量实验室使用。
1TUNEL Apoptosis Detection Kit Cat. No. L00297(For Paraffin-embedded Tissue Sections, Biotin labeled POD ) Technical Manual No.0263 Version 01132011I Description ......................................................................... ..................... 1 II Key Features ...................................................................... . .................... 1 III Kit Contents ................................................................ . ........................... 1 IV Storage..................................................................................................... 2 V Protocol ....................................................... ............................................ 2 VI Related Products ........................................................................................ 5 VII Troubleshooting ........................................................................................ 6 VIII Ordering Information .......................................................................... . (7)I. DESCRIPTIONThe TUNEL Apoptosis Detection Kit for Paraffin-embedded Tissue Sections (Biotin labeled POD ) (Cat. No. L00297) is one of GenScrip ’s newly introduced products. The kit can detect fragmented DNA in the nucleus during apoptosis. In this modified TUNEL assay kit, biotinylated nucleotide is labeled at the DNA 3´-OH ends using the natural or recombinant terminal deoxynucleotidyl transferase (TdT or rTdT). Then, horseradish peroxidase-labeled streptavidin (streptavidin-HRP) is bound to these biotinylated nucleotides, which are detected using the peroxidase substrate, hydrogen peroxide, and 3,3’-diaminobenzidine (DAB), a stable chromogen. Using this procedure, apoptotic nuclei are stained dark brown .II. KEY FEATURESSimplified Procedure: The kit contains ready-to-use reagents, including proteinase K, DAB and DNase I Enhanced Sensitivity: This kit can assay the cells during the early stages of apoptosis. Enhanced Specificity: The kit can stain apoptotic cells.Streamlined Process: The entire procedure takes about three hours.Increased Convenience: The results can be observed by light microscope. High Veracity :The kit contains positive control reagent.III. KIT CONTENTSThe TUNEL Apoptosis Detection Kit (L00297) employs biotinylated nucleotides (Biotin-11-dUTP), horseradish peroxidase-labeled streptavidin, TdT, and DAB.2IV. STORAGEStore streptavidin-HRP at 4°C, and do not expose it to light. Store the rest of the kit at -20°C. It will remain stable for one year.V . TUNEL Apoptosis Detection Kit PROTOCOLBefore use, order or prepare the following:Fixation Solution: 4% paraformaldehyde in PBS, pH 7.4, freshly prepared. Blocking Solution: 3% H 2O 2 in methanol. e.g. 1 ml 30% H 2O 2 + 9ml methanol.Permeabilization Solution: 0.1% Triton X-100 and 0.1% sodium citrate in water, freshly prepared.Note :1. Please centrifuge the reagents in the kit before use.2. Please prepare the proper amount of TUNEL Reaction Mixture according to the amount of the samples to save reagent.3. The DAB is powder, please dissolve the DAB powder in PBS to make 20×DAB buffer (10 mg/ml DBA buffer) before use.1. Preparing Conventional Paraffin-embedded Tissue Sections3﹡ Alternative TreatmentsThere