purification of GST-tagged proteins
- 格式:doc
- 大小:27.00 KB
- 文档页数:1

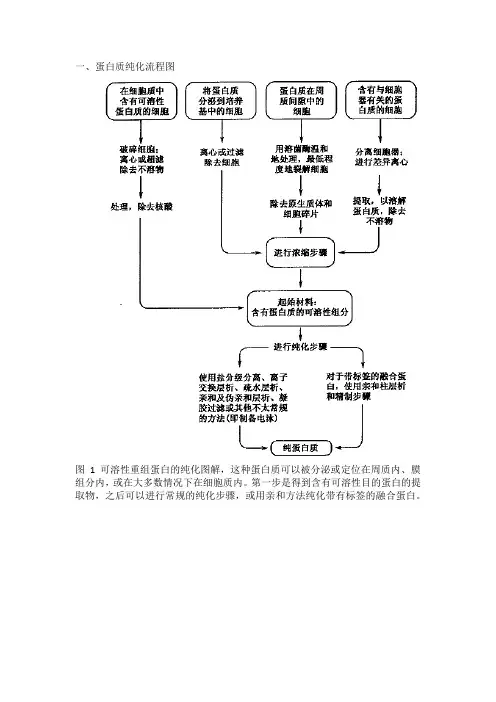

蛋白异源表达纯化那些事儿研究蛋白质功能、生产功能型蛋白质基本都离不开蛋白质的异源表达及纯化,今天小编将从以下几个方面与大家聊一聊蛋白质表达、纯化的那些事。
1、标签选择目前常见的表达标签有His-tag(组氨酸标签)、GST-tag(谷胱甘肽巯基转移酶标签)、MBP-tag(麦芽糖结合蛋白标签)、CBD-tag(几丁质结合区标签)等。
每种标签都具有独特的性质,比如标签的分子量、对蛋白可溶性的调节能力、对蛋白折叠的影响、是否需切除等。
根据自己要纯化的蛋白的特性,选择合适的标签。
标签选的好,后续的实验事半功倍。
2、载体构建选好要用的标签后,就要准备构建表达载体了。
目前已有很多商品化的载体可供选择,例如带His-tag的pET28系列,带MBP-tag的pMAL系列等。
绝大多数的载体系列,都包含了标签插入N端或C端的形式,可以根据自己的蛋白的特质进行灵活选择。
插入蛋白的编码基因时需要注意,不要出现移码。
如果想要自己从头构建载体,别忘了插入合适的启动子和转录终止序列。
启动子序列一般使用诱导型启动子,组成型启动子会导致外源蛋白过早表达积累,不利于宿主增殖。
3、宿主选择常见的宿主有大肠杆菌、哺乳动物细胞、酵母以及昆虫。
以大肠杆菌来说,最适合做异源表达的菌株为BL21及其衍生菌株,但仍需要根据载体中选择的转录、翻译元件来挑选。
BL21中内源性的蛋白酶基因缺失,因此是一种适合于外源蛋白表达的菌株,但是它没有T7 RNA 聚合酶,所以不能用于含T7启动子蛋白表达;BL21(DE3),在BL21的基础上整合了T7噬菌体基因组,适合T7表达系统,如pET系列;BL21(DE3)PLySs,在BL21(DE3)基础,附带了T7溶菌酶基因,可以更为严谨控制T7聚合酶的表达。
需要注意的是,由于核酸内切酶(endA1)重组酶(recA1)等基因的存在,质粒在BL21系列菌株中不那么稳定,可能出现整合至基因组、丢失、突变等现象,所以构建好的载体仍需保存在DH5α之类的菌株中。

GST bestarose 4FF说明书默认分类2010-01-18 09:32:34 阅读257 评论0 字号:大中小GST bestarose 4FF是专门用于纯化pGEX系列载体表达的GST标签蛋白,其它谷胱甘肽转移酶以及与谷胱甘肽有亲和作用蛋白的分离介质,操作简单,快速。
GST bestarose 4FF 可直接从预处理的细胞裂解物中纯化GST-tagged蛋白,纯化条件温和,可以保证蛋白的生物活性。
GST bestarose 4FF 具有高流动性,易于放大,GST bestarose 4FF有快速方便的预装柱,Ez-Fast bestarose 4FF 1ml和5ml。
介质性能谷胱甘肽是通过10碳原子的连接臂偶联到4%高度交联的琼脂糖上。
最优的偶联作用使介质具有高GST-tagged蛋白及与谷胱甘肽相互作用蛋白的结合能力。
总的蛋白结合能力约为每毫升填料结合10mg标签蛋白,动态结合能力随着外界因素改变,如不同的目标蛋白,流速等等。
如果需要去除GST部分(天然蛋白分子量为26KD),当蛋白吸附在柱上,或在洗脱后用位点专一的蛋白酶消化,选用前种方法,可减少额外分离目的蛋白与GST步骤,消化后,目的蛋白直接用结合液洗脱即可。
表1 GST bestarose 4FF性能配体谷胱甘肽与10碳原子连接臂配体浓度 120-320μmol谷胱甘肽/ml填料蛋白结合能力≈ 10mg重组GST-tagged蛋白/ml填料,GST,Mr26000动态结合能力≈11mgGST-tagged蛋白/ml填料,Mr43000平均颗粒大小90μm组成 4%高度交联的琼脂糖最大流速* 450cm/h(15ml/min),XK层析柱,柱高5cm,水溶性缓冲液,室温纯化建议的流速** 上样:<100cm/h(<3ml/min ,XK层析柱)洗涤与洗脱:100-300cm/h(3-10ml/min,XK层析柱)化学稳定性所有常用的水溶性缓冲液中稳定,如:1M醋酸盐,pH7.4,6M 盐酸胍,室温,1hpH稳定性 pH3-12贮存温度 +4-30℃贮存液 20%乙醇* 水是室温的。

1、Western blotting:Western blotting using ECL reagentused by:Laboratory of P.J. Hansen,Dept. of Animal Sciences, University of FloridaKamps's Western Blotting Protocolused by:Sefton Lab, Salk InstituteDry Transfer 干法蛋白转膜used by:The Preuss Lab,The Division of Biological Sciences,The University of ChicagoWestern Blot of TBP from TBP-GST bacteriaused by:Steven Finkbeiner, Departments of Neurology and Physiology, UCSFWestern Blot/Anti-P-CREBused by:Steven Finkbeiner, Departments of Neurology and Physiology, UCSFWestern blotting protocol for 1C2, 3B5H10, and 4C8 Antibodiesused by:Steven Finkbeiner, Departments of Neurology and Physiology, UCSFProtocol for anti-HA antibody Western Blottingused by:Steven Finkbeiner, Departments of Neurology and Physiology, UCSFProbing and Stripping of Western Blotused by:Hahn Lab,The Fred Hutchinson Cancer Research Center and Howard Hughes Medical Institute Western Blotting Protocolused by:Mirmira Laboratory at the University of VirginiaWestern Blotting and Immunostainingused by:Chazin Lab, Center for Structural Biology, Vanderbilt UniversityWestern Blotsused by:Lab of Dr. Mark Barton Frank, Oklahoma Medical Research FoundationWestern Blotting - Antibodiesused by:The Minion Lab, College of Veterinary Medicine at Iowa State UniversityWestern Blottingused by:Cepko/Tabin Lab, Harvard UniversityCarbonate Solution For Western Blotused by:Dr. DE Koshland, Carnegie Institution of WashingtonWestern Blot with BSAused by:Dr. DE Koshland, Carnegie Institution of WashingtonMETHOD for Western Blotsused by:Dr. DE Koshland, Carnegie Institution of WashingtonWestern Blotsused by:Michael Koelle's Lab,Department of Molecular Biophysics and BiochemistryYale University2、蛋白分离/提取/浓缩Lysis Buffersused by:Steven Finkbeiner, Departments of Neurology and Physiology, UCSFYeast Protein Isolation "Yaffe-Schatz"used by:Amberg Lab ,Upstate Medical UniversityGuanidine Hydrochloride Purification of Proteins From SDS PAGEused by:Gottschling Lab,The Fred Hutchinson Cancer Research Center and Howard Hughes Medical Institute Yeast protein prep for SDS PAGE and western (rapid)used by:Gottschling Lab,The Fred Hutchinson Cancer Research Center and Howard Hughes Medical Institute Yeast protein prep for SDS PAGE and western (glass bead)used by:Hahn Lab,The Fred Hutchinson Cancer Research Center and Howard Hughes Medical Institute Purification of TFIIIC from yeastused by:Hahn Lab,The Fred Hutchinson Cancer Research Center and Howard Hughes Medical Institute Purification of TFIIA from E. coliused by:Hahn Lab,The Fred Hutchinson Cancer Research Center and Howard Hughes Medical InstituteTCA protein precipitationused by:Hahn Lab,The Fred Hutchinson Cancer Research Center and Howard Hughes Medical Institute Purification of MLCK ( Myosin Light Chain Kinase )from Rabbit Skeletal Muscle肌球蛋白轻链激酶used by:Stull Lab,University of Texas, Southwestern Medical CenterPurification of Calmodulin from Bovine Brainused by:Stull Lab,University of Texas, Southwestern Medical CenterMyosin Light Chain Preparation from Skeletal and Cardiac Muscleused by:Stull Lab,University of Texas, Southwestern Medical CenterPurifying Protein from Inclusion Bodiesused by:Chazin Lab, Center for Structural Biology, Vanderbilt UniversityProtocol for Protein Extraction from Plantused by:The University of Nebraska-Lincoln Protein Core Facility (PCF)Taq Polymerase Purificationused by:Cepko/Tabin Lab, Harvard UniversityYeast Protein Prep (No Boil)used by:Dr. DE Koshland, Carnegie Institution of WashingtonYeast Protein Prep (Boil)used by:Dr. DE Koshland, Carnegie Institution of WashingtonGST Fusion Protein Prepused by:Vesicle Trafficking, Stanford University3、蛋白质检测分析:AMIDO BLACK STAINused by:Hancock Laboratory Methods,Department of Microbiology and Immunology,University of British Columbia, British Columbia, CanadaJames Hardwick's angiotensin assay protocolused by:Sefton Lab,Salk InstitutePhosphoamino acid analysis:Mark Kamps's methodused by:Sefton Lab,Salk InstituteULTIMATE HIS-UB ASSAY FORused by:the Tansey Lab at Cold Spring Harbor LaboratoryULTIMATE HIS-UB ASSAY FOR YEASTused by:the Tansey Lab at Cold Spring Harbor LaboratoryProtocol for Beta-Hexosaminidase RBL-2H3 Secretion Assayused by:meyer lab,Stanford UniversityMeasurement of Myosin Regulatory Light Chain (RLC) Phosphorylationused by:Stull Lab,University of Texas, Southwestern Medical CenterBicinchoninic Acid (BCA) Protein Assayused by:David R. Caprette, Rice UniversityBiuret Protein Assayused by:David R. Caprette, Rice UniversityBradford protein assayused by:David R. Caprette, Rice UniversityHartree-Lowry and Modified Lowry Protein Assaysused by:David R. Caprette, Rice UniversityColorimetric Assaysused by:David R. Caprette, Rice UniversityKd measurement, fitting, calculation, and simulationused by:Chazin Lab, Center for Structural Biology, Vanderbilt University4、蛋白标记修饰DSP Crosslinkingused by:Peter Novick Lab, Department of Cell Biology Yale University School of Medicine Labeling Gc-Globulin with Carboxyfluorescein Succinimidyl Esterused by:Yu-Li Wang Lab,University of massachusetts Medical School(UMASS)Labeling Tubulin with Tetramethylrhodamine Succinimidyl Esterused by:Yu-Li Wang Lab,University of massachusetts Medical School(UMASS)Labeling Gizzard Vinculin with Tetramethylrhodamine Iodoacetamideused by:Yu-Li Wang Lab,University of massachusetts Medical School(UMASS)Labeling Gizzard Alpha-actinin with Tetramethylrhodamine Iodoacetamideused by:Yu-Li Wang Lab,University of massachusetts Medical School(UMASS)Labeling Gizzard Myosin with Tetramethylrhodamine Iodoacetamide (primarily 17kD MLC) used by:Yu-Li Wang Lab,University of massachusetts Medical School(UMASS)Labeling Gizzard Myosin with Tetramethyorhodamine Iodoacetamide (MHC and 17kD MLC)used by:Yu-Li Wang Lab,University of massachusetts Medical School(UMASS) Labeling Muscle Actin with N-(1-pyrene) Iodoacetamideused by:Yu-Li Wang Lab,University of massachusetts Medical School(UMASS) Labeling Muscle Actin with Tetramethylrhodamine Iodoacetamideused by:Yu-Li Wang Lab,University of massachusetts Medical School(UMASS) Labeling Muscle Actin with Carboxyfluorescein Succinimidyle Esterused by:Yu-Li Wang Lab,University of massachusetts Medical School(UMASS) Labeling Muscle Actin with 5-Iodoacetamidofluoresceinused by:Yu-Li Wang Lab,University of massachusetts Medical School(UMASS) ultimate mammalian cell pulse-chaseused by:the Tansey Lab at Cold Spring Harbor Laboratoryultimate yeast pulse-chaseused by:the Tansey Lab at Cold Spring Harbor LaboratoryGluteraldehyde Conjugation of Oligopeptides to Proteinsused by:Lab of Dr. Mark Barton Frank, Oklahoma Medical Research Foundation Fluorescein labeling of proteinsused by:David Chambers,Salk instituteBiosynthetic labelingused by:Sefton Lab, Salk Institute , San Diego, California5、免疫沉淀(IP/CoIP/ChIP)Immunoprecipitation buffers and protocolsused by:Steven Finkbeiner, Departments of Neurology and Physiology, UCSFCell Lysis/Western/IPused by:Steven Finkbeiner, Departments of Neurology and Physiology, UCSF Amplification and Labelling with Cy dyesused by:Yale Genome Analysis Center, Yale UniversityChromatin Immunoprecipitation from Mammalian Cell Extractsused by:Yale Genome Analysis Center, Yale UniversityChromatin Immunoprecipitation from Yeast Whole Cell Extractsused by:Yale Genome Analysis Center, Yale UniversityImmunoprecipitationused by:Hancock Laboratory Methods. Department of Microbiology and Immunology, University of British Columbia, British Columbia, CanadaUltimate yeast denaturing IPused by:the Tansey Lab at Cold Spring Harbor LaboratoryULTIMATE FREEZE-THAW LYSIS FORused by:the Tansey Lab at Cold Spring Harbor LaboratoryUltimate mammalian nondenaturing IPused by:the Tansey Lab at Cold Spring Harbor LaboratoryUltimate mammalian denaturing IPused by:the Tansey Lab at Cold Spring Harbor LaboratoryUltimate yeast ChIP assayused by:the Tansey Lab at Cold Spring Harbor Laborat oryChromatin Immunoprecipitation Assay and PCRused by:Hahn Lab,The Fred Hutchinson Cancer Research Center and Howard Hughes Medical Institute Yeast Chromatin Immunoprecipitation (ChIP)used by:Hahn Lab,The Fred Hutchinson Cancer Research Center and Howard Hughes Medical Institute Mirmira Lab ChIP Protocolused by:Mirmira Laboratory at the University of VirginiaChIP Assay Protocolused by:Mirmira Laboratory at the University of VirginiaImmunoprecipitation and Immune Complex kinase assayused by:Sefton Lab, Salk InstituteGeneral Principles of Immunoprecipitationused by:Sefton Lab, Salk Institute6、融合蛋白表达及纯化Use of B-PER lysis reagent to purify recombinant proteins tagged with His6 or maltose binding protein used by:Smith Lab,Division of Biological Sciences,University of MissouriBacterial Expression of Kinesin Motorsused by:Liz Greene and Steve HenikoffPurification of recombinant sBRF M166Lused by:Hahn Lab,The Fred Hutchinson Cancer Research Center and Howard Hughes Medical Institute Protein Expression and Purification of RPA70-ABused by:Chazin Lab, Center for Structural Biology, Vanderbilt UniversityCo-Expression and purification of human DNA primase p49-p58 subunits引发酶p49-p58亚基共表达纯化方法used by:Chazin Lab, Center for Structural Biology, Vanderbilt UniversityProtein Expression and Purification of MBP-T antigen MBP-T抗原表达纯化used by:Chazin Lab, Center for Structural Biology, Vanderbilt UniversityPerspectives on Baculovirus Expression Systems表达used by:Lab of Dr. Mark Barton Frank, Oklahoma Medical Research FoundationExpression & Purification of His Tagged Proteins in E. coliused by:Cepko/Tabin Lab, Harvard University7、相互作用分析Amplifying a large phage-display library without losing diversityused by:Smith Lab,Division of Biological Sciences,University of MissouriProtein Interaction Analysis Using an IASYS Biosensorused by:Kitto Lab, The University of Texas at AustinYeast Two-Hybrid Screen with Library and Baitused by:Mirmira Laboratory at the University of Virginia8、蛋白电泳/双向电泳SDS-PAGEused by:Steven Finkbeiner, Departments of Neurology and Physiology, UCSFGel Drying干胶used by:Steven Finkbeiner, Departments of Neurology and Physiology, UCSFNuPAGE Gel Electrophoresis 蛋白电泳used by:Kitto Lab, The University of Texas at AustinIsoelectric Focussing of Membrane Proteins by Slab Gel Methodused by:Hancock Laboratory Methods,Department of Microbiology and Immunology,University of British Columbia, British Columbia, CanadaSilver Staining SDS PAGE Gelsused by:Hahn Lab,The Fred Hutchinson Cancer Research Center and Howard Hughes Medical Institute E. coli total protein for SDS PAGEused by:Hahn Lab,The Fred Hutchinson Cancer Research Center and Howard Hughes Medical Institute Comassie Blue Protein gel stainused by:Hahn Lab,The Fred Hutchinson Cancer Research Center and Howard Hughes Medical Institute Coomassie & Silver Staining of Polyacrylamide Gelsused by:Lab of Dr. Mark Barton Frank, Oklahoma Medical Research Foundation2-D Polyacrylamide Gel Electrophoresisused by:Molecular Profiling Initiative, National Cancer Institute(NCI)Silver Staining of SDS-PAGE Gelsused by:The Minion Lab, College of Veterinary Medicine at Iowa State UniversitySDS-PAGE Gelsused by:The Minion Lab, College of Veterinary Medicine at Iowa State UniversityCuCl2 Staining of SDS-PAGE Gelsused by:Cepko/Tabin Lab, Harvard UniversityElectroelution of Proteins From SDS-PAGE Gelsused by:Drs. C Cepko and C Tabin, Harvard UniversitySilver Stain of Protein Gelsused by:Dr. DE Koshland, Carnegie Institution of WashingtonSDS-PAGE (BIO-RADused by:Dr. DE Koshland, Carnegie Institution of WashingtonSDS-PAGEused by:Michael Koelle's Lab,Department of Molecular Biophysics and Biochemistry Yale University9、质谱分析Polycrystalline thin films质谱样品准备used by:Prowl Lab,Rockefeller UniversitySeeded films (Vorm-Roepstorff)used by:Prowl Lab,Rockefeller UniversityDried dropletused by:Prowl Lab,Rockefeller UniversityIn-gel Tryptic Digest for Protein ID by Mass Spectrometryused by:Mitshison Lab, Department of Systems Biology, Harvard Medical School10、crystallizationPolycrystalline thin filmsused by:PROWL, Rockefeller UniversitySlow crystallizationused by:PROWL, Rockefeller UniversityCrystallization of Kinesin Family Motor Proteins驱动蛋白家族的结晶条件和晶体参数used by:Liz Greene and Steve HenikoffCrystallization Trialsused by:Liz Greene and Steve Henikoff11、in vitro protein synthesisProtein Syntheses in Cell Free Systemsused by:The Laboratory of William Heidcamp at Gustavus Adolphus University12、蛋白纯化/层析6xHis-tagged protein purification using Qiagen Ni-NTA Column under Native Conditionsused by:Sugden lab,McArdle Laboratory for Cancer Research, University of Wisconsin-Madison Medical School Purification of dnEBNA-1/Soft from E. coli BL21 LysSused by:Sugden lab,McArdle Laboratory for Cancer Research, University of Wisconsin-Madison Medical School Purification of GST fusion proteins in E.coli GST融合蛋白纯化(方法四)筛选表达株used by:Sugden lab,McArdle Laboratory for Cancer Research, University of Wisconsin-Madison Medical School Purification of GST fusion proteins in E.coli GST融合蛋白纯化,方法三,纯化小量蛋白used by:Sugden lab,McArdle Laboratory for Cancer Research, University of Wisconsin-Madison Medical School Purification of GST fusion proteins in E.coli GST融合蛋白纯化,方法二used by:Sugden lab,McArdle Laboratory for Cancer Research, University of Wisconsin-Madison Medical School Purification of GST fusion proteins in E.coli GST融合蛋白纯化,方法一used by:Sugden lab,McArdle Laboratory for Cancer Research, University of Wisconsin-Madison Medical School Purification of 6xHis epitope tagged proteins by Ni-NTA-Agarose His标签蛋白纯化used by:Sugden lab,McArdle Laboratory for Cancer Research, University of Wisconsin-Madison Medical School Fusion Protein Isolation 融合蛋白分离纯化used by:Peter Novick Lab, Department of Cell Biology Yale University School of MedicineAffinity Column Preparation 亲和层析柱制备used by:Peter Novick Lab, Department of Cell Biology Yale University School of MedicinePreparation of Affinity Column制备亲和层析柱used by:Steven Finkbeiner, Departments of Neurology and Physiology, UCSFColumn Buffersused by:Steven Finkbeiner, Departments of Neurology and Physiology, UCSF13、蛋白定量Glutathione - a microassay for determining glutathione content in cells 检测细胞谷胱苷肽含量used by:Laboratory of P.J. Hansen,Dept. of Animal Sciences, University of FloridaBradford Protein Assay - a microassay for determining protein content in a 96-well microtiter plate format used by:Laboratory of P.J. Hansen,Dept. of Animal Sciences, University of FloridaLowry Protein Assayused by:Laboratory of P.J. Hansen,Dept. of Animal Sciences, University of FloridaBradford Assayused by:Kitto Lab, The University of Texas at Austin欢迎您的下载,资料仅供参考!致力为企业和个人提供合同协议,策划案计划书,学习资料等等打造全网一站式需求精品文档。

