halohydrin dehalogenase are structurally pdf
- 格式:pdf
- 大小:201.59 KB
- 文档页数:9

脱卤酶发展史
卤醇脱卤酶(halohydrin dehalogenases,HHDHs)是微生物代谢卤代醇的重要酶类,它既能催化邻卤代醇脱卤成环又能催化逆反应环氧化物开环。
近年来,通过基于数据库的基因挖掘,HHDHs酶家族扩增至70多种,亚类也由A类发展至G类。
通过酶表征,发现酶的底物范围不一,而且酶活力、稳定性以及立体选择性难以满足工业生产需求,通过分子改造改善了酶的性能,拓展了 HHDHs在生物催化方面的应用。
本文综述了近年来HHDHs的结构机制、来源分布、酶分子改造以及催化应用等方面的研究。
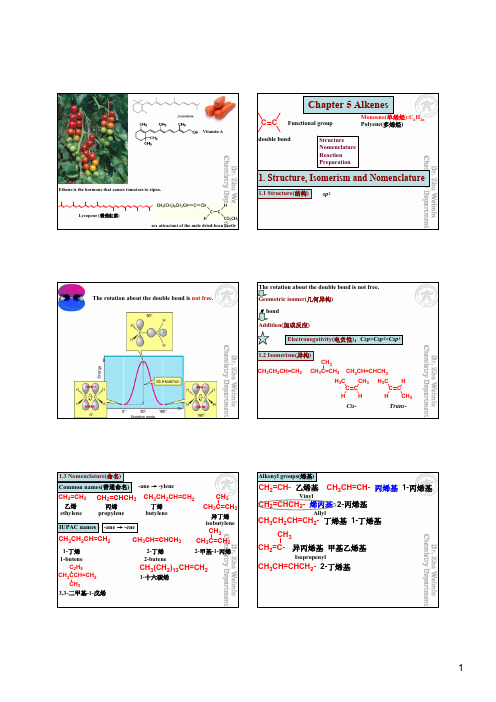
Ethene is the hormone that causes tomatoes to ripen.Lycopene (番茄红素)sex attractant of the male dried-bean beetleVitamin AChapter 5 AlkenesC CFunctional groupMonoene(单烯烃):C n H 2n Polyene(多烯烃)Structure Nomenclature Reaction Preparationdouble bond1. Structure, Isomerism and Nomenclature1.1 Structure(结构)sp 2The rotation about the double bond is not free .The rotation about the double bond is not free.Geometric isomer(几何异构)πbondAddition(加成反应)Electronegativity(电负性):C sp >C sp 2>C sp 31.2 Isomerism(异构)CH 3CH 2CH=CH 2CH 3C=CH 2CH 3CH 3CH=CHCH 3C C C C H H 3C CH 3HH H 3C HCH 3Cis -Trans -1.3 Nomenclature(命名)Common names(普通命名)-ane →-ylene乙烯ethylene 丙烯propylene CH 2=CH 2CH 2=CHCH 3丁烯butylene异丁烯isobutyleneCH 3CH 2CH=CH 2CH 3C=CH 2CH 3IUPAC names-ane →-eneCH 3CH 2CH=CH 2CH 3C=CH 2CH 3CH 3CH=CHCH 3CH 3CCH=CH 2C 2H 5CH 33,3-二甲基-1-戊烯CH 3(CH 2)13CH=CH 21-十六碳烯1-丁烯1-butene2-丁烯2-butene 2-甲基-1-丙烯Alkenyl groups(烯基)CH 2=CH- 乙烯基CH 2=CHCH 2- 烯丙基 2-丙烯基CH 3CH=CH- 丙烯基 1-丙烯基CH 3CH 2CH=CH 2- 丁烯基 1-丁烯基CH 3CH=CHCH 2- 2-丁烯基CH 2=C-CH 3异丙烯基 甲基乙烯基VinylAllylIsopropenylHH HCH 3HC 2H 5trans -2,2,5-三甲基-3-己烯Geometric isomers(几何异构体)Cis -/ Trans -(顺-/ 反-)Z -/ E -?Z -/ E -(Z )-3-甲基-4-丙基-3-辛烯12345612345687cis -2-戊烯ClH ClBr H 3C Cl DBrHO 2NH 3CCl HBr (Z )-1-氯-2-溴丙烯?Cis -/ Trans -(顺-/ 反-)Z -/ E -cis -1,2-二氯-1-溴乙烯1-烯丙基-4-乙烯基环己烷12345(1E ,3Z )-3-氘-2-硝基-4-氯-1-溴-1,3-戊二烯(E )-1,2-二氯-1-溴乙烯H 3C HCH 2CH 2CH 3CH 2CC 2H 5CH 3H1234567*(2E ,5R )-5-甲基-3-丙基-2-庚烯Cycloalkenes(环烯)H HH H HHHHH H H H HH HH H HHHHH If the ring is large enough, a trans stereoisomer is also possible.C CHHC CHH H HH H H H*4-甲基环己烯(R )-~ (S)-~反-3,4-二甲基环己烯×**1243(3S ,4R )-~12345687910二环[4.4.0]-1-癸烯*HH(R )-~1234567二环[2.2.1]-1-庚烯*(R )-(S )-~(S )-~Stability of alkenes(烯的稳定性)RRR RR HR RH HR RH RR HR HR HHHR HH HH HTetrasubstitutedTrisubstituted Monosubstituted Unsubstituted>>>>>>Disubstituted The most important factors governing alkene stability are:1. Degree of substitution (alkyl substituents stabilize a double bond)2. Van der Waals strain (destabilizing when alkyl groups are cis to each other)2. Physical PropertiesDipole moment(μ)μ= q X dMethyl group is a electron-donating substituent. (甲基是一个给电子基)Electronic effect HCH 3H 3CHH 3CH CH 3Hμ= 0 C .mBp. 1 ℃Mp. -105.6 ℃μ= 1.100 X10-30C.mBp. 4℃Mp. -138.9 ℃Debye won Nobel Prize in Chemistry , 1936, “for his contributions to our knowledge of molecular structure through his investigations on dipole moments and on the diffraction of X-rays and electrons in gases”.Z-/E-Physical properties Chemical properties Biological activitiesOCOCH 3OCOCH 3(Z )-dodec-9-enyl acetate(E )-dodec-9-enyl acetateGrape berry month European pine shoot monthSex attractantsHHHHA comprehensive review of studies of trans fats was published in 2006 in the New England Journal of Medicine reports a strong and reliable connection between trans fat consumption and coronary heart disease (CHD).3. Electronic Effect (电子效应)An effect that results when two or more atoms or groups interact so as to alter the electron distribution in a system is called an electronic effect .Inductive effect (诱导效应)Conjugation (共轭效应)Hyperconjugation (超共轭效应)Field effect (场效应) Steric effect (位阻效应)3.1 Inductive effect(诱导效应)CH 3→CH 2→FOrigin: electronegativity (由于原子的电负性不同而引起)Successive transmission through the carbon chain (沿碳链传递)Short distance (短距离)δδ+δ+δ−Electron attracting (or electron withdrawing) inductive effect (吸电子诱导效应)-IElectron donating (or electron releasing) inductive effect (给电子诱导效应)+I Electron attractor (吸电子基)Electron donator (给电子基)-NO 2> -CN > -F > -Cl > -Br > -I > -C ΞCR > -OR > -NR 2> -SR > -Ph > -CH=CH 2-NH 3+-NR 3+ -COOH -COOR Alkyl(烷基)-C(CH 3)3> -CH(CH 3)2> -CH 2CH 3> -CH 33.2 Conjugation (共轭效应)Conjugated system(共轭体系): have a p orbital on an atom adjacent to a double bond (连在双键碳上的原子具有p 轨道)CH 2=CHCH 2π,π-共轭p ,π-共轭CH 2=CH 2CH 2p ,π-共轭N OOCH 2=CH-CH=CH-CH=CH-C ≡NCH 2=CH-CH=CH-CH=CH 2δ−δ+δ−δ+δ−δ+δ−δ+δ+δ−δ+δ−δ+δ−δ+δ−δ+δ−δ+δ−δ+δ−Existence in conjugated system (存在于共轭体系)Unlimitedly far (无限远)Alternately polarization (交替极化)Averaging of bond lengths (键长平均化)Delocalization of πelectrons (电子离域)Stability being increased (稳定性增强) Conjugation (共轭效应)-C(吸电子共轭效应)+C(给电子共轭效应)-NO 2-CN -COOH -COOR -CHO -COR -SO 3H -NH 2-NHCOR -OH -OR -OCOR -XHXHHH 2C CH-XH 2C CH-XH 2C CH-Xδδ+δ+δ−δ−δ+H 2C CH-NH 2H 2C CH-NH 2H 2C CH-NH 2δδ+δ+δ−δ−δ+3.3 Hyperconjugation (超共轭效应)C-H σ键与p 轨道发生部分重叠CH 3CH=CH 2σ,π-超共轭C C H CCH 3CH 2σ,p-超共轭C CH H H HHHHClHOOC C COOH XCOO -H 2CCOOH3.4 Field effect (场效应)GCOOHG=H p Ka =6.04G=Cl p Ka =6.25OCO OCO2-OOCO O -O -CO 32-R CO O -R CO O-RCOO -Inductive effect(诱导效应)CH 3→CH 2→F由于原子的电负性不同而引起沿碳链传递短距离吸电子诱导效应-I -NH 2多数电负性较强-CN 含有重键-NR 3+带正电给电子诱导效应+I 烷基带负电Conjugation (共轭效应)共轭体系: 连在双键碳上的原子具有p 轨道(可以被π电子或者p 电子占据,也可以是空的p 轨道)CH 2=CHCH 2CH 2=CH 2CH 2存在于共轭体系无限远交替极化键长平均化电子离域稳定性增强吸电子共轭效应-C多具有重键给电子共轭效应+C -NH 2等多具有孤对电子带负电δ++δ+δ−有些基团的诱导效应和共轭效应是一致的,还有些基团的诱导效应和共轭效应是相反的。