are other methods of preparing Paraffin-embedded Tissue Sections:1. Incubate the dewaxed and rehydrated tissue sections with Permeabilization solution for 8-10minutes. Permeabilization Solution contains 0.1% TritonX –100 and 0.1% sodium citrate in water, freshly prepared.2. Incubate the dewaxed and rehydrated tissue sections with Pepsin Buffer * or Trypsin Buffer *, for 8-10 minutes.Pepsin Buffer* contains 0.25%-0.5% pepsin in HCl buffer, pH 2.0 Trypsin Buffer * contains 0.25%-0.5% trypsin in 0.01M HCl buffer.3. Incubate the dewaxed and rehydrated tissue sections with 200 ml 0.1 M Citrate Buffer (pH 6.0) in a plastic jar. Irradiate with 350 W microwaves for five minutes.42. Preparing Particular Paraffin-embedded Tissue Sections (e.g. cardiac muscle and brain tissue)Controls:Negative control: Employ the cells or sections as described the labeling protocol. Label solution but do not addany Terminal Deoxynucleotidyl Transferase (TdT) to the TUNEL Reaction Mixture.Positive control: Before beginning the labeling procedures, incubate the fixed and permeabilized cells or sectionswith 100 μl DNase I Solution for 10 minutes at 15-25°C to induce DNA strand degradation.(DNase I Solution contains 30000 U/ml-50000 U/ml DNase I (grade I) depending on the sample to be stained in 1X DNase I buffer. One example of 1X DNase I buffer is 10 mM CaCl 2, 6 mM MgCl 2, and 10 mM NaCl in 40 mM Tris-HCl, pH 7.9)5Labeling Protocol:*20X DAB buffer (10 mg/mL DAB buffer ) contains 10 mg DAB dissolved in1.0 ml PBS buffer.VI. RELATED PRODUCTSTUNEL Universal Apoptosis Detection Kit (Biotin labeled POD), Cat. No. L00290TUNEL Apoptosis Detection Kit for Adherent Cells (Biotin labeled POD), Cat. No. L002966TUNEL Apoptosis Detection Kit for Adherent Cells (FITC labeled POD), Cat. No. L00299TUNEL Apoptosis Detection Kit for Paraffin-embedded Tissue Sections (FITC labeled POD), Cat. No. L00300 TUNEL Apoptosis Detection Kit for Cryopreserved Tissue Sections (FITC labeled POD), Cat. No. L00301VII. TROUBLESHOOTING7TdT Dilution Buffer* contains 150 mM KCl, 1 mM 2-mercaptoethanol, and 50% glycerol in 60 mM KPB, pH7.2VIII. ORDER INFORMATIONTUNEL Apoptosis Detection Kit for Paraffin-embedded Tissue Sections (Biotin labeled POD), Cat. No. L00297GenScript USA Inc. 860 Centennial Ave., Piscataway, NJ 08854 Tel: 1-877-436-7472 Fax: 1-732-210-0262Email:*********************Web: For Research Use Only.。
MTT 细胞增殖及细胞毒性检测试剂盒产品编号 产品名称包装 C0009S MTT 细胞增殖及细胞毒性检测试剂盒 500次 C0009MMTT 细胞增殖及细胞毒性检测试剂盒2500次产品简介:MTT 细胞增殖及细胞毒性检测试剂盒(MTT Cell Proliferation and Cytotoxicity Assay Kit)是一种非常经典的细胞增殖和细胞毒性检测试剂盒,被广泛应用于细胞增殖和细胞毒性的检测。
MTT 可以被线粒体内的一些脱氢酶还原生成结晶状的深紫色产物formazan (图1A)。
在特定溶剂存在的情况下,可以被完全溶解(图1B)。
然后通过酶标仪可以测定570nm 波长附近的吸光度(图2)。
细胞增殖越多越快,则吸光度越高;细胞毒性越大,则吸光度越低。
图1. MTT 细胞增殖及细胞毒性检测试剂盒实测效果图。
A. HeLa 细胞加入使用本试剂盒配制的MTT 溶液,在细胞培养箱内孵育4小时,显微镜下可见大量结晶状的深紫色产物formazan 生成。
B. 不同数量HeLa 细胞在MTT溶液(MTT solution)加入后4小时的效果图(上图)及深紫色产物formazan 生成后加入Formazan 溶解液(Formazan solvent),充分溶解后的效果图(下图)。
图2. 本试剂盒检测不同数量HeLa 细胞的效果图。
不同的检测条件下,实际读数会因标准品的配制、检测仪器等的不同而存在差异,图中数据仅供参考。
本试剂盒采用了独特的Formazan 溶解液配方,无需去除原有的培养液,可以直接加入Formazan 溶解液溶解formazan 。
从而避免了由于去除培养液时formazan 被部分去除而引起的误差。
本试剂盒本底低,灵敏度高,线性范围宽,使用方便。
碧云天各种细胞增殖和细胞毒性检测试剂盒的比较和选择,请参考/support/cell-proliferation.htm 。
本试剂盒C0009S 包装可以测定500个样品,C0009M 包装可以测定2500个样品。