GE HealthcareInstructions 71-5016-96 AK HiTrap affinity columns GSTrap FF,1 ml and 5 mlGSTrap™ FF columns are prepacked 1 ml and 5 ml HiTrap™ columns for convenient, one-step purification of glutathione S-transferase (GST) tagged proteins produced using the pGEX series of expression vectors, other glutathione S-transferases and glutathione binding proteins.GST-tagged proteins can be purified directly from pretreated bacterial lysates using GSTrap FF. Tagged proteins are eluted under mild, nondenaturing conditions that preserve protein antigenicity and function.The medium, Glutathione Sepharose™ 4 Fast Flow, is also available as lab packages and is an excellent choice for scale-up.The columns can be operated with a syringe, peristaltic pump or liquid chromatography system such as ÄKTAdesign™ or FPLC™ System.Code No. Product No. supplied 17-5130-01 GSTrap FF 5 × 1 ml17-5130-02 GSTrap FF 2 × 1 ml17-5130-05 GSTrap FF 100 × 1 ml* 17-5131-01 GSTrap FF 1 × 5 ml17-5131-02 GSTrap FF 5 × 5 ml17-5131-05 GSTrap FF 100 × 5 ml* * Special package delivered on specific customer order.ConnectorkitConnectors supplied Usage No. supplied 1/16” male/luer female Connection of syringe to top ofHiTrap column 1 Tubing connector Connection of tubing (e.g. Peristalticflangeless/M6 female Pump P1) to bottom of HiTrap column* 1 Tubing connector Connection of tubing (e.g. Peristalticflangeless/M6 male Pump P1) to top of HiTrap column** 1 Union 1/16” female/ Connection to original FPLC SystemM6 male through bottom of HiTrap column 1 Union M6 female/ Connection to original FPLC System1/16” male through top of HiTrap column 1 Stop plug female, 1/16” Sealing bottom of HiTrap column 2, 5 or 7 * Union 1/16” female/M6 male is also needed.** Union M6 female/1/16” male is also needed.Tables of contents1. Description 32. Operation 53. Scaling up 74. Storage 75. Cleavage of GST-tagged proteins 76. Troubleshooting guide 127. References 178. Ordering information 18p.p.1. DescriptionMedium propertiesGlutathione Sepharose 4 Fast Flow is designed for purification ofglutathione S-transferase (GST) tagged proteins produced using the pGEX series of expression vectors (1), other glutathione S-transferases andglutathione binding proteins. GST-tagged proteins can be purified directly from pretreated bacterial lysates with a one-step method using GSTrap FF. The tagged proteins are eluted under mild, non-denaturing conditions that preserve protein antigenicity and function. The glutathione ligand is coupled via a 10-carbon linker to highly cross-linked 4% agarose. The coupling is optimized to give high binding capacity for GST-tagged proteins and other glutathione binding proteins.The total binding capacity is approximately 10 mg recombinant GST/mlmedium. The dynamic binding capacity will vary depending on the flow rate and the sample. If removal of the GST-tag (a naturally occurring M r 26 000 protein) is required, the tagged protein can be digested with the appropriate site-specific protease while bound to GSTrap FF or, alternatively, after elution. Cleavage on GSTrap FF eliminates the extra step of separating the released protein from GST, since the GST-tag remains bound. The target protein is eluted using binding buffer.ColumnThe columns are made of polypropylene, which is biocompatible and non-interactive with biomolecules. The columns have porous top and bottom frits that allow high flow rates. The columns are delivered with a stopper on the inlet and a snapoff end on the outlet. The separation can be easily achieved using a syringe together with the supplied adaptor, a pump, or a chromatography system such as ÄKTA ™ or FPLC.Note: To prevent leakage it is essential to ensure that the adaptor is tight.Several columns can be connected in series to increase binding capacity. (Backpressure will increase).The column cannot be opened or refilled.The characteristics of GSTrap FF are summarized below.Table . GSTrap FF characteristicsColumn dimensions (i.d. × h) 0.7 × 2.5 cm (1 ml) and 1.6 × 2.5 cm (5 ml)Column volumes 1 ml and 5 ml respectivelyLigand Glutathione and 10-carbon linker armLigand concentration 120–320 µmol glutathione/ml mediumBinding capacity* ≈ 10 mg recombinant glutathioneS-transferase/ml medium GST, M r 26 000 Dynamic binding capacity* ≈ 11 mg GST-tagged protein/ml mediumM r 43 000 (GSTrap FF 1 ml at 1 ml/min) Average particle size 90 µmBead structure Highly cross-linked 4% agaroseMaximum back pressure 0.3 MPa, 3 barMaximum flow rate 4 ml/min and 15 ml/min for 1 and 5 mlcolumns respectivelyRecommended flow rates* Sample loading: 0.2–1 ml/min (1 ml ) and1–5 ml (5 ml)Washing and elution: 1–2 ml/min (1 ml) and 5–10 ml/min (5 ml)Chemical stability All commonly used aqueous buffers, e.g.1 M acetate pH 4.0 and 6 M guanidinehydrochloride for 1 hour at room temperature pH stability pH 3–12Storage temperature + 4 to + 30 °CStorage 20 % ethanol*Note:Binding of GST to glutathione is flow dependent and lower flow rates often increase the binding capacity. This is important during sample loading and elution. Proteincharacteristics, pH and temperature may also affect the binding capacity.p.p. 52. OperationThe columns can be operated with a syringe, peristaltic pump or a liquid chromatography system.Buffer preparationWater and chemicals used for buffer preparation should be of high purity. We recommend filtering the buffers by passing them through a 0.45 µm filter before use.Binding buffer: PBS, pH 7.3 (140 mM NaCl, 2.7 mM KCl, 10 mMNa 2HPO 4, 1.8 mM KH 2PO 4, pH 7.3)Elution Buffer:50 mM Tris-HCl, 10 mM reduced glutathione, pH 8.0Note: 1–10 mM DTT can be included in the binding and elution buffers.Sample preparationThe sample should be centrifuged and/or filtered through a 45 µm filter immediately before it is applied to the column. If the sample is too viscous, dilute it with binding buffer to prevent clogging the column.Purification1. Fill the pump tubing or syringe with binding buffer. Connect the columnto the syringe (use the adaptor supplied) or pump tubing “drop to drop” to avoid introducing air into the column.2. Remove the snap-off end at the column outlet.3. Equilibrate the column with 5 column volumes of binding buffer.4. Apply the sample using a syringe fitted to the luer adaptor or bypumping it onto the column. For best results, use a flow rate of0.2–1 ml/min (1 ml column) and 1–5 ml/min (5 ml column) during sample application.p. 65. Wash with 5–10 column volumes of binding buffer or until no materialappears in the effluent. A flow rate of 1–2 ml/min (1 ml column) and 5–10 ml/min (5 ml column) is recommended for washing.6. Elute with 5–10 column volumes of elution buffer. A flow rate of1–2 ml/min (1 ml column) and 5–10 ml/min (5 ml column) is recommended for elution.Note: • One of the most important parameters affecting the bindingof GST-tagged proteins or other glutathione binding proteins to GSTrap FF is the flow rate. Due to the relatively slow binding kinetics between GST and glutathione, it is important to keep the flow rate low during sample application for maximum binding capacity. Protein characteristics, pH and temperature are other factors that may affect the binding capacity.• Volumes and times used for elution may vary among fusionproteins. Additional elutions with higher concentrations of glutathione may be required. Flowthrough, wash and eluted material from the column should be monitored for GST-tagged proteins using SDS-PAGE in combination with Western Blot if necessary.• The GST Detection Module can be used to optimize conditions forelution or to trace steps in the purification of a GST-tagged protein. The Module is designed to identify GST-tagged proteins using either a biochemical or an immunological assay.• The concentration of GST-tagged protein can be estimatedby measuring the absorbance at 280 nm. The GST-tag can be approximated using the conversion; A 280 ≈ 1 corresponds to ~ 0.5 mg/ml.• The concentration of GST-tagged protein may also be determinedby standard chromogenic methods (e.g. Lowry, BCA, and Bradford assays). If Lowry or BCA assays are to be used, the sample must first be buffer exchanged using a HiTrap Desalting 5 ml column, a HiPrep ™ 26/10 Desalting column or dialysed against PBS to remove glutathione, which can interfere with the protein measurement. The Bradford method can be used in the presence of glutathione.• The reuse of GSTrap FF depends on the nature of the sample andshould only be performed with identical samples to prevent cross-contamination.Cleaning GSTrap FFIf the medium appears to be losing binding capacity, it may be due to an accumulation of precipitate, denatured or nonspecifically bound proteins. Removal of precipitated or denatured substances:• Wash with 2 column volumes of 6 M guanidine hydrochloride, immediately followed by 5 column volumes of PBS.Removal of hydrophobically bound substances:• Wash with 3–4 column volumes of 70% ethanol or 2 column volumes of 1% Triton™ X-100 immediately followed by 5 column volumes of PBS. 3. Scaling upFor quick scale-up of purifications, two or three GSTrap FF can be connected in series (backpressure will increase). Further scaling up is easy using the20 ml prepacked GSTPrep™ FF 16/10 column or bulk media packages.4. StorageStore the column at +4 to 30 °C in 20% ethanol.5. Cleavage of GST-tagged proteinsIf removal of the GST-tag is necessary, tagged proteins containing a PreScission™ Protease recognition site, a thrombin recognition site or afactor Xa recognition site may be cleaved either while bound to GSTrap FF or in solution after elution. Cleavage after elution is suggested if optimizationof cleavage conditions is necessary. Samples can easily be removed at various time points and analyzed by SDS-PAGE to estimate the yield, purity and extent of digestion. The amount of protease used, the temperatureand the length of incubation required for complete digestion may varyp. 7p.depending on the fusion partner. Optimal conditions for each fusion should be determined in pilot experiments, e.g. incubation time may be reduced by using higher concentrations of proteolytic enzyme.1. PreScission ProteasePreScission Protease, M r 46 000PreScission cleavage buffer: 50 mM Tris-HCl, 150 mM NaCl,1 mM EDTA, 1 mM dithiothreitol (DTT), pH 7.5PreScission Protease cleavage of GST-tagged protein bound to GSTrap FFAssumption: 8 mg GST-tagged protein bound/ml medium 1. Follow steps 1–5 under “Purification”, (see p. 5).2. Wash GSTrap FF with 10 column volumes of PreScission cleavage buffer.3. Prepare the PreScission Protease mix:GSTrap FF 1 ml column (8 mg GST-tagged protein bound): Mix 80 µl (160 units) of PreScission Protease with 920 µl of PreScission cleavage buffer at +4 °C.GSTrap FF 5 ml column (40 mg GST-tagged protein bound): Mix 400 µl (800 units) of PreScission Protease with 4.6 ml of PreScission cleavage buffer at +4 °C.4. Load the PreScission Protease mix onto the column using a syringe andthe adaptor supplied.Seal the column with the top and bottom stop plugs supplied.5. Incubate the column at +4 °C for 4 hours.6. Fill a syringe with 3 ml (1 ml column) or 15 ml (5 ml column) of PreScissioncleavage buffer. Remove the top and bottom stop plugs. Avoidintroducing air into the column. Elute the column and collect the eluate (0.5 ml-1 ml/tube). The eluate will contain the protein of interest, while the GST moiety of the tagged protein and the PreScission Protease will remain bound to GSTrap FF.PreScission Protease cleavage of eluted GST-tagged protein Assumption: 8 mg GST-tagged protein bound/ml medium1. Follow steps 1–6 under “Purification”, (see page 5).2. Remove the reduced glutathione from the eluate using a quick bufferexchange on HiTrap Desalting, a PD-10 column or HiPrep 26/10 Desalting depending on the sample volume, or dialyse against PreScissioncleavage buffer.3. Add 1 µl (2 U) of PreScission Protease for each 100 µg of tagged proteinin the eluate. If the amount of tagged protein in the eluate has not been determined, add 80 µl (160 units) of PreScission Protease (tagged protein eluted from GSTrap FF 1 ml column) or 400 µl (800 units) of PreScissionProtease (tagged protein eluted from GSTrap FF 5 ml column).4. Incubate at +4 °C for 4 hours.5. Once digestion is complete, apply the sample to an equilibrated GSTrapFF column to remove the GST moiety of the tagged protein and thePreScission Protease. The protein of interest will be found in the flow-through, while the GST moiety of the tagged protein and the PreScission Protease will remain bound to GSTrap FF.. Thrombin37 000Thrombin, MrThrombin cleavage buffer: PBS, pH 7.3Preparation of thrombin solution:1. Dissolve 500 U thrombin in cold 500 µl PBS (1 U/µl).2. Swirl gently to dissolve.3. Freeze as 80 µl aliqouts and keep at –80 °C.Thrombin cleavage of GST-tagged protein bound to GSTrap FF Assumption: 8 mg GST-tagged protein bound/ml medium1. Follow steps 1–5 under “Purification”, (see p. 5).2. Prepare the thrombin mix: GSTrap FF 1 ml column (8 mg GST fusionprotein bound): Mix 80 µl thrombin solution (1 U/µl ) with 920 µl PBS.p. 9GSTrap FF 5 ml column (40 mg GST-tagged protein bound): Mix 400 µlthrombin solution with 4.6 ml PBS.3. Load the thrombin solution onto the column using a syringe and theadaptor supplied. Seal the column with the top and bottom plugssupplied.4. Incubate the column at room temperature (+22 to +25 °C) for 2–16 hours.5. Fill a syringe with 3 ml (1 ml column) or 15 ml (5 ml column) PBS. Removethe top and bottom stop plugs from the column. Avoid introducing airinto the column. Elute the column and collect the eluate (0.5 ml-1 ml/tube). The eluate will contain the protein of interest and thrombin, while the GST moiety of the tagged protein will remain bound to GSTrap FF. Note:After cleavage using thrombin the enzyme can be removed from eluted protein using HiTrap Benzamidine FF (high sub), see orderinginformation.Thrombin cleavage of eluted GST-tagged proteinAssumption: 8 mg GST-tagged protein bound/ml medium1. Follow steps 1–6 under “Purification”, (see p. 5).2. Add 10 µl (10 units) of thrombin solution for each mg of tagged proteinin the eluate. If the amount of tagged protein in the eluate has not been determined, add 80 µl (80 U) thrombin solution (tagged protein elutedfrom GSTrap FF 1 ml column) or 400 µl (400 U) thrombin solution (tagged protein eluted from GSTrap FF 5 ml column).3. Incubate at room temperatue (+22 to 25 °C) for 2–16 hours.4. Once digestion is complete, GST can be removed by first removingglutathione using a quick buffer exchange on HiTrap Desalting, a PD-10 column or HiPrep 26/10 Desalting depending on the sample