The ChromoTek Halo-Trap Agarose consists of an anti-Halo-tag Nanobody (VHH), which is covalently bound to agarose beads. Halo-Trap Agarose is used to immunoprecipitate Halo-tagged fusion proteins from cell extracts of various organisms like mammals, plants, bacteria, yeast, insects etc. in the presence or absence of a covalently bound ligand. The interaction between Halo-Trap and the Halo-tagged fusion protein is reversible.Ligand: Anti-Halo-tag single domain antibody fragment (VHH, Nanobody)Reactivity: Specifically binds to Halo-tag (modified variant of the bacterial haloalkane dehalogenase enzyme from Rhodococcus rhodochrous) in the absence or presence of covalently bound chloralkane-based ligands. Binding capacity: 7.5-10 µg of recombinant Halo-tag per 25 µL bead slurryBead size: 90 µm (cross-linked 4 % agarose beads)Buffer compatibility: See Wash buffer compatibility table.Storage buffer: 20 % ethanolStorage conditions: Upon receipt store at +4°C. Do not freeze!Stability: Stable for 1 year upon receipt.Shipment: Shipped at ambient temperature.RRID: AB_2827595Required buffer solutionsNEW: Update of Wash buffer components.Buffer CompositionLysis buffer10 mM Tris/Cl pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 0.5 % Nonidet™ P40 Substitute (adjust the pH at +4°C)RIPA buffer10 mM Tris/Cl pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 0.1 % SDS, 1 % Triton™ X-100, 1 %deoxycholate (adjust the pH at +4°C)Dilution buffer10 mM Tris/Cl pH 7.5, 150 mM NaCl, 0.5 mM EDTA (adjust the pH at +4°C)Wash buffer10 mM Tris/Cl pH 7.5, 150 mM NaCl, 0.05 % Nonidet™ P40 Substitute, 0.5 mM EDTA (adjust the pH at +4°C)2x SDS-sample buffer120 mM Tris/Cl pH 6.8, 20 % glycerol, 4 % SDS, 0.04 % bromophenol blue, 10 % β-mercaptoethanolAcidic elution buffer 200 mM glycine pH 2.5 or 100 mM citric acid pH 3.0 (adjust the pH at +4°C)Neutralization buffer 1 M Tris pH 10.4 (adjust the pH at +4°C)Note: Use your equivalent cell lysis buffer for other cell types like yeast, plants, insects, bacteria.Note: Consider using a Wash buffer without detergent for co-immunoprecipitation.Buffer ingredients Max. concentrationDTT10 mMNaCl 1 MNonidet™ P40 Substitute tested up to 2 %SDS0 %Triton™ X-100tested up to 1 %Urea 4 MProduct Product code SizeHalo-Trap Agarose ota-1010 reactions (250 µL slurry)ota-2020 reactions (500 µL slurry)ota-100100 reactions (2.5 mL slurry)ota-200200 reactions (5 mL slurry)ota-400400 reactions (10 mL slurry)Halo-Trap Agarose Kit otak-2020 reactions (500 µL slurry) including buffersCell materialThe following protocol describes the preparation of mammalian cell lysate!For other type of cells, we recommend using 500 µg of cell extract and start the protocol with step Bead equilibration.Mammalian cell lysisNote: Harvesting of cells and cell lysis should be performed with ice-cold buffers. We strongly recommend to add protease inhibitors to the Lysis buffer to prevent degradation of your target protein and its binding partners.For one immunoprecipitation reaction, we recommend using ~106-107 cells.1.Choice of lysis buffer:·For cytoplasmic proteins, resuspend the cell pellet in 200 µL ice-cold Lysis buffer by pipetting up and down. Supplement Lysis buffer with protease inhibitor cocktail and 1 mM PMSF (not included).·For nuclear/chromatin proteins, resuspend cell pellet in 200 µL ice-cold RIPA buffer supplemented(f.c. 2.5 mM), protease inhibitor cocktail and PMSF (f.c. 1with DNaseI (f.c. 75-150 Kunitz U/mL), MgCl2mM) (not included).2.Place the tube on ice for 30 min and extensively pipette the suspension every 10 min.3.Centrifuge cell lysate at 17,000x g for 10 min at +4°C. Transfer cleared lysate (supernatant) to a pre-cooled tube and add 300 µL Dilution buffer supplemented with 1 mM PMSF and protease inhibitor cocktail (not included). If required, save 50 µL of diluted lysate for further analysis (input fraction).Bead equilibration1.Resuspend the beads by gently pipetting up and down or by inverting the tube. Do not vortex the beads!2.Transfer 25 µL of bead slurry into a 1.5 mL reaction tube.3.Add 500 µL ice-cold Dilution buffer.4.Sediment the beads by centrifugation at 2,500x g for 5 min at +4°C. Discard the supernatant.Note: Alternatively, Spin columns (sct-10; -20; -50) can be used to equilibrate the beads.Protein binding1.Add diluted lysate to the equilibrated beads.2.Rotate end-over-end for 1 hour at +4°C.Washing1.Sediment the beads by centrifugation at 2,500x g for 5 min at +4°C.2.If required, save 50 µL of supernatant for further analysis (flow-through/non-bound fraction).3.Discard remaining supernatant.4.Resuspend beads in 500 µL Wash buffer.5.Sediment the beads by centrifugation at 2,500x g for 5 min at +4°C. Discard remaining supernatant.6.Repeat this step at least twice.7.During the last washing step, transfer the beads to a new tube.Optional: To increase stringency of the Wash buffer, test various salt concentrations e.g. 150-500 mM, and/or add a non-ionic detergent e.g. Triton™ X-100 (see Wash buffer compatibility table for maximal concentrations). Note: Alternatively, Spin columns (sct-10; -20; -50) can be used to wash the beads.Elution with 2x SDS-sample buffer (Laemmli)1.Remove the remaining supernatant.2.Resuspend beads in 80 µL 2x SDS-sample buffer.3.Boil beads for 5 min at +95°C to dissociate immunocomplexes from beads.4.Sediment the beads by centrifugation at 2,500x g for 2 min at +4°C.5.Analyze the supernatant in SDS-PAGE / Western Blot.Note: For Western blot detection we recommend Halo antibody [28A8] (28a8-20; -100).Elution with Acidic elution buffer1.Remove the remaining supernatant.2.Add 50-100 µL Acidic elution buffer and constantly pipette up and down for 30-60 sec at +4°C or roomtemperature.3.Sediment the beads by centrifugation at 2,500x g for 2 min at +4°C.4.Transfer the supernatant to a new tube.5.Immediately neutralize the eluate fraction with 5-10 µL Neutralization buffer.6.Repeat this step at least once to increase elution efficiency.Note: Elution at room temperature is more efficient than elution at +4°C. Prewarm buffers for elution at room temperature.Note: Alternatively, Spin columns (sct-10; -20; -50) can be used to separate the beads.Halo-tag toolbox Product code Halo-Trap Agarose ota-10; -20; -100 Halo-Trap Agarose Kit otak-20Binding Control Agarose bab-20Spin columns sct-10; sct-20; sct-50 Halo VHH, recombinant binding protein ot-250Halo antibody [28A8]28a8-20; -100For product details, information, and ordering visit .*********************ChromoTek GmbHAm Klopferspitz 1982152 Planegg-Martinsried Germanyphone: +49 89 124 148 80 fax: +49 89 124 148 811ChromoTek Inc.62-64 Enter Lane Islandia, NY 11749 USAphone: 631 501 1058 fax: 631 501 1060Only for research applications, not for diagnostic or therapeutic use!ChromoTek and GFP-Trap, RFP-Trap, Myc-Trap, Spot-Trap, Spot-Tag, Spot-Label, Spot-Cap, Nano-Secondary, F2H Kit, and Chromobody are registered trademarks of ChromoTek GmbH, part of Proteintech group. Nano-CaptureLigand and V5-Trap are trademarks of ChromoTek GmbH, part of Proteintech group. Nanobody is a registered trademark of Ablynx, a Sanofi company. Alexa Fluor is a registered trademark of Life Technologies Corporation, a part of Thermo Fisher Scientific Inc. Dynabeads is a trademark of Life Technologies AS, a part of Thermo Fisher Scientific Inc. SNAP-tag is a registered trademark and CLIP-tag is a trademark of New England Biolabs, Inc. Octet is a registered trademark of FortéBio, a Sartorius brand. Other suppliers’ products may be trademarks or registered trademarks of the corresponding supplier each. Statements on other suppliers’ products are given according to our best knowledge.。
113海安凹陷是位于苏北盆地东南缘的一个重要的地质凹陷。
它北邻小海凸起,西接泰州低凸起,西北与溱潼凹陷以梁垛低凸起相隔,南至通扬隆起,东到勿南沙隆起。
海安凹陷在苏北盆地中属于重要的含油气凹陷之一。
海安凹陷具有复杂的断裂结构。
受区域构造格局和基底断裂活动的影响与控制明显。
由于其特殊的区域构造位置,与苏北盆地的其他凹陷相比,海安凹陷具有独特的构造特征。
不同构造位置形成的断裂形成了多样的构造样式组合[1]。
在海安凹陷的地质演化过程中,构造运动是主导力量之一。
构造活动导致断裂的发育,形成了复杂的断层网络。
这些断裂对沉积层和岩性产生了强烈影响,控制了油气的运移和富集。
因此,海安凹陷是油气勘探和开发的重要区域,也是地质科学研究的热点之一[2]。
1 区域构造特征苏北盆地是在苏鲁造山带南缘的印支期前陆变形带上发育而来的新生代陆相盆地,盆地基底经历了印支造山运动的前陆褶断作用和燕山期的区域挤压作用,基底构造对盆地格局影响较大[3]。
盆地在新生代时期总体上处于拉张伸展状态,但期间经历了若干次挤压反转事件,各沉积成盆期基金项目:江苏石油勘探局(JS22001-3)海安凹陷断裂特征与构造样式杨小军 季红军 刘俊成 张弛 韦祥中国石化股份有限公司江苏油田分公司物探研究院 江苏 南京 210046摘要:海安凹陷断裂复杂,构造多解性强、落实难度大,以地质模式为指导是提高构造解释精度的重要手段。
构造样式是同一期构造运动、在同一应力环境下所产生的构造变形组合,是构造解释地质模式建立的基础。
以基底构造和成盆期应力场研究为基础,分析区域构造特征及形成演化机制,总结海安凹陷结构及断裂特征;综合不同基底断裂特征、盆地性质和盆地结构特征分析,开展海安凹陷构造样式特征研究。
研究表明:海安凹陷结构、断裂特征及构造样式具有典型分区性,海中断隆以北为南断北超的半地堑结构,断裂走向NEE、NNW倾为主,主要发育走滑构造样式;海中断隆以南为北断南超的半地堑结构,断裂走向近EW、S倾为主,主要发育伸展构造样式。
复旦大学1999-2006年硕士研究生入学考试微生物学试题复旦大学1999年硕士研究生入学微生物学试题一、名词解释(30分)1. Koch's postulates2. negative stains3. RC(respiratory chain)4. stationary phase5. semi-synthetic antibiotics6. extremophiles(extremc-microorganisms)7. heterolactic fermentation8. biomass9. McAb(monoclonal antibody)10.BCDs二、什么是缺壁细菌?试简述4类缺壁细菌的形成、特点及实践意义。
(10分)三、发酵工业为何常遭噬菌体的危害?如何检验、预防和治理它?(10分)四、什么是EMB培养基?试述其主要成分、作用原理及实用价值。
(10分)五、试列表比较低频转导(LFT)和高频转导(HFT)的异同。
(10分)六、试图示并简介IgG的构造(10分)七、当今在国内市场上大量流行的“微生态口服液”主要含哪两类菌(写出其拉丁属名)?试从微生物学家的角度设计一项辩别其质量高低和真伪的实验方案。
(10分)八、试写出以下5种微生物的拉丁学名(不可简写)。
(10分)1. 苏云金芽孢杆菌2. 酿酒酵母3. 产朊假丝酵母4. 脆弱拟杆菌5. 运动发酵单胞菌复旦大学2000年硕士研究生入学微生物学试题一、名词解释(15分)1. 化能自养菌2.富集培养3. 生物氧化4. 厌氧罐5.鲎试剂法二、试写出下列几个重要的数据(15分)(1)典型的细菌的大小、重量(2)霉菌、酵母菌、放线菌、细菌、病毒个体直径间的大致比例。
(3)每克较肥沃的土壤中原生动物、藻类、霉菌、酵母菌、放线菌和细菌的大体细胞数。
(4)当今已知的微生物总数是多少?哪一大类最多、哪一大类最少?(5)大肠杆菌和酿酒酵母的代时各是多少?三、试述革兰氏染色的主要步骤及其染色原理?(10分)四、四大类微生物的菌落各有什么特点?原因是什么?掌握这些知识有何实用意义?(10分)五、何谓烈性噬菌体(举两种代表)?试作图并阐明它的裂解性生活史。
专英重点一、Parapharyngeal 咽旁 Septicemia 败血病 Sialolithiasis 涎石病Periostitis 骨膜炎Sialoductitis 涎管炎Fracture 骨折Comminution 粉碎 Hyperplasia 增生Reparative 修复性Mucoperiosteum 黏骨膜Radiolucent X光透射Space 间隙Infection 感染Prosthesis义齿Oblique 倾斜Scquestrum腐骨死骨Biopsy 切片检查法Sialogram 涎管X线造影片 Giant巨大Nonmalignant良性的Pyogenic 化脓性Mole 胎块Devoid 缺少的 Laceration撕裂 Hyperpyrexia高热Self-reduce 自行使脱臼复位句子翻译If proper preparation of solution, syringes, needles and technic has been carried out, untoward incidents should seldom occur during or after the injection of the local anesthetic.However, one should be in a position to cope with complications in the rare cases when they arise.若药液注射剂, 针头及技术准备妥当, 在局麻注射过程中或之后都将很少出现, 但是, 医生仍应做好应对罕见并发症的准备。
Postoperative pain which the patient experiences after the second and third postoperative day should be carefully examined, since this is not a normal postoperative course.It is caused by dry socket or sharp bone spine.患者于术后二三日之后的疼痛, 也许为非正常情况, 需特别仔细检查, 其有也许由于干槽症或是锋利骨刺引起。