volume, or dialysing against PBS. Then apply the sample to an equilibrated GSTrap FF column. The purified protein of interest and thrombin will be found in the flow-through.Note:After cleavage using thrombin the enzyme can be removed from eluted protein using HiTrap Benzamidine FF (high sub), see orderinginformation.p. 10. Factor Xa48 000Factor Xa, MrNote:Factor Xa consists of two subunits linked by disulfide bridges. As glutathione can disrupt disulfide bridges, it should be removed fromthe sample prior to the cleavage reaction., pH 7.5 Factor Xa cleavage buffer: 50 mM Tris-HCl, 150 mM NaCl, 1 mM CaCl2 Preparation of factor Xa solution:1. Dissolve 400 U factor Xa in 400 µl cold water (1 U/µl).2. Swirl gently to dissolve.3. Freeze as 80 µl aliqouts and keep at –80°C.Factor Xa cleavage of GST-tagged protein bound to GSTrap FF Assumption: 8 mg GST-tagged protein bound/ml medium1. Follow steps 1–5 under “Purification”, (see p. 5).2. Wash GSTrap FF with 10 column volumes of factor Xa cleavage buffer.3. Prepare the factor Xa mix:GSTrap FF 1 ml column (8 mg GST-tagged protein bound): Mix 80 µl factor Xa solution with 920 µl factor Xa cleavage buffer.GSTrap FF 5 ml column (40 mg GST-tagged protein bound): Mix 400 µlfactor Xa solution with 4.6 ml factor Xa cleavage buffer.4. Load the mix onto the column using a syringe and the adaptor supplied.Seal the column with the top and bottom stop plugs supplied.5. Incubate the column at room temperature (+22 to 25 °C) for 2–16 hours.6. Fill a syringe with 3 ml (1 ml column) or 15 ml (5 ml column) factorXa cleavage buffer. Remove the top and bottom stop plugs from thecolumn. Avoid introducing air into the column. Elute the column andcollect the eluate (0.5 ml-1 ml/tube). The eluate will contain the proteinof interest and factor Xa, while the GST moiety of the tagged protein will remain bound to GSTrap FF.p. 11Note:After cleavage using factor Xa the enzyme can be removed from eluted protein using HiTrap Benzamidine FF (high sub), see orderinginformation.Factor Xa cleavage of eluted GST-tagged proteinAssumption: 8 mg GST-tagged protein bound/ml medium1. Follow steps 1–6 under “Purification”, (see p. 5).2. Remove the reduced glutathione from the eluate using a quick bufferexchange on HiTrap Desalting, a PD-10 column or HiPrep 26/10 Desalting depending on sample volume, or dialyse against factor Xa cleavagebuffer.3. Add 10 units of factor Xa solution for each mg tagged protein in theeluate. If the amount of tagged protein in the eluate has not beendetermined, add 80 µl (80 units) of factor Xa solution (eluted taggedprotein from GSTrap FF 1 ml column) or 400 µl (400 units) of factor Xasolution (eluted tagged protein from GSTrap FF 5 ml column).4. Incubate the column at room temperature (+22 to 25 °C) for 2–16 hours.5. Once digestion is complete, apply the sample to an equilibrated GSTrapFF column to remove the GST moiety . The protein of interest will befound in the flow-through together with factor Xa.Note:After cleavage using factor Xa the enzyme can be removed from eluted protein using HiTrap Benzamidine FF (high sub), see orderinginformation.6. Troubleshooting guideConsult the GST Gene Fusion System Handbook (1) for more detailedinformation and pGEX instructions regarding troubleshootingrecommendations for expression, fermentation and solubilization.GST-tagged protein does not bind to GSTrap FF• GST-tagged protein denatured by sonication: Too extensive sonication can denature the tagged protein and prevent it binding to GSTrap FF. Use mild sonication conditions during cell lysis.p. 1• Add DTT prior to cell lysis: Adding DTT to a final concentration of 1–10 mM prior to cell lysis may significantly increase binding of someGST-tagged proteins to GSTrap FF.•Test the binding of GST from parental pGEX: Prepare a sonicate of cells harboring the parental pGEX plasmid and check binding to the matrix.If GST produced from the parental plasmid binds with high affinity,the fusion partner may have altered the conformation of GST, therebyreducing its affinity. Adequate results may be obtained by reducing the temperature used for binding to +4°C, and by limiting column washing.•Equilibrate GSTrap FF before use: Binding of GST-tagged proteins to GSTrap FF is not efficient at pH less than 6.5 or greater than 8. Check that the GSTrap FF column has been equilibrated with a buffer pH 6.5 to 8.0(e.g. PBS) before the cell lysate is applied.• Use a fresh GSTrap FF: If the GSTrap FF column has already been used several times, it may be necessary to use a new GSTrap FF column. Seealso “Cleaning GSTrap FF”.• Decrease flow rate during sample load, see note p. 6.GST-tagged protein is not eluted efficiently from GSTrap FF• Increase the time used for elution: Decrease the flow during elution.•Increase the volume of elution buffer: Sometimes, especially after on-column cleavage of fusion protein, a larger volume of buffer may be necessary to elute the fusion protein.• Increase the concentration of glutathione in the elution buffer: The10 mM recommended in this protocol should be sufficient for mostapplications, but exceptions exist. Try 50 mM Tris- HCl, 20–40 mMreduced glutathione, pH 8.0 as elution buffer.•Increase the pH of the elution buffer: A low pH may limit elution from GSTrap FF. Increasing the pH of the elution buffer to pH 8–9 may improve elution without requiring an increase in the concentration of glutathione used for elution.• Increase the ionic strength of the elution buffer: Adding 0.1–0.2 M NaCl to the elution buffer may also improve results.p. 1p. 1• Add a non-ionic detergent to the elution buffer: Non-specifichydrophobic interactions may prevent solubilization and elution of fusion proteins from GSTrap FF. Adding a non-ionic detergent may improve results. Adding 0.1% Triton X-100 or 2% N-octylglucosid can significantly improve elution of some GST-tagged proteins.Multiple bands are observed after electrophoresis/Western Blotting analysis of eluted target protein• M r 70 000 protein co-purifies with the GST-tagged protein:The M r 70 000 protein is probably a protein product of the E. coli genednaK. This protein is involved in protein folding in E. coli . It has beenreported that this association can be disrupted by incubating the fusion protein in 50 mM Tris-HCl, 2 mM ATP, 10 mM MgSO 4, pH 7.4 for 10 min. at +37 °C prior to loading on GSTrap FF.Alternatively, remove the DnaK protein by passing the tagged protein solution through ATP-agarose or by ion exchange.• Add a protease inhibitor: Multiple bands may be a result of partialdegradation of tagged proteins by proteases. Adding 1 mM PMSF to the lysis solution may improve results. A nontoxic, water-soluble alternative to PMSF is 4-(2-aminoethyl)-benzenesulfonyl fluoride hydrochloride (AEBSF), commercially available as Pefabloc ™ SC from Boehringer Mannheim.Note: Serine protease inhibitors must be removed prior to cleavage bythrombin or factor Xa. PreScission Protease is not a consensus serine protease and is insensitive to many of the protease inhibitors tested at GE Healthcare.• Use a protease-deficient host: Multiple bands may be the result ofproteolysis in the host bacteria. If this is the case, the use of a host-deficient strain may be required (e.g. lon - or ompT ). E. coli BL21 is provided with the pGEX vectors. This strain is ompT.• Decrease sonication: Cell disruption is apparent by partial clearing ofthe suspension and can be checked by microscopic examination. Adding lysozyme (0.1 volume of a 10 mg/ml lysozyme solution in 25 mM Tris-HCl, pH 8.0) prior to sonication may improve results. Avoid frothing as thisp. 15may denature the fusion protein. Over-sonication can also lead to the co-purification of host proteins with the GST-tagged protein.• Include an additional purification step: Additional bands maybe caused by the co-purification of a variety of proteins known as chaperonins, which are involved in the correct folding of nascent proteins in E. coli . These include, but may not be limited to: DnaK (M r ~ 70 000), DnaJ (Mr ~ 37 000), GrpE (M r ~ 40 000), GroEL (M r ~ 57 000) and GroES (M r ~ 10 000). Several methods for purifying GST-tagged proteins from these co-purifying proteins have been described.• Cross-adsorb antibody with E. coli proteins: Depending on the sourceof the anti-GST antibody, it may contain antibodies that react with various E. coli proteins that may be present in your fusion protein sample. Cross-adsorb the antibody with an E. coli sonicate to remove anti-E. coli antibodies from the preparation. Anti-GST antibody from GE Healthcare has been cross-adsorbed against E. coli proteins and tested for its lack of non-specific background binding in Western Blots.Incomplete cleavage of GST-tagged proteins• The PreScission Protease, thrombin or factor Xa to tagged proteinratios are incorrect: Check the amount of tagged protein in thedigestion. Note that the capacity of GSTrap FF for GST is approximately 10 mg/ml medium. In most purifications, however, the matrix is not saturated with tagged protein.Ratios: PreScission protease, at least 10 units/mg tagged protein.Thrombin, at least 10 units/mg tagged protein. One cleavage unit of thrombin from GE Healthcare digests ≥ 90% of 100 µg of a test tagged protein in 16 hours at +22 °C.Factor Xa, at least 1% (w/w) tagged protein. For some tagged proteins, up to 5% factor Xa can be used. The optimum amount must be determined empirically.In some cases, a tagged protein concentration of 1 mg/ ml has been found to give optimal results. Adding ≤ 0.5% (w/v) to the reaction buffer can significantly improve factor Xa cleavage with some tagged proteins. Various concentrations of SDS should be tested to find the optimum concentration.p. 16• Increase incubation time and enzyme concentration: For PreScissionProtease, thrombin or factor Xa, increase the reaction time to 20 hours or more if the tagged protein is not degraded by extensive incubation. The amount of enzymes can also be increased.• Verify the presence of specific cleavage sites: Check the DNA sequenceof the construct. Compare it with a known sequence and verify that the different specific cleavage sites for the enzyme used have not been altered during the cloning of your tagged protein.Ensure that cleavage enzyme inhibitors are absent• PreScission Protease: Buffer exchange or dialyse the tagged proteinagainst 50 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 1 mM DTT, pH 7.5 before cleavage.• Factor Xa: Buffer exchange on HiTrap Desalting, a PD-10 column orHiPrep 26/10 Desalting depending on the sample volume, or dialyse against 50 mM Tris-HCl, 150 mM NaCl, 1 mM CaCl 2, pH 7.5.• Factor Xa is not properly activated: Functional factor Xa requiresactivation of factor X with Russell’s viper venom. Activation conditions are a ratio of Russell’s viper venom to factor Xa of 1% in 8 mM Tris-HCl, 70 mM NaCl, 8 mM CaCl 2, pH 8.0. Incubate at +37 °C for 5 min. Factor Xa from GE Healthcare has been preactivated by this procedure.• The first amino acid after the factor Xa recognition sequence is Argor Pro: Check the sequence of the fusion partner to be sure that the first three nucleotides after the factor Xa recognition sequence do not code for Arg or Pro.Multiple bands are observed on SDS gels following enzyme cleavage:• Determine when the bands appear: Test to be certain that additionalbands are not present prior to PreScission Protease, thrombin or factor Xa cleavage. Such bands may be the result of proteolysis in the host bacteria.• Tagged partner may contain recognition sequences for PreScissionProtease, thrombin or factor Xa: Check the sequences. See the GST Gene Fusion System Handbook (1) for details.7. References1. GST Gene Fusion System Handbook, GE Healthcare, Code No. 18-1157-582. Rapid purification of GST-fusion proteins from large sample volumes.Miniposter, 18-1139-51, GE Healthcare.3. Efficient, rapid protein purification and on-column cleavage usingGSTrap FF columns. Application Note, 18-1146-70, GE Healthcare.4. Purification of GST-fusion proteins, on-column cleavage and sampleclean-up. Miniposter, 18-1150-20, GE Healthcare.5. Dian, C., et al, J of chromatography B, 769 (1), 133–144 (2002)6. Strategies for the purification and on-column cleavage of glutathioneS-transferase fusion target proteins. J of Chromatography B, 2002. 769(1): 133–144. Dian, C., et al.For more information about HiTrap columns and updated reference list forthe use of GSTrap FF columns, (Code No. 18-1156-67) visit/hitrapp. 17。

Instruction ManualProBond TM Purification SystemFor purification of polyhistidine-containing recombinant proteinsCatalog nos. K850-01, K851-01, K852-01, K853-01, K854-01,R801-01, R801-15Version K2 September200425-0006iiTable of ContentsKit Contents and Storage (iv)Accessory Products (vi)Introduction (1)Overview (1)Methods (2)Preparing Cell Lysates (2)Purification Procedure—Native Conditions (7)Purification Procedure—Denaturing Conditions (11)Purification Procedure—Hybrid Conditions (13)Troubleshooting (15)Appendix (17)Additional Protocols (17)Recipes (18)Frequently Asked Questions (21)References (22)Technical Service (23)iiiKit Contents and StorageTypes of Products This manual is supplied with the following products:Product CatalogNo.ProBond™ Purification System K850-01ProBond™ Purification System with Antibodywith Anti-Xpress™ Antibody K851-01with Anti-myc-HRP Antibody K852-01with Anti-His(C-term)-HRP Antibody K853-01with Anti-V5-HRP Antibody K854-01ProBond™ Nickel-Chelating Resin (50 ml) R801-01ProBond™ Nickel Chelating Resin (150 ml) R801-15ProBond™Purification System Components The ProBond™ Purification System includes enough resin, reagents, and columns for six purifications. The components are listed below. See next page for resin specifications.Component Composition Quantity ProBond™ Resin 50% slurry in 20% ethanol 12 ml5X NativePurification Buffer250 mM NaH2PO4, pH 8.02.5 M NaCl1 × 125 ml bottleGuanidinium LysisBuffer6 M Guanidine HCl20 mM sodium phosphate, pH 7.8500 mM NaCl1 × 60 ml bottleDenaturingBinding Buffer8 M Urea20 mM sodium phosphate, pH 7.8500 mM NaCl2 × 125 ml bottlesDenaturing WashBuffer8 M Urea20 mM sodium phosphate, pH 6.0500 mM NaCl2 × 125 ml bottlesDenaturing ElutionBuffer8 M Urea20 mM NaH2PO4, pH 4.0500 mM NaCl1 × 60 ml bottleImidazole 3 M Imidazole,20 mM sodium phosphate, pH 6.0500 mM NaCl1 × 8 ml bottlePurificationColumns10 ml columns 6Continued on next pageivKit Contents and Storage, ContinuedProBond™Purification System with Antibody The ProBond™ Purification System with Antibody includes resin, reagents, and columns as described for the ProBond™ Purification System (previous page) and 50 µl of the appropriate purified mouse monoclonal antibody. Sufficient reagents are included to perform six purifications and 25 Western blots with the antibody.For more details on the antibody specificity, subclass, and protocols for using the antibody, refer to the antibody manual supplied with the system.Storage Store ProBond™ resin at +4°C. Store buffer and columns at room temperature.Store the antibody at 4°C. Avoid repeated freezing and thawing of theantibody as it may result in loss of activity.The product is guaranteed for 6 months when stored properly.All native purification buffers are prepared from the 5X Native PurificationBuffer and the 3 M Imidazole, as described on page 7.The Denaturing Wash Buffer pH 5.3 is prepared from the Denaturing WashBuffer (pH 6.0), as described on page 11.Resin and ColumnSpecificationsProBond™ resin is precharged