J OURNAL OF B ACTERIOLOGY ,0021-9193/01/$04.00ϩ0DOI:10.1128/JB.183.17.5058–5066.2001Sept.2001,p.5058–5066Vol.183,No.17Copyright ©2001,American Society for Microbiology.All Rights Reserved.Halohydrin Dehalogenases Are Structurally and MechanisticallyRelated to Short-Chain Dehydrogenases/ReductasesJOHAN E.T.VANHYLCKAMA VLIEG,†LIXIA TANG,JEFFREY H.LUTJE SPELBERG,TIM SMILDA,‡GERRIT J.POELARENDS,TJIBBE BOSMA,ANNET E.J.VAN MERODE,MARCO W.FRAAIJE,AND DICK B.JANSSEN*Biochemical Laboratory,Groningen Biomolecular Sciences and Biotechnology Institute,University of Groningen,NL-9747AG Groningen,The NetherlandsReceived 16January 2001/Accepted 1March 2001Halohydrin dehalogenases,also known as haloalcohol dehalogenases or halohydrin hydrogen-halide lyases,catalyze the nucleophilic displacement of a halogen by a vicinal hydroxyl function in halohydrins to yield epoxides.Three novel bacterial genes encoding halohydrin dehalogenases were cloned and expressed in Escherichia coli ,and the enzymes were shown to display remarkable differences in substrate specificity.The halohydrin dehalogenase of Agrobacterium radiobacter strain AD1,designated HheC,was purified to homoge-neity.The k cat and K m values of this 28-kDa protein with 1,3-dichloro-2-propanol were 37s ؊1and 0.010mM,respectively.A sequence homology search as well as secondary and tertiary structure predictions indicated that the halohydrin dehalogenases are structurally similar to proteins belonging to the family of short-chain dehydrogenases/reductases (SDRs).Moreover,catalytically important serine and tyrosine residues that are highly conserved in the SDR family are also present in HheC and other halohydrin dehalogenases.The third essential catalytic residue in the SDR family,a lysine,is replaced by an arginine in halohydrin dehalogenases.A site-directed mutagenesis study,with HheC as a model enzyme,supports a mechanism for halohydrin dehalogenases in which the conserved Tyr145acts as a catalytic base and Ser132is involved in substrate binding.The primary role of Arg149may be lowering of the pK a of Tyr145,which abstracts a proton from the substrate hydroxyl group to increase its nucleophilicity for displacement of the neighboring halide.The proposed mechanism is fundamentally different from that of the well-studied hydrolytic dehalogenases,since it does not involve a covalent enzyme-substrate intermediate.Halogenated aliphatics constitute an important class of en-vironmental pollutants.Various microorganisms have evolved that are able to degrade some of these compounds and use them as sole sources of carbon and energy.Such organisms are of importance for bioremediation of polluted soil,groundwa-ter,and wastewater.In most cases,specialized enzymes,des-ignated dehalogenases,catalyze the cleavage of the carbon-halogen bonds,which is a key detoxification reaction.Hydrolytic dehalogenases have been studied extensively,which has resulted in detailed insight into the structure and mecha-nism of several enzymes of this class (8,33).For other deha-logenases,structural and mechanistic data are hardly available.Halohydrin dehalogenases,also referred to as haloalcohol dehalogenases or halohydrin hydrogen-halide lyases,occur in the degradation pathways of halopropanols and 1,2-dibromo-ethane,where they catalyze the nucleophilic displacement of a halogen by a vicinal hydroxyl group in halohydrins,yielding an epoxide,a proton,and a halide ion (7,22,30,31).These enzymes also efficiently catalyze the reverse reaction,the ha-logenation of epoxides,and the dehalogenation of vicinal chlo-rocarbonyls to hydroxycarbonyls (2,14,31).The interest in halohydrin dehalogenases increased when it was found that the dehalogenation of halohydrins may proceed with high enan-tioselectivity,making these enzymes useful catalysts for the production of optically pure epoxides and halohydrins (1,14–16).In this study,we report the cloning of three bacterial halo-hydrin dehalogenase genes.Sequence analysis suggested that these proteins are similar to proteins of the short-chain dehy-drogenase/reductase (SDR)family.The amino acids Ser132,Tyr145,and Arg149were identified as the catalytic residues and are proposed to play a role highly similar to that of the conserved residues involved in the redox reaction catalyzed by the SDR family proteins.MATERIALS AND METHODSMaterials.All chemicals were purchased from Acros Chimica,Merck,Aldrich,or Sigma.Molecular biology enzymes were purchased from Boehringer.Oligo-nucleotide primers were supplied by Eurosequence BV,Groningen,The Neth-erlands.Strains and growth conditions.Agrobacterium radiobacte r strain AD1and Arthrobacter sp.strain AD2were maintained on nutrient broth at 30°C.Myco-bacterium sp.strain GP1was maintained on selective plates with 1-propanol or 1,2-dibromoethane as a carbon source as described before (22).Escherichia coli strains HB101(6),JM101(35),and BL21(DE3)(28)were grown at 37°C in Luria-Bertani medium.For selection of recombinants carrying plasmids,the appropriate antibiotic was added at the following concentrations:50g ml Ϫ1for kanamycin,50g ml Ϫ1for ampicillin,and 12.5g ml Ϫ1for tetracycline.E.coli BL21(DE3)grown at 17°C was used for high-level expression of recombinant halohydrin dehalogenase driven by the T7promoter in plasmid pGEF ϩ(25).Construction and screening of genomic libraries of halohydrin dehalogenase-producing bacteria.Procedures for the isolation and manipulation of DNA were*Corresponding author.Mailing address:Biochemical Laboratory,Groningen Biomolecular Sciences and Biotechnology Institute,Uni-versity of Groningen,Nijenborgh 4,NL-9747AG Groningen,The Netherlands.Phone:31-50-3634209.Fax:31-50-3634165.E-mail:d.b.janssen@chem.rug.nl.†Present address:NIZO Food Research,6710BA Ede,The Neth-erlands.‡Present address:Friedrich Miescher-Institut,Maulbeerstrasse,4058Basel,Switzerland.5058performed essentially as described by Sambrook et al.(24).The construction of a genomic library of Mycobacterium sp.strain GP1was described before(22). The same procedure was used to construct a genomic library of A.radiobacter strain AD1in the cosmid vector pLAFR3(27).Restriction analysis of plasmids isolated from16transduced E.coli HB101clones showed that all plasmids contained inserts.About1,000transductants were screened for halohydrin de-halogenase activity by monitoring halide production with1,3-dichloro-2-propa-nol as a substrate as described before(22).PCR and construction of expression vectors for halohydrin dehalogenase genes.For overexpression,the halohydrin dehalogenase genes were amplified by PCR under conditions described before(32).The hheC gene was amplified from a plasmid preparation of pAD1–9B2with the forward primer PFHheC(5Ј-AT CTGA CC AT G GCAACCGCAATTG-3Ј)and the reverse primer PRHheC(5Ј-CCCAACG G A T CCACGAACCACGGC-3Ј)(Nco I and Bam HI sites under-lined,start codon shown in boldface,and substituted nucleotides shown in italics).The hheB GP1gene starting with the second possible start codon was amplified from recombinant cosmid pGP1–4B5(22)with the forward primer PF1HheB GP1(5Ј-AAAA CC ATG GCTAACGGAAGACTGGCAGGC-3Ј)and reverse primer PRHheB GP1(5Ј-GGGCTGTG GATC CTCTCAGGTGGCCCA GCCGCC-3Ј).The hheA AD2gene was directly amplified from cells of Ar-throbacter sp.strain AD2with the forward primer PFHheA AD2(5Ј-GAAC C AT G GT GATCGCCCTCGTGAC-3Ј)and the reverse primer PRHheA AD2(5Ј-TG GCTATCTGCCCTAACC AT GGCC-3Ј).The hheA AD2gene was cloned behind the T7promoter in the Nco I site,and hheC and hheB GP1were cloned between the Nco I and Bam HI sites of the expression vector pGEFϩ(25). Nucleotide sequencing.Sequencing on double-stranded DNA was performed with the Amersham Thermo Sequenase cycle sequencing kit(Amersham BV, Roosendaal,The Netherlands),with7-deaza-dGTP and5ЈCy5fluorescent prim-ers.Sequence reactions were run on the Pharmacia ALF-Express automatic sequencing machine(Uppsala,Sweden)at the BioMedical Technology Centre (Academic Hospital,Groningen,The Netherlands).Both strands were se-quenced to ensure accuracy.Homology searches and structure prediction.The BLAST program was used to screen DNA and protein databases for similar proteins.Multiple sequence alignments were made in ClustalW v1.7.Secondary structures were predicted with the programs SopM(10)and SSP(26).Tertiary structure modeling was done by comparative protein modeling to known three-dimensional structures of members of the SDR protein family with the program SWISS-MODEL(12)by using the structures of2HSD,1AE1,2AE1,1AHH,and1A4U(Protein Data Base codes)as templates.Overexpression and purification of the halohydrin dehalogenases.Both wild-type and mutant halohydrin dehalogenase genes were expressed in E.coli BL21(DE3)as described before(23).A1-liter culture of E.coli BL21(DE3) (pGEFHheC)was harvested by centrifugation for purification of HheC.Cells were resuspended in10mM Tris-sulfate buffer(pH7.5),and all further steps were carried out at0to4°C.Cells were washed twice with this buffer before they were resuspended in10mM Tris-sulfate buffer containing1mM EDTA and1 mM-mercaptoethanol(TEM buffer)or TEM buffer containing3mM NaN3 (TEMA buffer).After sonication,a crude extract was obtained by centrifugation (200,000ϫg,60min).The crude extract was applied to a Resource Q anion-exchange column(6ml; Pharmacia Biotech,Uppsala,Sweden)that was connected to an LCC500type fast protein liquid chromatography system(Pharmacia Biotech).The buffer system consisted of TEMA buffer(buffer A)and TEMA buffer with0.45M (NH4)2SO4(buffer B).Retained protein was eluted with a three-step increasing linear gradient:0to5%buffer B in15ml,15to45%buffer B in100ml,and45 to100%buffer B in35ml(flow rate,5ml minϪ1;fraction volume,5ml).The dehalogenase eluted at110to150mM(NH4)2SO4,and active fractions were pooled.Ammonium sulfate was added to a concentration of1.5M,and the protein was applied to a Resource Phe column(1ml;Pharmacia Biotech).Retained protein was eluted with a20-ml decreasing linear gradient of1.5to0M ammonium sulfate in buffer A(flow rate,0.5ml minϪ1;fraction volume,0.5ml).The dehalogenase eluted at1.0to0.8M(NH4)2SO4,and active fractions were pooled,yielding a pure protein as judged by sodium dodecyl sulfate-polyacryl-amide gel electrophoresis(SDS-PAGE).The purified protein was dialyzed against TEM buffer to remove azide,filtered with a0.2-m-pore-diameterfilter, and stored at4°C.Construction of HheC mutants.Site-directed mutagenesis was done by using the Quickchange site-directed mutagenesis kit of Stratagene(La Jolla,Calif.) with pGEFHheC as a template.Ser132was mutated to Cys with the primer set PCODS132C(5Ј-GGACATATTATCTTTATTACC T G T GCAACGCCCTTCG GGCCTTGG-3Ј)and PNONS132C(5Ј-CCAAGGCCCGAAGGGCGTTGCA C AGGTAATAAAGATAATATGTCC-3Ј)(codon of substituted residue shown in boldface,substituted nucleotides shown in italics).Ser132was mutated to Ala with the primer set PCODS132A(5Ј-GGACATATTATCTTTATTACC G CT G CAACGCCCTTCGGGCCTTGG-3Ј)and PNONS132A(5Ј-CCAAGGCCCGA AGGGCGTTGCAG C GGTAATAAAGATAATATGTCC-3Ј).Tyr145was mu-tated to Phe with the primer set PCODY145F(5Ј-CCTTGGAAGGAACTTTC TACC T T C ACGTCAGCCCGAGCAGGTGC-3Ј)and PNONY145F(5Ј-GCAC CTGCTCGGGCTGACGTG A AGGTAGAAAGTTCCTTCCAAGG-3Ј).Arg149 was mutated to Lys with the primer set PCODR149K(5Ј-CCTACACGTCAG CC AA A GCAGGTGCATGCACCTTGGC-3Ј)and PNONR149K(5Ј-GCCAAG GTGCATGCACCTGCT TT GGCTGACGTGTAGG-3Ј).Arg149was mutated to Gln with the primer set PCODR149Q(5Ј-CCTACACGTCAGCC C AG GCA GGTGCATGCACCTTGGC-3Ј)and PNONR149Q(5Ј-GCCAAGGTGCATG