with Ni2+ ions and appears blue in color. It isprovided as a 50% slurry in 20% ethanol.ProBond™ resin and purification columns have the following specifications:• Binding capacity of ProBond™ resin: 1–5 mg of protein per ml of resin• Average bead size: 45–165 microns• Pore size of purification columns: 30–35 microns• Recommended flow rate: 0.5 ml/min• Maximum flow rate: 2 ml/min• Maximum linear flow rate: 700 cm/h• Column material: Polypropylene• pH stability (long term): pH 3–13• pH stability (short term): pH 2–14ProductQualificationThe ProBond™ Purification System is qualified by purifying 2 mg of myoglobinprotein on a column and performing a Bradford assay. Protein recovery mustbe 75% or higher.vAccessory ProductsAdditionalProductsThe following products are also available for order from Invitrogen:Product QuantityCatalogNo.ProBond™ Nickel-Chelating Resin 50 ml150 mlR801-01R801-15Polypropylene columns(empty)50 R640-50Ni-NTA Agarose 10 ml25 ml R901-01 R901-15Ni-NTA Purification System 6 purifications K950-01 Ni-NTA Purification Systemwith Antibodywith Anti-Xpress™ Antibody with Anti-myc-HRP Antibody with Anti-His(C-term)-HRP Antibodywith Anti-V5-HRP Antibody 1 kit1 kit1 kit1 kitK951-01K952-01K953-01K954-01Anti-myc Antibody 50 µl R950-25 Anti-V5 Antibody 50 µl R960-25 Anti-Xpress™ Antibody 50 µl R910-25 Anti-His(C-term) Antibody 50 µl R930-25 InVision™ His-tag In-gel Stain 500 ml LC6030 InVision™ His-tag In-gelStaining Kit1 kit LC6033Pre-Cast Gels and Pre-made Buffers A large variety of pre-cast gels for SDS-PAGE and pre-made buffers for your convenience are available from Invitrogen. For details, visit our web site at or contact Technical Service (page 23).viIntroductionOverviewIntroduction The ProBond™ Purification System is designed for purification of 6xHis-tagged recombinant proteins expressed in bacteria, insect, and mammalian cells. Thesystem is designed around the high affinity and selectivity of ProBond™Nickel-Chelating Resin for recombinant fusion proteins containing six tandemhistidine residues.The ProBond™ Purification System is a complete system that includespurification buffers and resin for purifying proteins under native, denaturing,or hybrid conditions. The resulting proteins are ready for use in many targetapplications.This manual is designed to provide generic protocols that can be adapted foryour particular proteins. The optimal purification parameters will vary witheach protein being purified.ProBond™ Nickel-Chelating Resin ProBond™ Nickel-Chelating Resin is used for purification of recombinant proteins expressed in bacteria, insect, and mammalian cells from any 6xHis-tagged vector. ProBond™ Nickel-Chelating Resin exhibits high affinity and selectivity for 6xHis-tagged recombinant fusion proteins.Proteins can be purified under native, denaturing, or hybrid conditions using the ProBond™ Nickel-Chelating Resin. Proteins bound to the resin are eluted with low pH buffer or by competition with imidazole or histidine. The resulting proteins are ready for use in target applications.Binding Characteristics ProBond™ Nickel-Chelating Resin uses the chelating ligand iminodiacetic acid (IDA) in a highly cross-linked agarose matrix. IDA binds Ni2+ ions by three coordination sites.The protocols provided in this manual are generic, and may not result in 100%pure protein. These protocols should be optimized based on the bindingcharacteristics of your particular proteins.Native VersusDenaturingConditionsThe decision to purify your 6xHis-tagged fusion proteins under native ordenaturing conditions depends on the solubility of the protein and the need toretain biological activity for downstream applications.• Use native conditions if your protein is soluble (in the supernatant afterlysis) and you want to preserve protein activity.• Use denaturing conditions if the protein is insoluble (in the pellet afterlysis) or if your downstream application does not depend on proteinactivity.• Use hybrid protocol if your protein is insoluble but you want to preserveprotein activity. Using this protocol, you prepare the lysate and columnsunder denaturing conditions and then use native buffers during the washand elution steps to refold the protein. Note that this protocol may notrestore activity for all proteins. See page 14.1MethodsPreparing Cell LysatesIntroduction Instructions for preparing lysates from bacteria, insect, and mammalian cellsusing native or denaturing conditions are described below.Materials Needed You will need the following items:• Native Binding Buffer (recipe is on page 8) for preparing lysates undernative conditions• Sonicator• 10 µg/ml RNase and 5 µg/ml DNase I (optional)• Guanidinium Lysis Buffer (supplied with the system) for preparing lysatesunder denaturing conditions• 18-gauge needle• Centrifuge• Sterile, distilled water• SDS-PAGE sample buffer• Lysozyme for preparing bacterial cell lysates• Bestatin or Leupeptin, for preparing mammalian cell lysatesProcessing Higher Amount of Starting Material Instructions for preparing lysates from specific amount of starting material (bacteria, insect, and mammalian cells) and purification with 2 ml resin under native or denaturing conditions are described in this manual.If you wish to purify your protein of interest from higher amounts of starting material, you may need to optimize the lysis protocol and purification conditions (amount of resin used for binding). The optimization depends on the expected yield of your protein and amount of resin to use for purification. Perform a pilot experiment to optimize the purification conditions and then based on the pilot experiment results, scale-up accordingly.Continued on next page2Preparing Bacterial Cell Lysate—Native Conditions Follow the procedure below to prepare bacterial cell lysate under native conditions. Scale up or down as necessary.1. Harvest cells from a 50 ml culture by centrifugation (e.g., 5000 rpm for5 minutes in a Sorvall SS-34 rotor). Resuspend the cells in 8 ml NativeBinding Buffer (recipe on page 8).2. Add 8 mg lysozyme and incubate on ice for 30 minutes.3. Using a sonicator equipped with a microtip, sonicate the solution on iceusing six 10-second bursts at high intensity with a 10-second coolingperiod between each burst.Alternatively, sonicate the solution on ice using two or three 10-secondbursts at medium intensity, then flash freeze the lysate in liquid nitrogen or a methanol dry ice slurry. Quickly thaw the lysate at 37°C andperform two more rapid sonicate-freeze-thaw cycles.4. Optional: If the lysate is very viscous, add RNase A (10 µg/ml) andDNase I (5 µg/ml) and incubate on ice for 10–15 minutes. Alternatively,draw the lysate through a 18-gauge syringe needle several times.5. Centrifuge the lysate at 3,000 ×g for 15 minutes to pellet the cellulardebris. Transfer the supernatant to a fresh tube.Note: Some 6xHis-tagged protein may remain insoluble in the pellet, and can be recovered by preparing a denatured lysate (page 4) followed bythe denaturing purification protocol (page 12). To recover this insolubleprotein while preserving its biological activity, you can prepare thedenatured lysate and then follow the hybrid protocol on page 14. Notethat the hybrid protocol may not restore activity in all cases, and should be tested with your particular protein.6. Remove 5 µl of the lysate for SDS-PAGE analysis. Store the remaininglysate on ice or freeze at -20°C. When ready to use, proceed to theprotocol on page 7.Continued on next page3Preparing Bacterial Cell Lysate—Denaturing Conditions Follow the procedure below to prepare bacterial cell lysate under denaturing conditions:1. Equilibrate the Guanidinium Lysis Buffer, pH 7.8 (supplied with thesystem or see page 19 for recipe) to 37°C.2. Harvest cells from a 50 ml culture by centrifugation (e.g., 5000 rpm for5 minutes in a Sorvall SS-34 rotor).3. Resuspend the cell pellet in 8 ml Guanidinium Lysis Buffer from Step 1.4. Slowly rock the cells for 5–10 minutes at room temperature to ensurethorough cell lysis.5. Sonicate the cell lysate on ice with three 5-second pulses at high intensity.6. Centrifuge the lysate at 3,000 ×g for 15 minutes to pellet the cellulardebris.Transfer the supernatant to a fresh tube.7. Remove 5 µl of the lysate for SDS-PAGE analysis. Store the remaininglysate on ice or at -20°C. When ready to use, proceed to the denaturingprotocol on page 11 or hybrid protocol on page 13.Note: To perform SDS-PAGE with samples in Guanidinium Lysis Buffer, you need to dilute the samples, dialyze the samples, or perform TCAprecipitation prior to SDS-PAGE to prevent the precipitation of SDS.Harvesting Insect Cells For detailed protocols dealing with insect cell expression, consult the manual for your particular system. The following lysate protocols are for baculovirus-infected cells and are intended to be highly generic. They should be optimized for your cell lines.For baculovirus-infected insect cells, when the time point of maximal expression has been determined, large scale protein expression can be carried out. Generally, the large-scale expression is performed in 1 liter flasks seeded with cells at a density of 2 × 106 cells/ml in a total volume of 500 ml and infected with high titer viral stock at an MOI of 10 pfu/cell. At the point of maximal expression, harvest cells in 50 ml aliquots. Pellet the cells by centrifugation and store at -70°C until needed. Proceed to preparing cell lysates using native or denaturing conditions as described on the next page.Continued on next page4Preparing Insect Cell Lysate—Native Condition 1. Prepare 8 ml Native Binding Buffer (recipe on page 8) containingLeupeptin (a protease inhibitor) at a concentration of 0.5 µg/ml.2. After harvesting the cells (previous page), resuspend the cell pellet in8 ml Native Binding Buffer containing 0.5 µg/ml Leupeptin.3. Lyse the cells by two freeze-thaw cycles using a liquid nitrogen or dryice/ethanol bath and a 42°C water bath.4. Shear DNA by passing the preparation through an 18-gauge needle fourtimes.5. Centrifuge the lysate at 3,000 ×g for 15 minutes to pellet the cellulardebris.Transfer the supernatant to a fresh tube.6. Remove 5 µl of the lysate for SDS-PAGE analysis. Store remaining lysateon ice or freeze at -20°C. When ready to use, proceed to the protocol on page 7.Preparing Insect Cell Lysate—Denaturing Condition 1. After harvesting insect cells (previous page), resuspend the cell pellet in8 ml Guanidinium Lysis Buffer (supplied with the system or see page 19for recipe).2. Pass the preparation through an 18-gauge needle four times.3. Centrifuge the lysate at 3,000 ×g for 15 minutes to pellet the cellulardebris. Transfer the supernatant to a fresh tube.4. Remove 5 µl of the lysate for SDS-PAGE analysis. Store remaining lysateon ice or freeze at -20° C. When ready to use, proceed to the denaturingprotocol on page 11 or hybrid protocol on page 13.Note: To perform SDS-PAGE with samples in Guanidinium Lysis Buffer, you need to dilute the samples, dialyze the samples, or perform TCAprecipitation prior to SDS-PAGE to prevent the precipitation of SDS.Continued on next pagePreparing Mammalian Cell Lysate—Native Conditions For detailed protocols dealing with mammalian expression, consult the manual for your particular system. The following protocols are intended to be highly generic, and should be optimized for your cell lines.To produce recombinant protein, you need between 5 x 106and 1 x 107 cells. Seed cells and grow in the appropriate medium until they are 80–90% confluent. Harvest cells by trypsinization. You can freeze the cell pellet in liquid nitrogen and store at -70°C until use.1. Resuspend the cell pellet in 8 ml of Native Binding Buffer (page 8). Theaddition of protease inhibitors such as bestatin and leupeptin may benecessary depending on the cell line and expressed protein.2. Lyse the cells by two freeze-thaw cycles using a liquid nitrogen or dryice/ethanol bath and a 42°C water bath.3. Shear the DNA by passing the preparation through an 18-gauge needlefour times.4. Centrifuge the lysate at 3,000 ×g for 15 minutes to pellet the cellulardebris. Transfer the supernatant to a fresh tube.5. Remove 5 µl of the lysate for SDS-PAGE analysis. Store the remaininglysate on ice or freeze at -20° C. When ready to use, proceed to theprotocol on page 7.Preparing Mammalian Cell Lysates—Denaturing Conditions For detailed protocols dealing with mammalian expression, consult the manual for your particular system. The following protocols are intended to be highly generic, and should be optimized for your cell lines.To produce recombinant protein, you need between 5 x 106and 1 x 107 cells. Seed cells and grow in the appropriate medium until they are 80–90% confluent. Harvest cells by trypsinization. You can freeze the cell pellet in liquid nitrogen and store at -70°C until use.1. Resuspend the cell pellet in 8 ml Guanidinium Lysis Buffer (suppliedwith the system or see page 19 for recipe).2. Shear the DNA by passing the preparation through an 18-gauge needlefour times.3. Centrifuge the lysate at 3,000 ×g for 15 minutes to pellet the cellulardebris. Transfer the supernatant to a fresh tube.4. Remove 5 µl of the lysate for SDS-PAGE analysis. Store the remaininglysate on ice or freeze at -20° C until use. When ready to use, proceed to the denaturing protocol on page 11 or hybrid protocol on page 13.Note: To perform SDS-PAGE with samples in Guanidinium Lysis Buffer, you need to dilute the samples, dialyze the samples, or perform TCAprecipitation prior to SDS-PAGE to prevent the precipitation of SDS.Purification Procedure—Native ConditionsIntroduction In the following procedure, use the prepared Native Binding Buffer, NativeWash Buffer, and Native Elution Buffer, columns, and cell lysate preparedunder native conditions. Be sure to check the pH of your buffers before starting.Buffers for Native Purification All buffers for purification under native conditions are prepared from the5X Native Purification Buffer supplied with the system. Dilute and adjust the pH of the 5X Native Purification Buffer to create 1X Native Purification Buffer (page 8). From this, you can create the following buffers:• Native Binding Buffer• Native Wash Buffer• Native Elution BufferThe recipes described in this section will create sufficient buffers to perform one native purification using one kit-supplied purification column. Scale up accordingly.If you are preparing your own buffers, see page 18 for recipe.Materials Needed You will need the following items:• 5X Native Purification Buffer (supplied with the system or see page 18 forrecipe)• 3 M Imidazole (supplied with the system or see page 18 for recipe)• NaOH• HCl• Sterile distilled water• Prepared ProBond™ columns with native buffers (next page)• Lysate prepared under native conditions (page 2)Imidazole Concentration in Native Buffers Imidazole is included in the Native Wash and Elution Buffers to minimize the binding of untagged, contaminating proteins and increase the purity of the target protein with fewer wash steps. Note that, if your level of contaminating proteins is high, you may add imidazole to the Native Binding Buffer.If your protein does not bind well under these conditions, you can experiment with lowering or eliminating the imidazole in the buffers and increasing the number of wash and elution steps.Continued on next page1X Native Purification Buffer To prepare 100 ml 1X Native Purification Buffer, combine:• 80 ml of sterile distilled water• 20 ml of 5X Native Purification Buffer (supplied with the system or see page 18 for recipe)Mix well and adjust pH to 8.0 with NaOH or HCl.Native Binding Buffer Without ImidazoleUse 30 ml of the 1X Native Purification Buffer (see above for recipe) for use as the Native Binding Buffer (used for column preparation, cell lysis, and binding).With Imidazole (Optional):You can prepare the Native Binding Buffer with imidazole to reduce the binding of contaminating proteins. (Note that some His-tagged proteins may not bind under these conditions.).To prepare 30 ml Native Binding Buffer with 10 mM imidazole, combine: • 30 ml of 1X Native Purification Buffer• 100 µl of 3 M Imidazole, pH 6.0Mix well and adjust pH to 8.0 with NaOH or HCl.Native Wash Buffer To prepare 50 ml Native Wash Buffer with 20 mM imidazole, combine:• 50 ml of 1X Native Purification Buffer• 335 µl of 3 M Imidazole, pH 6.0Mix well and adjust pH to 8.0 with NaOH or HCl.Native Elution Buffer To prepare 15 ml Native Elution Buffer with 250 mM imidazole, combine:• 13.75 ml of 1X Native Purification Buffer• 1.25 ml of 3 M Imidazole, pH 6.0Mix well and adjust pH to 8.0 with NaOH or HCl.Continued on next pageDo not use strong reducing agents such as DTT with ProBond™ columns. DTTreduces the nickel ions in the resin. In addition, do not use strong chelatingagents such as EDTA or EGTA in the loading buffers or wash buffers, as thesewill strip the nickel from the columns.Be sure to check the pH of your buffers before starting.PreparingProBond™ ColumnWhen preparing a column as described below, make sure that the snap-off capat the bottom of the column remains intact. To prepare a column:1. Resuspend the ProBond™ resin in its bottle by inverting and gentlytapping the bottle repeatedly.2. Pipet or pour 2 ml of the resin into a 10-ml Purification Columnsupplied with the kit. Allow the resin to settle completely by gravity(5-10 minutes) or gently pellet it by low-speed centrifugation (1 minuteat 800 ×g). Gently aspirate the supernatant.3. Add 6 ml of sterile, distilled water and resuspend the resin byalternately inverting and gently tapping the column.4. Allow the resin to settle using gravity or centrifugation as described inStep 2, and gently aspirate the supernatant.5. For purification under Native Conditions, add 6 ml Native BindingBuffer (recipe on page 8).6. Resuspend the resin by alternately inverting and gently tapping thecolumn.7. Allow the resin to settle using gravity or centrifugation as described inStep 2, and gently aspirate the supernatant.8. Repeat Steps 5 through 7.Storing PreparedColumnsTo store a column containing resin, add 0.02% azide or 20% ethanol as apreservative and cap or parafilm the column. Store at room temperature.Continued on next pagePurification Under Native Conditions Using the native buffers, columns and cell lysate, follow the procedure below to purify proteins under native conditions:1. Add 8 ml of lysate prepared under native conditions to a preparedPurification Column (page 9).2. Bind for 30–60 minutes using gentle agitation to keep the resinsuspended in the lysate solution.3. Settle the resin by gravity or low speed centrifugation (800 ×g), andcarefully aspirate the supernatant. Save supernatant at 4°C forSDS-PAGE analysis.4. Wash with 8 ml Native Wash Buffer (page 8). Settle the resin by gravityor low speed centrifugation (800 ×g), and carefully aspirate thesupernatant. Save supernatant at 4°C for SDS-PAGE analysis.5. Repeat Step 4 three more times.6. Clamp the column in a vertical position and snap off the cap on thelower end. Elute the protein with 8–12 ml Native Elution Buffer (seepage 2). Collect 1 ml fractions and analyze with SDS-PAGE.Note: Store the eluted fractions at 4°C. If -20°C storage is required, addglycerol to the fractions. For long term storage, add protease inhibitors to the fractions.If you wish to reuse the resin to purify the same recombinant protein, wash the resin with 0.5 M NaOH for 30 minutes and equilibrate the resin in a suitable binding buffer. If you need to recharge the resin, see page 17.Purification Procedure—Denaturing ConditionsIntroduction Instructions to perform purification using denaturing conditions with prepareddenaturing buffers, columns, and cell lysate are described below.Materials Needed You will need the following items:• Denaturing Binding Buffer (supplied with the system or see page 19 forrecipe)• Denaturing Wash Buffer, pH 6.0 (supplied with the system or see page 19 forrecipe) and Denaturing Wash Buffer, pH 5.3 (see recipe below)• Denaturing Elution Buffer (supplied with the system or see page 20 forrecipe)• Prepared ProBond™ columns with Denaturing buffers (see below)• Lysate prepared under denaturing conditions (page 11)Preparing the Denaturing Wash Buffer pH 5.3 Using a 10 ml aliquot of the kit-supplied Denaturing Wash Buffer (pH 6.0), mix well, and adjust the pH to 5.3 using HCl. Use this for the Denaturing Wash Buffer pH 5.3 in Step 5 next page.Be sure to check the pH of your buffers before starting. Note that thedenaturing buffers containing urea will become more basic over time. PreparingProBond™ ColumnWhen preparing a column as described below, make sure that the snap-off capat the bottom of the column remains intact.If you are reusing the ProBond™ resin, see page 17 for recharging protocol.To prepare a column:1. Resuspend the ProBond™ resin in its bottle by inverting and gentlytapping the bottle repeatedly.2. Pipet or pour 2 ml of the resin into a 10-ml Purification Columnsupplied with the kit. Allow the resin to settle completely by gravity(5-10 minutes) or gently pellet it by low-speed centrifugation (1 minuteat 800 ×g). Gently aspirate the supernatant.3. Add 6 ml of sterile, distilled water and resuspend the resin byalternately inverting and gently tapping the column.4. Allow the resin to settle using gravity or centrifugation as described inStep 2, and gently aspirate the supernatant.5. For purification under Denaturing Conditions, add 6 ml of DenaturingBinding Buffer.6. Resuspend the resin by alternately inverting and gently tapping thecolumn.7. Allow the resin to settle using gravity or centrifugation as described inStep 2, and gently aspirate the supernatant. Repeat Steps 5 through 7.Continued on next pagePurification Procedure—Denaturing Conditions, ContinuedPurification Under Denaturing Conditions Using the denaturing buffers, columns, and cell lysate, follow the procedure below to purify proteins under denaturing conditions:1. Add 8 ml lysate prepared under denaturing conditions to a preparedPurification Column (page 11).2. Bind for 15–30 minutes at room temperature using gentle agitation (e.g.,using a rotating wheel) to keep the resin suspended in the lysatesolution. Settle the resin by gravity or low speed centrifugation (800 ×g), and carefully aspirate the supernatant.3. Wash the column with 4 ml Denaturing Binding Buffer supplied with thekit by resuspending the resin and rocking for two minutes. Settle theresin by gravity or low speed centrifugation (800 ×g), and carefullyaspirate the supernatant. Save supernatant at 4°C for SDS-PAGEanalysis. Repeat this step one more time.4. Wash the column with 4 ml Denaturing Wash Buffer, pH 6.0 supplied inthe kit by resuspending the resin and rocking for two minutes. Settle the resin by gravity or low speed centrifugation (800 ×g), and carefullyaspirate the supernatant. Save supernatant at 4°C for SDS-PAGEanalysis. Repeat this step one more time.5. Wash the column with 4 ml Denaturing Wash Buffer pH 5.3 (see recipeon previous page) by resuspending the resin and rocking for 2 minutes.Settle the resin by gravity or low speed centrifugation (800 ×g), andcarefully aspirate the supernatant. Save supernatant at 4°C for SDS-PAGE analysis. Repeat this step once more for a total of two washes with Denaturing Wash Buffer pH 5.3.6. Clamp the column in a vertical position and snap off the cap on thelower end. Elute the protein by adding 5 ml Denaturing Elution Buffersupplied with the kit. Collect 1 ml fractions and monitor the elution bytaking OD280readings of the fractions. Pool the fractions that contain the peak absorbance and dialyze against 10 mM Tris, pH 8.0, 0.1% Triton X-100 overnight at 4°C to remove the urea. Concentrate the dialyzedmaterial by any standard method (i.e., using 10,000 MW cut-off, low-protein binding centrifugal instruments or vacuum concentrationinstruments).If you wish to reuse the resin to purify the same recombinant protein, wash the resin with 0.5 M NaOH for 30 minutes and equilibrate the resin in a suitable binding buffer. If you need to recharge the resin, see page 17.。

Amicon® Pro AffinityConcentration Kit - GSTPurification of GST-tagged recombinant proteins.Catalog Nos. ACR5000GS, ACK5003GS, ACK5010GS, ACK5030GS,ACK5050GS, ACK5100GSFOR RESEARCH USE ONLYNot for use in diagnostic procedures.USA & Canada Phone: +1(800) 437-7500 Fax: +1 (951) 676-9209Australia +61 3 9839 2000IntroductionThe expression and purification of recombinant proteins is central to protein regulation, structure and function studies. Purification has been greatly facilitated by the addition of small affinity tags to protein sequences such as the glutathione S-transferase domain (GST). GST agarose resin is an affinity chromatography matrix, when used in concert with the Amicon®Pro Affinity Concentration device, offers rapid purification of GST fusion proteins, native GST, or glutathione-binding proteins. GST-tagged proteins bind specifically to reduced glutathione in near-neutral, non-denaturing conditions (e.g., phosphate buffer). Following resin capture of the target protein, unbound lysate components are removed by spin-based clearing and washing steps. Bound protein is recovered by centrifugation following competitive displacement with buffer containing free, reduced glutathione.By condensing the entire purification workflow into one device, the Amicon® Pro device eliminates the need for multiple sample transfers thereby minimizing protein loss. The large exchange device reservoir (up to 9ml) accommodates a range of sample capacities as well as reduces the need for multiple spin steps during the wash and elution phases. Direct coupling to an Amicon® Ultra-0.5 device further provides simultaneous concentration during the elution phase. Lastly, the Amicon® Pro device offers highly efficient diafiltration in a single 15 minute spin.The kit contains sufficient GST resin, optimized buffers, and Amicon® Pro devices for 12 standard reactions.Sample Preparation GuidelinesOptimizing lysis parameters is critical for protein extraction and downstream purification. The Amicon®Pro Affinity Concentration GST kit is compatible with a range of conditions, including reducing agents (β-mercaptoethanol, DTT), chelating agents (EDTA), non-ionic detergents, and BugBuster®Protein Extraction Reagent (Cat. No. 70584), a proprietary mixture of non-ionic detergents offering a rapid cost-effective alternative to mechanical cell lysis methods such as sonication.Irrespective of extraction method, we recommend inclusion of rLysozyme™ Solution (Cat. No.71110) and Benzonase®Nuclease (Cat. No. 70746) and during protein extraction for increased cell lysis (protein yield) and a reduction in sample viscosity (improved sample handling), respectively.Protease inhibitors may also be added to the lysis buffers to protect against degradative enzymes.A listing of individual protease inhibitors and cocktails is available at /search on key words “Protease Inhibitor.” Note: Serine proteases should be used with caution if the GST fusion tag is to be cleaved via Thrombin or Factor Xa digestion. If used during protein extraction, we strongly recommend the eluted fraction be buffer exchanged prior to performing the cleavage reaction.In certain systems, high protein expression can lead to aggregation in the form of inclusion bodies.Strong denaturants such as 6 M guanidine or 8 M urea can be used to solubilize aggregates greatly enhancing yield. However, only properly folded, functional GST is capable of binding GST resin. GST fractions recovered from denaturation of inclusion bodies must be refolded to reconstitute active GST fusion constructs. To a degree, this can be accomplished through use of the Protein Refolding Kit (Cat. No. 70123).Cat. No. BWEGSFor more information, consult the GST•Bind™ Resin User Guide available at /chemicals. Search using the Cat. No. 70541.Kit ComponentsCS211421-GST Resin (3 mL) – Crosslinked agarose with immobilized reduced glutathione supplied as 50% slurry in 50 mM Phosphate Buffer pH 7.5, 0.15 M NaCl, 0.1% NaN3. The resin utilizes an 11-atom spacer arm (~16 Å) for covalent attachment of reduced glutathione via a sulfide linkage. The binding capacity is typically 5-8 mg/mL of settled resin.Buffers:o CS211416-10X GST Bind/Wash Buffer (25 mL) – 43 mM Na2HPO4, 14.7 mM KH2PO4,1.37 M NaCl, 27 mM KCl pH7.2o CS211354-10X Glutathione Reconstitution buffer (5 mL) – 500 mM Tris, pH 8.0o CS211418-Glutathione, reduced, free acid (125 mg)Amicon®Pro Devices – The kit includes 12 complete assemblies. Each device consists of an exchange device, holder tube, 50 mL collection tube, and Amicon®Ultra-0.5 centrifugal filter device. A 2 mL collection tube is included for sample recovery from the AU-0.5 device by reverse spin. The kit is available in five formats based on the nominal molecular weight limit (NMWL) of the AU-0.5: 3 (ACK5003GS), 10 (ACK5010GS), 30 (ACK5030GS), 50 (ACK5050GS), and 100 kDa (ACK5100GS). Consult the Amicon®Pro (/psp, search keywords “Amicon Pro”) and Amicon®Ultra-0.5 centrifugal filter device User Guides (/catalogue/module/c82301) for proper assembly/disassembly and additional product information.All reagents should be stored at 2° to 8°C (do not freeze). The Amicon Pro devices can be stored separately at room temperature.Procedures for using the Amicon® Pro Affinity Concentration Kit - GSTThe protocol is based on purification of GST-tagged protein from 1 mL of E. coli lysate using 200 µl of GST resin slurry (100 µL packed resin). The protocol is linearly scalable for 50-1000 µL of resin slurry. Due to large variability among sample preps, parameters which may require optimization include bead input, binding time, wash, and elution parameters. This protocol includes steps for simultaneous concentration during the elution step as well as buffer exchange using the Amicon®Ultra-0.5 centrifugal filter device.Note: Given the collection tube’s capacity, it is not necessary to remove filtrate between the various centrifugation steps. However, if process samples need be retained for analytical purposes, the collection tube should be cleared.Buffer Preparation10X Bind/Wash Buffer should be diluted to 1X with sterile deionized H2O; 1X Bind/Wash solution may be stored at 4°C for 1 week.Prepare 10X GST Elution Buffer containing 100 mM reduced glutathione as follows: add 4ml 10X Reconstitution buffer directly to the glutathione powder vial. Pipette repeatedly then incubate for30 minutes at room temperature. Divide buffer into working volumes and store at -20o C (400Cat. No. BWEGSµl/standard reaction); 10X GST Elution Buffer is stable for 6 months at -20o C. We do not recommend repeated freeze/thaw cycles. To minimize glutathione oxidation, dilute 10X buffer to 1X using sterile deionized water and pH to 8.0 immediately before use. Discard any unused 1X Elution Buffer.Bead Preparation1. To ensure uniform suspension, vortex the GST resin thoroughly before adding it to the device.2. Remove the collection tube cap and open the