CACCTGC CT GGGCTGACGTGTAGG-3Ј).Arg149was mutated to Glu with the primer set PCODR149E(5Ј-CCTACACGTCAGCC GA A GCAGGTGCAT GCACCTTGGC-3Ј)and PNONR149E(5Ј-GCCAAGGTGCATGCACCTGCT TC GGCTGACGTGTAGG-3Ј).CD spectroscopy.Circular dichroism(CD)spectra were recorded with an AVIV62A DS spectrometer.The far-UV spectra of both the wild type and the HheC variants were recorded at25°C from190to250nm with a0.1-cm cuvette containing0.1mg of halohydrin dehalogenase(5mM potassium phosphate[pH 7.5])per ml.Enzyme assays.Halohydrin dehalogenase activities were assayed at30°C in50 mM Tris-sulfate buffer(pH8.0)containing5mM substrate by monitoring halide liberation or epoxide formation or with a colorimetric assay using the chromo-genic substrate p-nitro-2-bromo-1-phenylethanol.Protein concentration and ha-lide liberation were determined as described before(30).Halohydrins and ep-oxides were analyzed by gas chromatography.Samples(1.5ml)were extracted with1.5ml of diethyl ether containing0.05mM1-chlorohexane,1-bromohexane, or mesitylene as an internal standard.Extracts were analyzed by split injection of 2or4l on an HP5column(model HP19091J-413;Hewlett-Packard)with helium as a carrier gas.Separation of enantiomers of chiral compounds was carried out with chiral gas chromatography as described before(16). Nucleotide sequence accession numbers.The nucleotide sequences described in this article have been deposited in the EMBL/DDBJ/GenBank database under the following accession numbers:HheC,AF397296;HheA AD2,AF397297; HheB GP1,AY044094.RESULTSCloning and sequence analysis of the halohydrin dehaloge-nase gene from Agrobacterium radiobacter strain AD1.The gram-negative bacterium A.radiobacter strain AD1was iso-lated for its capability to use chloropropanols and epichloro-hydrin as growth substrates(30).The organism utilizes(R)-2,3-dichloro-1-propanol,whereas the(S)-enantiomer is not degraded(4).This is caused by the high enantioselectivity of thefirst enzyme in the degradation pathway,the halohydrin dehalogenase(16).To clone the gene encoding the halohydrin dehalogenase,a gene bank of strain AD1was constructed in E. coli with the cosmid vector pLAFR3.Three active clones were identified when1,000cosmid clones were screened for deha-logenase activity with1,3-dichloro-2-propanol.Sequence anal-ysis of one of the clones,pAD1–9B2,showed the presence of a complete open reading frame of765nucleotides,designated hheC.Thefirst104bp were identical to a fragment of a puta-tive halohydrin dehalogenase gene that was located on a genomic DNA segment of A.radiobacter strain CFZ11that carried the epoxide hydrolase gene(echA)(23).The deduced protein,HheC,has a predicted molecular mass of27,954Da and is highly similar to HalB of Agrobacterium tumefaciens (Table1),for which no biochemical characterization has been published.The hheC gene was amplified by PCR,and the start codon was fused into the Nco I site of pGEFϩ.The resulting expression vector was designated pGEFHheC.Cloning and sequence analysis of the halohydrin dehaloge-nase gene of Arthrobacter sp.strain AD2.Previously,Van denV OL.183,2001HALOHYDRIN DEHALOGENASES ARE RELATED TO SDR PROTEINS5059Wijngaard et al.(31)purified the halohydrin dehalogenase of the 3-chloro-1,2-propanediol-degrading Arthrobacter sp.strain AD2.Out of the 34N-terminal residues,32were identical to the halohydrin dehalogenase HheA of Corynebacterium sp.strain N-1074(36),which suggests that the halohydrin dehalo-genase genes of both strains are highly similar.Therefore,primers designed for the hheA gene were used to amplify the halohydrin dehalogenase gene of strain AD2.Three indepen-dent clones were sequenced and found to have identical DNA sequences.The gene encodes a 244-amino-acid protein that was designated HheA AD2,since it was identical to HheA ex-cept for 7amino acid substitutions (Table 1).The gene was fused into the start codon of pGEF ϩ,and the resulting plas-mid was designated pGEFHheA AD2.Cloning and sequence analysis of the halohydrin dehaloge-nase gene of Mycobacterium sp.strain GP1.At least two de-halogenases are produced by Mycobacterium sp.strain GP1when 1,2-dibromoethane serves as a growth substrate (22).A haloalkane dehalogenase,encoded by dhaA f ,catalyzes the hydrolytic dehalogenation of 1,2-dibromoethane to 2-bromo-ethanol,which is subsequently dehalogenated by a halohydrin dehalogenase to epoxyethane.No halohydrin dehalogenase-producing clones were identified when a gene library of strain GP1in E.coli was screened for dehalogenase activity with 1,3-dichloro-2-propanol,suggesting that transcription or trans-lation signals were not recognized in E.coli (22).However,when cosmid pGP1–4B5,which carries dhaA f ,was transferred to Pseudomonas sp.strain GJ1or Burkholderia cepacia G4,the resulting transconjugants rapidly dehalogenated 1,3-dichloro-2-propanol,indicating that active halohydrin dehalogenase was produced.Hence,pGP1–4B5also harbors the gene encoding the halohydrin dehalogenase of strain GP1.Sequencing showed that the halohydrin dehalogenase gene is located 2,637bp downstream of dhaA f and encodes a 235-amino-acid pro-tein.The gene was identical to dehalogenase gene hheB of Corynebacterium sp.strain N-1074,except for 4nucleotide substitutions that result in 4amino acid substitutions in the encoded protein (Table 1).As in strain N-1074,a duplication of a 27-nucleotide region (36)has resulted in a duplicate set of AGGA ribosome binding sites,each located 12nucleotides upstream of an ATG start codon.Hence,two polypeptides can be produced,which differ in the presence or absence of a MANGRKRE amino acid sequence extension at the N termi-nus,by translation starting from the first or the second start codon,respectively.Previously,Yu et al.