exchange device cap.3. Add 200 µL of resin slurry to the base of the exchange device. Close the exchange cap.Up to 500 µl packed resin (1000 µl slurry volume) may be added per device We recommend using wide-bore tips (Cat. No. 02-707-134, Fisher Scientific) for resin transfer.4. To remove storage buffer, centrifuge in a swinging bucket rotor at 1000 g X 1 min.5. Add 500 µL of 1X Bind Buffer. Centrifuge at 1000 g X 1 min.Protein Binding1. Add 500 µL of sample to the exchange device.Up to 9 mL of sample can be added. The volume loaded is determined by the target protein’s expression level and resin’s binding capacity.2. Incubate for 60 min at room temp with gentle agitation.We recommend upright agitation on a plate shaker at low setting.End-over-end mixing, particularly with small volumes or for extended time, may result in substantial bead loss to the sides of the feeder tube.The duration of binding time may vary with application.3. Centrifuge the device at 1000 g X 1 min in a swinging bucket rotor. Recover the sample flow-through from the 50 mL collection tube (optional).To ensure maximal protein capture, collect all resin into solution prior to centrifugation.4. Add1.5 mL of Wash Buffer. Centrifuge at 1000 g X 1 min. Recover the wash fraction from the 50mL collection tube (optional).Due to the large capacity of the exchange device, the volume of the wash can be increased for greater sample purity. There is no need for multiple wash steps.Sample ElutionSamples can be eluted without concentration by adding elution buffer and centrifuging (1000g X 2 minute) directly into a clean 50 ml collection tube. Given the limited volume processing capacity of the AU-0.5 device, we recommend this protocol if elution volumes > 1.5 ml are required.For simultaneous elution with concentration, attach the Amicon® Ultra-0.5 device and follow the steps outlined below.1. Remove the exchange device and insert it into the AU0.5 device.2. Place the exchange device/AU-0.5 assembly back in the holder and return the device to thecollection tube.3. Add up to 1.5 mL of Elution Buffer, gently resuspend the resin, and incubate for 5 min.Under standard conditions, one elution is sufficient for recovery of 90-95% of captured protein.4. Close the exchange device cap and screw on the collection tube cap to ensure a proper seal.5. Centrifuge at 4000 g X 15 min in a swinging bucket rotor. Concentrated samples can be bufferexchanged or recovered from the AU-0.5 device by reverse spin (see below).Cat. No. BWEGSDepending on the starting elution volume, NMWL of AU0.5 device employed, and the degree of concentration desired, the length of the spin time can range for 10-30 minutes. Please consult the Performance Characteristics section in the Amicon®Pro Affinity Concentration System User Guide (/psp, and search keywords "Amicon Pro”) for recommended guidelines., consult.6. Recover the concentrated fraction by reverse spin or proceed to Buffer Exchange (see below).Buffer Exchange (Optional if samples have been collected in the Amicon® Ultra-0.5 device)1. After sample concentration, add 1.5ml desired buffer to the exchange device/AU-0.5 assembly.2. Centrifuge device at 4000g X 15 minutes in a swinging bucket rotor. Concentrated samples canbe recovered from the AU-0.5 device by reverse spin (see below).Collect sample from the AU0.5 device by Reverse Spin(following Concentration or Buffer Exchange)1. Disassemble the exchange device/AU-0.5 assembly from the holder tube.2. Using a gentle twisting motion, detach the AU-0.5 from the exchange device.3. If there is residual sample in the exchange device tip, depress the exchange device cap to expelthe remaining sample volume into the AU-0.5.4. Hold AU-0.5 upright and slide the 2 ml collection tube on top of it.5. Invert the assembly and centrifuge (in a microcentrifuge) with a fixed angle rotor 1000g X 2min.Sample protein yield can be determined by Mid IR-based spectrometry using the DirectDetect™ biomolecular quantitation system and DirectDetect™ Assay Free Sample Cards.TroubleshootingCat. No. BWEGSIssue: Recombinant Protein is present in low amount in eluatePossible Cause SolutionProtein is insoluble and may have formed inclusion bodies. After lysate clearance, check both the supernatant and pellet for protein. Perform lysis and binding procedures under denaturing conditions.The GST fusion protein is not in the proper three dimensional conformation. Attempt to reconstitute native protein structure using Protein Refolding Kit (Cat. No. 70532).The GST-tag is not exposed for binding to the affinityresin.The protein may require denaturing conditions for binding.The GST-tag is not present. Sequence the ligation junctions to ensure that the readingframe is correct. Check for possible internal translation starts(N-terminal tag) or premature termination sites (C-terminal tag). Recombinant protein is degraded during cell lysis. Add protease inhibitors during cell lysis.Protein forms aggregates. Add solubilizing agents such as detergents (0.1% Triton® X-100, TWEEN®-20) or increase salt concentration.pH of the Lysis or Binding buffers is incorrect. Check buffer pH; the acceptable range is 7-8. Acidic bufferswill prevent binding.Protein expression is insufficient. Optimize the growth/induction conditions.Cell Lysis is incomplete. Optimize the Cell Lysis Protocol.Cell Lysate is too viscous. If possible, dilute the lysate in Bind Buffer. Alternatively,include Benzonase® Nuclease during lysis to remove freeRNA/DNA.Protein precipitates during sample concentration while using the AU0.5 device due to over-concentration. Reduce the duration of the centrifugation time during the elution/concentration step.Protein is lost during sample concentration while using AU0.5 device. Check the protein's expected size and MWCO of AU0.5 device used. AU0.5 device is offered in 5 different MWCO formats - 3, 10, 30, 50, and 100 kDa.Issue: High Non-specific bindingPossible Cause SolutionInsufficient washing. Increase the volume of the Wash Buffer used or the number ofwash steps. Alternatively, supplement the Bind/Wash Buffers.Contaminants interact directly with the GST fusion protein. Add reducing agents such as DTT or β-mercaptoethanol to reduce disulfide bonds. Add detergents to disrupt non-specific interactions.GST fusion protein is degraded. Degraded/truncated forms of the recombinant protein will stillbind to the GST resin and appear as contaminating bands inSDS-PAGE. Perform a lysis procedure on ice and includeprotease inhibitors.Cell lysate is too concentrated. Dilute the lysate in Bind Buffer before purification.Cat. No. BWEGSCat. No. BWEGSPerformanceProduct Ordering InformationNo Devices Amicon ® Pro + AU 0.5 ml with MWCO: 3k 10k 30k 50k 100k Amicon ® Pro Affinity Concentration Kit - GSTACR5000GS ACK5003GS ACK5010GS ACK5030GS ACK5050GS ACK5100GS Amicon ® Pro AffinityConcentration System 12PKACS500312 ACS501012 ACS503012 ACS505012 ACS510012 Amicon ® Pro AffinityConcentration System 24PK ACS500324 ACS501024 ACS503024 ACS505024 ACS510024The Amicon ® Pro Affinity Concentration Kit contains reagents and devices sufficient for 12 standard reactions. Amicon ® Pro devices are also sold separately in 12 and 24 packs.Description Catalogue Number Qty/Pack GST•Bind™ Resin 70541-3/4/5 10/50/25 mL GST•Bind™ Buffer Kit 70534 1 KitPurification using Amicon Pro Affinity Concentration Kit – GST purifications of a 50 kDa GST-tagged protein the Amicon ® Pro Affinity Concentration Kit numbered lanes are: 2 – flowthrough fraction 3 – wash fraction 4 – eluted fractionNoticeThe information in this document is subject to change without notice and should not be construed as a commitment by EMD Millipore Corporation (“Millipore”) or an affiliate. Neither EMD Millipore nor any of its affiliates assumes responsibility for any errors that may appear in this document. Technical AssistanceFor more information, contact the office nearest you. Up-to-date world-wide contact information is available on our web site at /offices. You can also visit the tech service page on our web site at /techserves.WarrantyEMD Millipore Corporation(“EMD Millipore”) warrants its products will meet their applicable published specifications when used in accordance with their applicable instructions for a period of one year from shipment of the products. EMD MILLIPORE MAKES NO OTHER WARRANTY, EXPRESSED OR IMPLIED. THERE IS NO WARRANTY OF MERCHANTABILITY OR FITNESS FOR A PARTICULAR PURPOSE. The warranty provided herein and the data, specifications and descriptions of EMD Millipore products appearing in EMD Millipore’s published catalogues and product literature may not be altered except by express written agreement signed by an officer of EMD Millipore. Representations, oral or written, which are inconsistent with this warranty or such publications are not authorized and if given, should not be relied upon.In the event of a breach of the foregoing warranty, EMD Millipore Corporation’s sole obligation shall be to repair or replace, at its option, the applicable product or part thereof, provided the customer notifies EMD Millipore Corporation promptly of any such breach. If after exercising reasonable efforts, EMD Millipore Corporation is unable to repair or replace the product or part, then EDM Millipore shall refund to the Company all monies paid for such applicable Product. EMD MILLIPORE CORPORATION SHALL NOT BE LIABLE FOR CONSEQUENTIAL, INCIDENTAL, SPECIAL OR ANY OTHER DAMAGES RESULTING FROM ECONOMIC LOSS OR PROPERTY DAMAGE SUSTAINED BY ANY COMPANY CUSTOMER FROM THE USE OF ITS PRODUCTS.Unless otherwise stated in our catalog or other company documentation accompanying the product(s), our products are intended for research use only and are not to be used for any other purpose, which includes but is not limited to, unauthorized commercial uses, in vitro diagnostic uses, ex vivo or in vivo therapeutic uses or any type of consumption or application to humans or animals.(c) 2009 - 2012: Merck KGaA, Darmstadt. All rights reserved. No part of these works may be reproduced in any form without permission in writing.Benzonase®, BugBuster®, and Amicon® are registered trademarks of Merck KGaA, Darmstadt, Germany. DirectDetect™ and GST•Bind™ is a trademark of Merck KGaA, Darmstadt, Germany. Triton® is a registered trademark of Dow Chemical Company. Tween® is a registered trademark of ICI Americas, Inc.Cat. No. BWEGS。


檵檵檵檵檵檵檵檵檵檵殝殝殝殝研究论文标签谷胱甘肽S-转移酶的二价亲和标记试剂的制备与应用卢进鹏a 夏超a 周伟a 李亚平a 胡小蕾a 廖飞b 杨晓兰a *(a 重庆医科大学检验医学院,临床检验诊断学教育部重点实验室重庆400016;b 重庆理工大学药学与生物工程学院重庆401135)摘要设计合成融合表达标签谷胱甘肽S-转移酶(GST )的二价亲和标记试剂,用于功能化磁珠后位点选择性固定化标签GST ,为磁分离筛选配体混合物库提供固定化融合靶蛋白的候选方案。
为减少疏水配体在标签GST 活性位点的结合,需同时占据标签GST 双活性中心内疏水结合位点并发生共价修饰的二价亲和标记试剂。
以双苯环为疏水定位基、溴乙酰基为巯基修饰基团、羧基为连接官能团得单价标记试剂,以二乙基三胺为连接臂将单价标记试剂与连接臂两端伯胺连接得标签GST 的对称二价亲和标记试剂,再以线性三胺连接臂中间的氨基与羧基磁珠偶联得功能化磁珠。
表征目标化合物对标签GST 的标记动力学、结合比;功能化磁珠对标签GST 的不可逆固定化动力学和固载容量,及将磁珠表面二价亲和标记试剂转变成还原型谷胱甘肽(GSH )加合物后对标签GST 可逆固定化的效果;以碱性磷酸酶及疏水荧光配体为模型考察磁珠固定化标签GST 后的非特异结合。
目标化合物对标签GST 半抑制浓度为(22ʃ0.2)μmol /L ,其与GSH 的饱和加合物半抑制浓度为(0.41ʃ0.06)μmol /L ,二者与标签GST 二聚体结合比接近1ʒ1。
功能化磁珠对标签GST 不可逆及可逆固定化的容量均接近25mg /g 磁珠。
偶联GST 的磁珠对蛋白非特异吸附很弱,再进一步用单价亲和标记试剂和GSH 加合物封闭固定化标签GST 剩余的活性位点后对疏水小分子也无显著结合。
结果表明,所设计二价亲和标记试剂功能化磁珠适合用于标签GST 及其融合表达蛋白的位点选择性固定化。
关键词谷胱甘肽S-转移酶;亲和标记试剂;磁珠;固定化中图分类号:O621文献标识码:A 文章编号:1000-0518(2019)04-0402-12DOI :10.11944/j.issn.1000-0518.2019.04.1802692018-08-17收稿,2018-11-22修回,2018-12-05接受国家自然科学基金(31570862,81773625)资助项目通讯联系人:杨晓兰,教授;Tel :023-68485240;Fax :023-********;E-mail :xiaolanyang666@yeah.net ;研究方向:蛋白配体设计合成、高通量筛选表面固定化特定蛋白的微纳米材料广泛应用在生物分子高通量筛选、蛋白相互作用分析和微流体催化等领域[1-4]。

Expression, purification and identification of GST fusion proteinProtein Purification by Affinity ChromatographyProtein Separation by SDS-PAGESpecific Protein Identification by Western-blottingContents1. Induced Expression of GST fusion protein in E coli. P1142. Protein Separation of by SDS-PAGE. P383. Protein Identification by Western-blotting. P114工程菌中外源GST融合蛋白的分离纯化及鉴定1、E coli菌中外源GST融合蛋白的诱导表达(IPTG诱导)2、特异外源GST融合蛋白表达的鉴定(Western-blotting)3、细菌的裂解(溶菌酶法lysozyme)4、Glu-Agarose beads亲和层析分离GST fusion蛋白5、SDS-PAGE鉴定GST fusion蛋白的纯度Determining Protein Expression by Western-blottingGST (Glutathione S-transferases) protein expression in E. coliWestern blotting ( Immunoblotting )1. DefinitionA technique for detecting specific proteins separated by electrophoresis by use of labeled antibodies. So called since it has some similarity to a Southern blotting.2. Why we have to use it?To identify a target protein in a complex mixture, and measure its expression level.Pre-Procedure of Western blottingProtein extraction:Sample come from cells, serum/plasma, tissues and culture supernatantMeasurement of Protein concentration(For Western Blotting: 20-60 g/well)Steps of Western blottingStep1. Use gel electrophoresis to separate proteins PAGE ( polyacryamide gel electrophoresis ) or SDS-PAGE -- Sodium dodecyl sulfate PAGE.Step2. Transfer the proteins in the gel onto a NC (nitrocellulose) or PVDF membraneStep3. Staining: use the specific antibody to identify the target proteinProcedure of Western blotting1. Gel preparation2. loading Sample:Sample (50~100 μg/well ) + equal volume of 2×SDS-PAGE loading buffer,100℃for 3~5 min,loading Sample and prot. Marker.3. Running the gel: Apply 8V/cm gel until the proteins are well into the stacking gel, then 15V/cm gel until the tracking dye reaches the bottom of the gel.Membrane transfer: soak the NC membrane and filter papers in transfer buffer before assembling the transfer apparatus. Assemble the transfer apparatus according to the following sequence. Avoid air bubbles between any of the layers .4°C,60mA, overnight.Blocking membrane: block the membrane in 10~15 ml 5% BSA in TBS-T for 1 hour of (rotation)Primary antibody incubation: Remove the blocking solution and incubate the membrane with 10 ml primary antibody for 2 hours at room temperature(rotation) or overnight at 4°CWash the membrane: Remove the antibody and perform 3 room temperature washes (15 minutes of rotation each) with 30ml TBS-T.Secondary antibody incubation: Incubate the membrane with 10ml secondary antibody for 1 hour at room temperature (rotation)Wash the membrane: emove the antibody and perform 3 room temperature washes (15 minutes of rotation each) with 30ml TBS-T.Developing: (Use of chromomeric substrate- 3,3’-diaminobenzidine, DAB) DAB is most sensitive for immunocoupled horseradish peroxidase. It is converted in situ a brown precipitate. -Transfer the membrane to a shallow tray.Procedure of Western blottingTransfer the membrane to a shallow tray.-Add 10ul H2O2 (30%) to 10 ml of 0.05%DAB in PBS, mix well immediately;-Pour the DAB to the membrane, incubate at room temperature with gentle shaking in the dark if possible.-Monitor the progress of the reaction carefully. When the bands are of the desired intensity (2-10min), wash the filter briefly in water, and in PBS.-Dry the membrane and photograph it to provide a permanent record of the experiment.Western blottingStripping Membranes:*Put the membrane in PBS or TBS with 7ul/ml β-mercaptoethanol and 2%SDS for 30' at Room temperature in agitation.* Wash the membrane for 30' in PBS or TBS.* Put the membrane in blocking buffer注意:硝酸纤维素(NC)膜是通过疏水作用和蛋白质相联,反复洗几次后,蛋白易掉下来,结果较差。