(36)have shown that HheB is active as a tetramer and that all possible combinations of the two slightly different subunits occur.However,this sub-unit composition had little effect on substrate specificity,since enzyme variants exclusively composed of either the long or the short protein were kinetically indistinguishable.The hheB GP1gene was amplified by PCR,and the second possible start codon was fused into the Nco I site of pGEF ϩto yield the expression vector pGEFHheB GP1.In this way,a homotet-rameric protein consisting only of the shorter 245-amino-acid polypeptide was produced.Substrate range of halohydrin dehalogenases.All proteins were expressed in a soluble and active form up to 15to 25%of the total cellular protein content of E.coli BL21(DE3)as judged by SDS-PAGE.Further experiments focused on HheC,because it is enantioselective with various valuable halohydrinsT A B L E 1.P a i r w i s e s e q u e n c e i d e n t i t i e s o f h a l o h y d r i n d e h a l o g e n a s e s a n d S D R sE n z y m e a c r o n y m a n d f u n c t i o nO r g a n i s mS o u r c e o r r e f e r e n c eI d e n t i t y (%)H h e AH H e A A D 2H h e BH h e B G P 1H h e CH a l B1A E 12A E 11A H H2H S D 1A U 4H a l o h y d r i n d e h a l o g e n a s e s G r o u p A H h e A C o r y n e b a c t e r i u m s p .s t r a i n N -10743610097.124.124.032.029.224.324.125.524.219.9H h e A A D 2A r t h r o b a c t e r s p .s t r a i n A D 2T h i s s t u d y10024.524.232.429.624.624.125.123.520.2G r o u p B H h e B C o r y n e b a c t e r i u m s p .s t r a i n N -10743610098.425.423.920.918.722.224.218.0H h e B G P 1M y c o b a c t e r i u m s p .s t r a i n G P 1T h i s s t u d y10025.823.920.218.721.523.818.2G r o u p C H h e C A .r a d i o b a c t e r A D 1T h i s s t u d y 10080.722.223.125.523.720.4H a l BA .t u m e f a c i e n s—a10017.620.622.621.720.2S D R o x i d o r e d u c t a s e s 1A E 1(t r o p i n o n e r e d u c t a s e I )D a t u r a s t r a m o n i u m 810061.529.727.421.92A E 1(p r o p i n o n e r e d u c t a s e I I )D .s t r a m o n i u m 810032.427.620.41A H H (7␣-h y d r o x y s t e r o i d d e h y d r o g e n a s e )E .c o l i 2910032.219.72H S D (20--h y d r o x y s t e r o i d d e h y d r o g e n a s e )S t r e p t o m y c e s e x f o l i a t u s 1110022.61A 4U (a l c o h o l d e h y d r o g e n a s e )D r o s o p h i l a l e b a n o n e n s i s3100a—,u n p u b l i s h e d (a c c e s s i o n n o .A A D 34609).5060VANHYLCKAMA VLIEG ET AL.J.B ACTERIOL .(16).Moreover,its sequence is very different from that of halohydrin dehalogenases that have previously been character-ized.HheC was purified by anion-exchange chromatography followed by hydrophobic interaction chromatography as de-scribed in Materials and Methods.Purified HheC displayed optimal activity at around pH8.0to9.0,and the temperature optimum for activity was50°C.Purified HheC and crude extracts of E.coli BL21(DE3) overexpressing HheA AD2or HheB GP1were used to study the substrate range of the three halohydrin dehalogenases(Table 2).Similar to the known HheA and HheB halohydrin dehalo-genases,all three enzymes were active with all chlorinated and brominated C2and C3vicinal halohydrins tested.The only exception was HheA AD2,for which no activity was detected with2,3-dichloro-1-propanol.The activities with brominated substrates were in most cases higher than with their chlori-nated analogs.The substrate range of recombinant HheA AD2 was in agreement with that of the enzyme isolated from strain AD2(31).The substrate range of HheB GP1was similar to those reported for HheB(20,21)(Table2)and DehA(2). Halohydrin dehalogenase HheC displayed a relatively high level of activity with chloroacetone,which clearly distinguishes this enzyme from HheA AD2and HheB GP1.The kinetics of 1,3-dichloro-2-propanol conversion by HheC were evaluated by measuring initial degradation rates at various substrate con-centrations(Table3).The enzyme followed Michaelis-Menten kinetics with a k cat value of37sϪ1and a K m value of0.010mM. The specific activity of HheC with this substrate is similar to that reported for HheA AD2(31).However,the K m value of HheC for1,3-dichloro-2-propanol is2to3orders of magnitude lower than those of HheA,HheA AD2,and HheB(17,21,31). Recently,it was shown that aromatic halohydrins can also be dehalogenated by HheC(16).To explore the substrate speci-ficity of HheC,the steady-state kinetic parameters of HheC with a range of aliphatic and aromatic substrates were deter-mined(Table3).It was found that HheC can efficiently convert aliphatic and aromatic halohydrins.Purified HheC has k cat and K m values for(R)-2-chloro-1-phenylethanol of48.5sϪ1and 0.37mM,respectively,and for the(S)-enantiomer,these val-ues are8.9sϪ1and4.2mM(Table3).From these steady-state kinetic data,it can be calculated that the enantioselectivity (E-value)of HheC for this substrate is73,which is in close agreement with the value calculated from kinetic resolutions by Lutje Spelberg et al.(16).We tested whether HheA AD2and HheB GP1could also convert2-chloro-1-phenylethanol,and found that both enzymes were active with both enantiomers. The highest activities were observed with the(S)-enantiomer (Table2).The enantioselectivity of2-chloro-1-phenylethanol conversion by HheB GP1was evaluated by means of kinetic resolution(data not shown),which showed that the(S)-enan-tiomer was preferentially converted with an E-value of8. Hence,HheC and HheB GP1exhibit opposite enantioselectivi-ties for2-chloro-1-phenylethanol conversion.Sequence similarities of halohydrin dehalogenases with SDRs.The pairwise sequence identities of the three halohydrin dehalogenases described in this paper are between24.2and 32.4%(Table1).Interestingly,similarity searches with the amino acid sequences of HheA AD2,HheB GP1,and HheC in various protein and DNA databases showed that these en-zymes are similar to proteins belonging to the family of SDR enzymes.Sequence similarities of up to25.5%were observed with well-characterized SDR family members(Table1).The SDR family constitutes a large number of proteins that cata-lyze oxidation or reduction reactions with NAD(H)or NADP(H)as a cofactor(13).They are active as dimers or tetramers,where each monomer consists of approximately250 residues.Halohydrin dehalogenases consist of subunits with similar sizes,and HheA(17)and HheB(20)also appear to be active as tetramers.In Fig.1is depicted an alignment of six halohydrin dehalo-genase sequences together withfive members of the SDRTABLE2.Substrate range of halohydrin dehalogenases HheC,HheA AD2,and HheB GP1Substrate%of activity aHheC HheA AD2HheA HheB GP1HheB1,3-Dichloro-2-propanol100100100100100 (R,S)-2,3-Dichloro-1-propanol10.1Ͻ0.050.150.330.12 3-Chloro-1,2-propanediol18.89.3035.1 1.760.98 2-Chloroethanol 5.300.310.190.730.10 1,3-Dibromo-2-propanol80.26,68020,00076.31662-Bromoethanol12870.6 5.871.89.15 Chloroacetone1657.89 1.78(R)-2-Chloro-1-phenylethanol13.2 3.62 1.73(S)-2-Chloro-1-phenylethanol24.215.516.9a Activity was measured with purified HheC and crude extracts of E.coli BL21(DE3)overexpressing HheA AD2or HheB GP1.With purified HheC,a rate of100% corresponds to20.7mol of halide released minϪ1mg of proteinϪ1with1,3-dichloro-2-propanol as the substrate.With crude extracts of E.coli BL21(DE3) overexpressing HheA AD2or HheB GP1,these values were0.480and8.52mol minϪ1mg of proteinϪ1,respectively.Relative activities of HheA and HheB were obtained from reference21.TABLE3.Steady-state kinetic parameters of purified HheCSubstrate k cat(sϪ1)K m(mM)k cat/K m(sϪ1MϪ1)1,3-Dichloro-2-propanol370.010 3.7ϫ106(R,S)-2,3-Dichloro-1-propanol 6.50.827.9ϫ1032-Chloroethanol 3.90.84 4.6ϫ103Chloroacetone235 2.49.8ϫ1042-Bromoethanol26.5Ͻ0.2Ͼ1.3ϫ105(R)-2-Chloro-1-phenylethanol48.50.37 1.3ϫ105(S)-2-Chloro-1-phenylethanol8.9 4.2 2.2ϫ103(R)-p-Nitro-2-bromo-1-phenylethanol75Ͻ0.01Ͼ7.5ϫ106V OL.183,2001HALOHYDRIN DEHALOGENASES ARE RELATED TO SDR PROTEINS5061protein family for which the three-dimensional structure is known.The highest degree of conservation between members of the SDR protein family is observed within the N-terminal part of these proteins,which contains a typical (G/A)-(G/A)-X-X-(G/A)-X-G fingerprint.This fingerprint is characteristic for the Rossman fold of the cofactor binding site (13),but it is not conserved in the halohydrin dehalogenases.The absence of a cofactor-binding motif in halohydrin dehalogenases is in agreement with the fact that the dehalogenation reaction that is catalyzed by these enzymes is not a redox reaction.The C-terminal part of members of the SDR family is in-volved in substrate binding and is much less conserved within the SDR protein family.However,it contains three highly conserved residues (a Ser,Tyr,and Lys)that play a critical role in catalysis (11,13,29).The conserved serine and tyrosine residues can also be identified in the halohydrin dehalogenase sequences (Ser132and Tyr145in HheC).However,halohydrin dehalogenases differ from members of the SDR family by the presence of an arginine residue at a position at which a lysine is conserved in the SDR family (Fig.1).The implications of the partial conservation of the active site residues for the catalytic mechanism of halohydrin dehalogenases will be discussed be-low.Conservation in several other sequence regions alsosuggestsFIG.1.Sequence alignment of halohydrin dehalogenases with SDRs with known three-dimensional structures.The sequences were aligned by using the multiple alignment program ClustalW and are shown in decreasing sequence similarity to HheC.Amino acids that are identical in six or more sequences are depicted below the sequence.Positions at which six or more similar residues occur are indicated by an asterisk.The position of the G/A-G/A-X-X-G/A-X-G/A fingerprint typical of the Rossman fold in the SDR family is indicated by $on top of the alignment.The proposed active site residues in halohydrin dehalogenases are indicated by #.The structural elements identified in three-dimensional structures of SDR family members are underlined.The first line below the sequence alignment shows the secondary structure elements and the nomenclature of 7␣-hydroxysteroid dehydrogenase (29).-Strands are indicated as double broken lines,and ␣-helices are indicated as single broken lines.The second,third,and fourth lines show the predicted secondary structure elements for HheC,HheA AD2,and HheB GP1,respectively.The nomen-clature of the sequences is explained in Table 1.5062VANHYLCKAMA VLIEG ET AL.J.B ACTERIOL .。