A Method for the Rapid and Efficient Elution of NativeAffinity-Purified Protein A Tagged ComplexesCaterina Strambio-de-Castillia,†Jaclyn Tetenbaum-Novatt,†Brian S.Imai,‡Brian T.Chait,§andMichael P.Rout*,†The Rockefeller University,1230York Avenue,New York New York 10021-6399Received May 24,2005A problem faced in proteomics studies is the recovery of tagged protein complexes in their native and active form.Here we describe a peptide,Bio-Ox,that mimics the immunoglobulin G (IgG)binding interface of Staphylococcus aureus Protein A,and competitively displaces affinity-purified Protein A fusion proteins and protein complexes from IgG-Sepharose.We show that Bio-Ox elution is a robust method for the efficient and rapid recovery of native tagged proteins,and can be applied to a variety of structural genomics and proteomics studies.Keywords:Staphylococcus aureus •Protein A •affinity purification •proteomics •fusion proteinIntroductionProtein -protein interactions are central to the maintenance and control of cellular processes.The study of such protein -protein interactions has been greatly enhanced by fusion protein technology,wherein specific peptide or protein domain “tags”are fused to the protein of interest (generally at either its carboxyl-terminus or amino-terminus).These tags can facilitate the detection,increase the yield,and enhance the solubility of their associated proteins.1-3Most importantly,these fusion domains have been exploited to allow the single-step purification of the test protein either alone or in complexes with its in vivo binding partners.4-6The yield of these purifica-tion methods is often high enough to allow the identification of such binding partners by mass spectrometry.A commonly used affinity tagging method generates ge-nomically expressed Protein A (PrA)fusion proteins by modify-ing the coding sequence of the protein under study via PCR-directed approaches.7-9This method takes advantage of the ∼10nM binding affinity of PrA from S taphylococcus aureus for the constant region (Fc)of immunoglobulin G (IgG).10After purification on IgG-conjugated resins,PrA-tagged proteins or protein complexes are most commonly eluted from the resin using high or low pH conditions.These elution methods typically lead to the denaturation of the isolated proteins,the dissociation of complexes,and concomitant loss of activity.However,it is often desirable to recover soluble native protein or protein complexes.One method by which this can be achieved is by constructing a cleavable tag.Such tags carry a specific cleavage site for a protease placed proximal to the tagged protein,allowing the tag to be removed from the fusion protein.Proteases that are widely used for this purpose includeblood coagulation factors X (factor Xa),enteropeptidase (en-terokinase),alpha-thrombin,and the tobacco etch virus (TEV)protease.Nevertheless,this method has drawbacks.First,the literature is replete with reports of fusion proteins that were cleaved by these proteases at sites other than the canonical cleaving site.11-14Second,the removal of the tag destroys the ability to detect or further purify the protein of interest,necessitating the encumbrance of a second,tandem tag.15Here,we describe a rapid single step method for the efficient recovery of native and active PrA fusion proteins and protein complexes from IgG-Sepharose.This technique avoids the complications of having to use a protease and in addition has the advantage of retaining the original tag on the target protein after elution,permitting further purification steps and detection of the fusion protein in subsequent experiments.Our method takes advantage of a previously described peptide,termed FcIII,16which mimics the protein -protein binding interface of PrA for the hinge region on the Fc domain of human IgG.We modified FcIII by the addition of a biotin moiety to its amino-terminus to increase the peptide’s solubility while leaving its affinity for Fc intact s making it a more effective elution reagent.We termed this modified peptide,Bio-Ox.To investigate the properties of Bio-Ox,PrA-tagged proteins were isolated in their native state from yeast on an IgG-conjugated Sepharose resin,either alone or in combination with their in vivo interacting partners;the Bio-Ox peptide was then used to competitively displace the tagged proteins and elute them from the resin.The efficiency of elution was monitored by quantitatively comparing the amounts of proteins eluted to the amounts remaining on the resin under a variety of test conditions.We show that Bio-Ox elution is a robust method for the efficient and rapid recovery of native tagged proteins that can be applied to a variety of structural genomics and proteomics studies.*To whom correspondence should be addressed.Tel:+1(212)327-8135.E-mail:rout@.†Laboratory of Cellular and Structural Biology,Box 213.‡Proteomics Resource Center,Box 105.§Laboratory of Mass Spectrometry and Gaseous Ion Chemistry,Box 170.2250Journal of Proteome Research 2005,4,2250-225610.1021/pr0501517CCC:$30.25©2005American Chemical SocietyPublished on Web10/08/2005Experimental SectionPeptide Synthesis,Oxidation and Cyclization.Peptides were synthesized using standard Fmoc protocols.Typical deprotec-tion times with20%piperidine were2times10min and typical coupling times with4-10-fold excess of amino acids over resin were2to6h.Small batches of peptides were made on a Symphony synthesizer(Protein Technologies,Inc.),while larger batches were made manually.Peptides were cleaved from the resin using94.5%trifluoroacetic acid, 2.5%water, 2.5% ethanedithiol and1%triisopropylsilane for3h at25°C.The solubilized peptides were precipitated with10volumes of cold tert-butyl methyl ether and the precipitated peptide was washed several times with ether prior to air-drying.The air-dried peptide was dissolved in20%acetonitrile in water to approximately0.5mg/mL,the pH was adjusted to8.5using sodium bicarbonate and the peptide was allowed to air oxidize overnight to promote cyclization.The progress of cyclization was monitored by mass spectrometry.The cyclized crude peptide was purified using standard preparative reversed phase HPLC using a Vydac218TP1022C18column.Peptide Solubility.Eluting peptides were suspended at a concentration of440µM(0.77mg/mL for BioOx;0.67mg/mL for FcIII),in peptide buffer by extensive vortexing.The peptide concentration was verified by measuring the OD280nm of each solution(extinction coefficient:1OD280nm)0.13mg/mL).The peptide solutions/suspensions were then combined with equal amounts of a100mM buffer to obtain∼220µM peptide at a range of pH values(buffers:Na-Acetate pH4.8,Na-Citrate pH 5.4,Na-Succinate pH5.8,Na-MES pH6.2,BisTris-Cl pH6.5, Na-HEPES pH7.4,Na-TES pH7.5,Tris-Cl pH8.3,Na-CAPSO pH9.6).Samples were incubated at room temperature with gentle agitation for20min,and then insoluble material was removed by centrifugation at21000×g max for20min at25°C.The concentration of peptide in each remaining superna-tant was determined by measuring its OD280nm.To determine the maximum solubility of each peptide,the peptides were dissolved to saturation in peptide buffer by extensive vortexing and incubation with stirring at25°C overnight.Insoluble material was removed by centrifugation at15000×g for15min at25°C and the amount of dissolved peptide was measured directly by amino acid analysis.Peptide Competitive Displacement of Bound Recombinant PrA from IgG-Sepharose.Recombinant PrA(280µg;6.7nmol) from S.aureus(Pierce)was dissolved in1mL TB-T[20mM HEPES-KOH pH7.4,110mM KOAc,2mM MgCl2,0.1%Tween-20(vol/vol)]and added to280µL of packed pre-equilibrated Sepharose4B(Amersham Biosciences)conjugated with affinity-purified rabbit IgG(ICN/Cappel; 1.87nmoles IgG).After incubation on a rotating wheel overnight at4°C,the resin was washed twice with1mL TB-T,twice with1mL TB-T containing 200mM MgCl2,and twice with1mL TB-T.After the final wash, the resin was divided evenly into14equal aliquots.The peptide was dissolved in peptide buffer at concentrations ranging between0and440µM peptide.Aliquots of400µL of the appropriate peptide solution was added to each PrA-IgG-Sepharose containing tube,and the tubes were then incubated on a rotating platform for3h at4°C followed by1h at25°C. After displacement of bound PrA from the IgG-Sepharose,the resin was recovered by centrifugation on a Bio-Spin column (BioRad),and resuspended in one-bed volume of sample buffer.Samples were separated by SDS-PAGE.Yeast Strains.Strains are isogenic to DH5alpha unless otherwise specified.All yeast strains were constructed using standard genetic techniques.C-terminal genomically tagged strains were generated using the PCR method previously described.7,17Affinity Purification of Proteins and Protein Complexes on IgG-Sepharose.The protocol for the purification of PrA-containing complexes was modified from published methods.18-20For the purification of Kap95p-PrA,yeast cytosol was prepared essentially as previously described.21,22Kap95p-PrA cytosol was diluted with3.75volumes of extraction buffer 1[EB1:20mM Hepes/KOH,pH7.4,0.1%(vol/vol)Tween-20, 1mM EDTA,1mM DTT,4µg/mL pepstatin,0.2mg/mL PMSF]. The diluted cytosol was cleared by centrifugation at2000×g av for10min in a Sorvall T6000D tabletop centrifuge and at 181000×g max for1h in a Type80Ti Beckman rotor at4°C.10µL bed volume of IgG-Sepharose pre-equilibrated in EB1was added per0.5mL of cytosol and the binding reaction was incubated overnight at4°C on a rotating wheel.The resin was recovered by centrifugation at2000×g av for1min in a Sorvall T6000D tabletop centrifuge,transferred to1.5mL snap-cap tubes(Eppendorf),and washed6times with EB1without DTT. For the purification of Nup82p-PrA,cells were grown in Whickerham’s medium21to a concentration of4×107cells/ mL,washed with water and with20mM Hepes/KOH pH7.4, 1.2%PVP(weight/vol),4µg/mL pepstatin,0.2mg/mL PMSF, and frozen in liquid N2before being ground with a motorized grinder(Retsch).Ground cell powder(1g)was thawed into10 mL of extraction buffer2[EB2;20mM Na-HEPES,pH7.4,0.5% TritonX-100(vol/vol),75mM NaCl,1mM DTT,4µg/mL pepstatin,0.2mg/mL PMSF].Cell lysates were homogenized by extensive vortexing at25°C followed by the use of a Polytron for25s(PT10/35;Brinkman Instruments)at4°C.Clearing of the homogenate,binding to IgG-Sepharose,resin recovery and washing was done as above except that10µL of IgG-Sepharose bed volume was used per1g of cell powder and EB2without DTT was used for all the washes.Elution of the PrA tagged complexes was performed as described below.Peptide Elution of Test Proteins and Protein Complexes and Removal of Peptide by Size Exclusion.Kap95p-PrA or Nup82p-PrA bound IgG-Sepharose resin was recovered over a pre-equilibrated Bio-Spin column(BioRad)by centrifugation for1min at1000×g max.Three bed-volumes of440µM(unless otherwise indicated in the text)of eluting peptide in peptide buffer were added per volume of packed IgG Sepharose resin. The elution was carried out for various times(as indicated in the text)at either4°C or at25°C.When elution was complete, the eluate was recovered over a Bio-Spin column.Finally,the resin was washed with one bed-volume of elution buffer to displace more eluted material from the resin and the wash was pooled with the initial eluate.The peptide was removed by filtration of the eluate over a micro spin G25column(Amer-sham Biosciences)as described by the manufacturer.Kap95p-Nup2p in Vitro Binding Experiments.To demon-strate in vitro binding of proteins after elution from the resin, Kap95p-PrA from0.3mL of yeast cytosol was affinity-purified on17.5µL of packed IgG-Sepharose and eluted with52.5µL of440µM Bio-Ox for2.5h at4°C followed by1h at25°C. The resulting sample(total volume88µL)was mixed with0.1µL of E.coli total cell lysate containing Nup2p-GST(generous gift from David Dilworth and John Aitchison23)and brought to a total volume of500µL with TB-T,1mM DTT,4µg/mL pepstatin,0.2mg/mL PMSF.Controls were set up in the absence of either Kap95p-PrA or Nup2p-GST.The samples were incubated at25°C for30min after which40µL of packed,pre-Native Elution of PrA-Tagged Proteins research articlesJournal of Proteome Research•Vol.4,No.6,20052251equilibrated glutathione-Sepharose 4B resin (Amersham Bio-sciences)was added per sample and the incubation was continued at 4°C for 1h.After nine washes with 1mL of TB-T,1mM DTT,4µg/mL pepstatin,0.2mg/mL PMSF,at 25°C,the resin was recovered on Bio-Spin columns as described above and bound material was eluted with 40µL of sample buffer.The samples were resolved on SDS-PAGE alongside an aliquot of input peptide-eluted Kap95p-PrA.To demonstrate the recovery of in vitro reconstituted protein complexes from the resin,Kap95p-PrA from 0.3mL of yeast cytosol was affinity-purified on 10µL of packed IgG-Sepharose and the washed resin was equilibrated in TB-T,1mM DTT,4µg/mL pepstatin,0.2mg/mL PMSF.This pre-bound Kap95p-PrA was mixed with 50µL of E.coli total cell lysate containing Nup2p-GST in a total volume of 1mL of TB-T,1mM DTT,4µg/mL pepstatin,0.2mg/mL PMSF.A mock control experiment was set up in the absence of Nup2p-GST.The binding reaction was carried out for 1h at 4°C and the resin was washed 2times with 1mL of TB-T,2times with 1mL of TB-T containing 100µM ATP and 3times with peptide buffer (all washed were without DTT).Bound material was eluted with 30µL of 440µM Bio-Ox in peptide buffer at 4°C for 2.5h at 4°C followed by 1h at 25°C.Samples were resolved by SDS-PAGE.Figure 1.Addition of a Biotin moiety to the FcIII peptide does not alter the ability of the peptide to competitively displace bound PrA from IgG-Sepharose.(a)Primary sequence and chemical structure of the biotinylated FcIII peptide,Bio-Ox.(b)220µM suspensions of peptides were prepared in buffers of different pHs,and allowed to solubilize.The material remaining in the buffer after centrifugation is plotted for Bio-Ox (closed triangles,black trend line )and FcIII (open circles,gray trend line ;dashed horizontal line represents the starting 220µM level .(c)Increasing amounts of Bio-Ox (closed triangles )and FcIII (open diamonds )were used to competitively displace recombinant PrA from IgG-Sepharose.The amounts of PrA left on the resin after elution were resolved by SDS-PAGE alongside known amounts of PrA standards.The data are displayed on logarithmic scale on both axes.Data are displayed as a %recovery relative to the input PrA amount (i.e.,PrA amount remaining bound in the absence of eluting peptide).Linear regression for both data sets was used to calculate the IC50.research articlesStrambio-de-Castillia et al.2252Journal of Proteome Research •Vol.4,No.6,2005Quantitation and Image Analyses.Band intensities were quantified with the Openlab software (Improvision),and the data was plotted using Excel (Microsoft).Results and DiscussionDesign of the PrA Mimicking Peptide.The hinge region on the Fc fragment of immunoglobulin G (IgG)interacts with Staphylococcus aureus Protein A (PrA).This region was also found to be the preferred binding site for peptides selected by bacteriophage display from a random library.16The specific Fc binding interactions of a selected 13amino acid peptide (termed FcIII),were shown to closely mimic those of natural Fc binding partners.We reasoned that this peptide could be used to efficiently displace PrA tagged proteins from IgG-conjugated affinity resins.Initial trials with FcIII determined that,although it functioned as an eluant,it exhibited a strong tendency to aggregate and its solubility under physiological conditions was not sufficient for many practical purposes,leading to low yields and nonreproducible results.As the high peptide concentrations needed for elution are outside the conditions for which the FcIII peptide was designed,we synthesized several modified peptides based on FcIII,with the specific aim of increasing their solubility and decreasing their degree of aggregation under conditions that would be useful for the isolation of proteins and protein complexes.Among the different alternatives,the most efficient in the displacement of bound PrA-tagged Kap95p from IgG-Sepharose was a peptide in which the amino-terminus of the original FcIII peptide wasFigure 2.Bio-Ox can be used to efficiently compete bound PrA-tagged proteins and protein complexes from IgG-Sepharose in a temperature-dependent fashion.(a )Kap95p-PrA/Kap60p was affinity-purified on IgG-Sepharose from logarithmically growing yeast cells.440µM Bio-Ox was used to competitively displace the bound tagged proteins from the IgG-Sepharose resin.The elution reaction was carried out for the times indicated.At the end of the incubation time eluted proteins (E )and proteins remaining bound to the resin (B )were resolved on SDS-PAGE.(b )Kap95p-PrA (closed squares)and Nup82p-PrA (open squares )were affinity-purified on IgG-Sepharose from logarithmically growing yeast cells and eluted as described above.The amounts of eluted versus resin-bound protein was quantified using the OpenLab software and the elution efficiency for each time point is presented as the percentage of eluted material over the total amount of bound plus eluted material (%eluted).(c )440µM Bio-Ox was used to elute Kap95p-PrA or Nup82p-PrA for 1h at 4°C or 25°C as indicated.Native Elution of PrA-Tagged Proteinsresearch articlesJournal of Proteome Research •Vol.4,No.6,20052253modified by the addition of a Biotin moiety (data not shown).We termed this peptide Bio-Ox (Figure 1,panel a).The solubility of Bio-Ox was measured directly by amino acid analysis and was shown to be ∼3-fold greater than the solubility of FcIII at pH 7.4.In addition,comparison of the solubility of both peptides over a range of pHs indicated that the Bio-Ox was considerably more soluble than FcIII at all but the most extreme pHs tested;importantly,Bio-Ox is very soluble across the full physiological range of pHs (Figure 1,panel b).To determine whether the addition of the Biotin moiety could have altered the inhibiting ability of the peptide,we measured the inhibition constant for Bio-Ox and found it to be comparable with the reported K i for FcIII (∼11nM;data not shown).We then measured the IC 50for competitive displacement for FcIII and Bio-Ox,under conditions in which both were soluble.For this test,commercially available recom-binant PrA from S.aureus was first bound to IgG-Sepharose and then increasing concentrations of the peptide were used to displace the bound PrA from the immobilized IgG (Figure 1,panel c).The apparent IC 50was found to be 10.4(3.2µM for FcIII and 9.8(2.6µM for Bio-Ox (mean value of four independent trials (standard deviation of the mean).Taken together,Bio-Ox appears to be as efficient as FcIII at binding to the F c portion of antibodies and competing for this site with Protein A,but is far more soluble in physiologically compatible buffers,a key requirement for an efficient elution peptide (Figure 3).Experimental Design of the Competitive Elution Procedure.The principle of the method is as follows;genomically PrA-tagged proteins of interest are expressed in yeast and affinity isolated on IgG-conjugated Sepharose resin.Depending on the conditions used for lysis and extraction,the test protein can be recovered in native form either in isolation or in complexes with protein partners.After binding,the resin is recovered by centrifugation and washed extensively to remove unbound material.The bound material is competitively displaced from the IgG-Sepharose resin by incubation with 440µM Bio-Ox peptide in peptide buffer for 2h at 4°C.Finally,the peptide is rapidly (<1min)removed from the eluted sample by fraction-ation over a size exclusion spin column.Given a typical protein of average abundance,1-10µg of pure protein can be recovered from 1g of cells using this method.Figure 3.Elution of Kap95-PrA/Kap60p is dose dependent.(a )Kap95p-PrA was affinity-purified on IgG-Sepharose from loga-rithmically growing yeast cells and eluted using increasing concentrations of Bio-Ox peptide as indicated.(b )The elution efficiency measured as described in Figure 2was plotted versus the peptide concentration in logarithmic scale as indicated.Figure 4.Eluted Kap95p-PrA/Kap60complex retains its biological activity.(a )Kap95p-PrA was prepared by affinity purification followed by Bio-Ox peptide elution (Kap95-PrA eluate ).Three binding reactions were then set up containing eluted Kap95p-PrA and Nup2p-GST bacterial lysate,Kap95p-PrA alone or Nup2p-GST alone.At the end of the incubation,Nup2p-GST was affinity-purified on glutathione-Sepharose and the immobilized material was eluted from the resin with sample buffer and resolved on SDS -PAGE (GST bound ).(b )Kap95p-PrA was immobilized on IgG-Sepharose and incubated with (+)or without (-)bacterial lysate containing Nup2p-GST.The resulting material was eluted using Bio-Ox.Eluate (E )and resin bound (B )material was resolved on SDS-PAGE.*,indicates a Nup2p breakdown product.Table 1.Elution Efficiency for PrA Tagged Nupsname of nup%yieldNup53p 56Nup59p 81Nup84p 88Nup85p 81Nic96p 76Nsp1p 99Nup1p 99Nup120p 69Nup157p 82Nup159p 53Nup170p 80Nup192p 76Gle2p 90research articlesStrambio-de-Castillia et al.2254Journal of Proteome Research •Vol.4,No.6,2005To explore the characteristics of Bio-Ox elution under conditions that preserve native protein complexes,we chose to work with the yeast karyopherin Kap95p-PrA/Kap60p com-plex,24and with the yeast nucleoporin Nup82p-PrA/Nsp1p/ Nup159p complex.25,26This choice was dictated by our interest in the structure and function of the yeast nuclear pore complex (NPC).17,27Optimization of the Elution Conditions.An elution time course for Kap95p-PrA/Kap60p and Nup82p-PrA from IgG-Sepharose at4°C is shown in Figure2,panels a and b.In both cases,the elution was virtually complete after2h at4°C.The largest difference in elution efficiency between the two test proteins was found at the earlier time points.Thus,more than 50%of initially bound Kap95p-PrA was displaced by10min, while it took∼1h to obtain the same result with Nup82p-PrA. We also determined the temperature dependence of the elution process(Figure2,panel c).Elutions of Kap95p-PrA and Nup82p-PrA with Bio-Ox,for1h were compared at4°C and 25°C(Figure2,panel c),showing that elution was improved at25°C over4°C for both test proteins.These various factors underscore the need to conduct appropriate test experiments to determine the optimal conditions for any given application. For example,elution for shorter periods and at4°C is preferable when the proteins under study are sensitive to denaturation,dissociation or proteolytic degradation.We also tested the dependence of elution efficiency upon Bio-Ox concentration.For this test,Kap95p-PrA bound to IgG-Sepharose was competitively displaced using increasing amounts of Bio-Ox peptide for4h at4°C.(Figure3).Bio-Ox peptide displaced IgG-Sepharose bound PrA tagged Kap95p with an apparent IC50of60.8µM.For practical purposes,the protocol we use in most cases takes advantage of the high solubility of Bio-Ox to obtain maximally efficient elutions,utilizing a concentration of440µM of Bio-Ox peptide for2h at4°C.To test the general applicability of the method,we performed peptide elution experiments using a series of PrA tagged proteins that were available in our laboratories.17The yield for these proteins was in all cases>50%and in most cases was >80%(average yield78%(14%;Table1).Eluted Proteins Retain their Biological Activity.The trans-location of macromolecules between the nucleus and cytosol of eukaryotic cells occurs through the NPC and is facilitated by soluble transport factors termed karyopherins(reviewed in ref28).Nucleoporins that contain FG peptide repeats(FG Nups)function as binding sites for karyopherins within the NPC.One example of an FG Nup-karyopherin interaction is represented by the binding of the Kap95p/Kap60p complex to Nup2p,29an interaction that requires both karyopherins to be natively folded.30,31We took advantage of this interaction to demonstrate that the Bio-Ox eluted Kap95p-PrA/Kap60p com-plex retains its biological activity and is able to bind Nup2p in vitro(Figure4,panel a).In this test,Kap95p-PrA was affinity-purified and eluted from IgG-Sepharose as described above. The eluate was incubated with whole cell lysate from E.coli expressing Nup2p-GST,23and GST-tagged Nup2p was isolated over gluthatione-Sepharose resin.As a control,the same experiment was performed either in the absence of Nup2p-GST containing bacterial lysate or in the absence of Kap95p-PrA eluate.As shown,Nup2p-GST binds specifically and directly to the peptide-eluted Kap95p-PrA/Kap60p complex. This result is consistent with reported data and demonstrates that elution with Bio-Ox does not alter the native state and biological activity of Kap95p-PrA.Moreover,the apparent equimolar stoichiometry of the Nup2-GST/Kap95p-PrA/Kap60p complex indicates that essentially all of the peptide eluted karyopherins were in their native,active conformation.This result underscores the usefulness of this method for the preparation of native protein samples.The method can also be used for in vitro reconstitution experiments of biologically relevant protein-protein interac-tions of interest.For this test,Kap95p-PrA was affinity isolated on IgG-Sepharose,Nup2p-GST was bound to the immobilized Kap95p-PrA and then the reconstituted complex was competi-tively displaced from the resin by Bio-Ox peptide elution(Figure 4,panel b).This shows that the method can be used in vitro to study protein-protein interactions using purified compo-nents.ConclusionWe have used the Bio-Ox technology extensively in our laboratories for a wide variety of applications including:(1) the semipreparative purification of∼30PrA-tagged natively folded Nups for the determination of their sedimentation coefficient over a sucrose velocity gradient(S.Dokudovskaya, L.Veenhoff,personal communication);(2)the isolation of yeast cyclins and cyclin-Cdk associated proteins;32(3)the semi-preparative purification of enzymatically active Dpb4p-PrA chromatin remodeling/histone complexes;33and(4)the study of the in vitro binding property of proteins of interest using blot and resin binding experiments.34Thus,this method should be generally applicable to the native purification of most other proteins and protein complexes.Acknowledgment.We are very grateful to David Dil-worth and John Aitchison for the generous gift of bacterially expressed Nup2p-GST.We are deeply indebted to Rosemary Williams for her skilled technical assistance throughout the course of this study and to all members of the Rout and Chait laboratories and of the Proteomic Research Center,past and present,for their continual help and unwavering support.We are particularly grateful to Markus Kalkum,Bhaskar Chan-drasekhar,Svetlana Dokudovskaya and Liesbeth Veenhoff.This work was supported by grants from the American Cancer Society(RSG-0404251)and the NIH(GM062427,RR00862,and CA89810).References(1)Uhlen,M.;Forsberg,G.;Moks,T.;Hartmanis,M.;Nilsson,B.Fusion proteins in biotechnology.Curr.Opin.Biotechnol.1992, 3(4),363-369.(2)Nygren,P.A.;Stahl,S.;Uhlen,M.Engineering proteins to facilitatebioprocessing.Trends Biotechnol.1994,12(5),184-188.(3)Baneyx,F.Recombinant protein expression in Escherichia coli.Curr.Opin.Biotechnol.1999,10(5),411-421.(4)LaVallie,E.R.;McCoy,J.M.Gene fusion expression systems inEscherichia coli.Curr.Opin.Biotechnol.1995,6(5),501-506.(5)Nilsson,J.;Stahl,S.;Lundeberg,J.;Uhlen,M.;Nygren,P.A.Affinity fusion strategies for detection,purification,and im-mobilization of recombinant proteins.Protein Expr.Purif.1997, 11(1),1-16.(6)Einhauer,A.;Jungbauer,A.The FLAG peptide,a versatile fusiontag for the purification of recombinant proteins.J.Biochem.Biophys.Methods2001,49(1-3),455-465.(7)Aitchison,J. D.;Blobel,G.;Rout,M.P.Nup120p:a yeastnucleoporin required for NPC distribution and mRNA transport.J.Cell Biol.1995,131(6Pt2),1659-1675.(8)Grandi,P.;Doye,V.;Hurt,E.C.Purification of NSP1revealscomplex formation with‘GLFG’nucleoporins and a novel nuclear pore protein NIC96.EMBO J.1993,12(8),3061-3071.(9)Stirling,D.A.;Petrie,A.;Pulford,D.J.;Paterson,D.T.;Stark,M.J.Protein A-calmodulin fusions:a novel approach for investigat-ing calmodulin function in yeast.Mol.Microbiol.1992,6(6),703-713.Native Elution of PrA-Tagged Proteins research articlesJournal of Proteome Research•Vol.4,No.6,20052255。
摘要:来自通用电气医疗集团(GE Healthcare)的谷胱甘肽硫转移酶(GST)基因融合系统是一种表达、纯化和检测大肠杆菌中所产生的GST标签蛋白的多功能系统。
系统包括三个主要成分:pGEX质粒表达载体、GST纯化的产品以及一系列GST检测产品。
一系列切割GST 标签的位点特异蛋白酶则是该系统的补充。
GST亲和标签实现了温和的纯化过程,不会影响蛋白的天然结构和功能。
GST基因融合系统的优势包括:•所有pGEX载体带有tac启动子,实现化学诱导的高水平表达•温和、非变性的缓冲液成分,以分离活性蛋白•基于Glutathione Sepharose™填料的亲和层析产品实现了从微克到克规模样品的一步纯化•方便的预装柱形式适合单个样品或平行筛选多个重组克隆•pGEX载体上的PreScission™蛋白酶、凝血酶或Xa因子识别位点能将目的蛋白从融合产物中切割下来•利用抗-GST抗体轻松检测融合蛋白在天然情况下,GST以分子量26,000的蛋白存在,它在大肠杆菌中的表达产物具有完全的酶活。
经过鉴定,pGEX载体的重组日本血吸虫(Schistosomajaponicum)GST的晶体结构与天然蛋白一致。
pGEX质粒载体将基因或基因片段与GST融合后,在细胞内高水平诱导表达。
重组蛋白很容易利用Glutathione Sepharose填料通过亲和层析从比如大肠杆菌细胞裂解液中纯化(图1),要么是预装柱,要么是分批纯化。
图 1. Glutathione Sepharose High Performance、Glutathione Sepharose 4 Fast Flow 以及GlutahioneSepharose 4B目前都有大包装填料,预装柱以及多孔板,用于GST标签蛋白的纯化。
利用位点特异的蛋白酶,可实现目的蛋白与GST的切割,此蛋白酶的识别位点位于pGEX载体上多克隆位点的上游。
GST标签的切除是在柱中进行的,作为纯化步骤或纯化后离线的一部分。
GST bestarose 4FF说明书默认分类2010-01-18 09:32:34 阅读257 评论0 字号:大中小GST bestarose 4FF是专门用于纯化pGEX系列载体表达的GST标签蛋白,其它谷胱甘肽转移酶以及与谷胱甘肽有亲和作用蛋白的分离介质,操作简单,快速。
GST bestarose 4FF 可直接从预处理的细胞裂解物中纯化GST-tagged蛋白,纯化条件温和,可以保证蛋白的生物活性。
GST bestarose 4FF 具有高流动性,易于放大,GST bestarose 4FF有快速方便的预装柱,Ez-Fast bestarose 4FF 1ml和5ml。
介质性能谷胱甘肽是通过10碳原子的连接臂偶联到4%高度交联的琼脂糖上。
最优的偶联作用使介质具有高GST-tagged蛋白及与谷胱甘肽相互作用蛋白的结合能力。
总的蛋白结合能力约为每毫升填料结合10mg标签蛋白,动态结合能力随着外界因素改变,如不同的目标蛋白,流速等等。
如果需要去除GST部分(天然蛋白分子量为26KD),当蛋白吸附在柱上,或在洗脱后用位点专一的蛋白酶消化,选用前种方法,可减少额外分离目的蛋白与GST步骤,消化后,目的蛋白直接用结合液洗脱即可。
表1 GST bestarose 4FF性能配体谷胱甘肽与10碳原子连接臂配体浓度 120-320μmol谷胱甘肽/ml填料蛋白结合能力≈ 10mg重组GST-tagged蛋白/ml填料,GST,Mr26000动态结合能力≈11mgGST-tagged蛋白/ml填料,Mr43000平均颗粒大小90μm组成 4%高度交联的琼脂糖最大流速* 450cm/h(15ml/min),XK层析柱,柱高5cm,水溶性缓冲液,室温纯化建议的流速** 上样:<100cm/h(<3ml/min ,XK层析柱)洗涤与洗脱:100-300cm/h(3-10ml/min,XK层析柱)化学稳定性所有常用的水溶性缓冲液中稳定,如:1M醋酸盐,pH7.4,6M 盐酸胍,室温,1hpH稳定性 pH3-12贮存温度 +4-30℃贮存液 20%乙醇* 水是室温的。
GST融合蛋白纯化--筛选表达株(英文)摘要:Purification of GST fusion proteins in E.coli GST Sugden lab,McArdle Laboratory for Cancer Research,University of Wisconsin-Madison Medical School Screen of GST-Fusion Protein ExpressionPurification of GST fusion proteins in E.coli GSTSugden lab,McArdle Laboratory for Cancer Research,University ofWisconsin-Madison Medical SchoolScreen of GST-Fusion Protein expression(pGEX system by Amersham: for check clones for expression of the desired fusion protein prior to large-scale purification)Pick several colonies of E.coli transformed with the pGEX reco mbi nants into separate tubes containing 2 ml of 2 x YTA medium.-Note: For comparison,it is advis ABL e to inoculate a control tube with bacteria transformed with the parental pGEX plasmid.Grow l IQ uid cultures to an A600 of 0.6-0.8 (3~5 h)with vigorous agitation at 20~37℃.inc ubate fusion protein expression by adding 2 μl of 100 mM IPTG (final concentration 0.1 mM).Continue incubation for an additional 1~2 hTransfer 1.5 ml of the liquid cultures to labeled 1.5 ml microcentrifuge tubes.Centrifuge in a microcentrifuge for 5 sec and discard the supernatant.Resuspend each pellet in 300 μl of ic e-cold 1 x PBS,remove 10 μl of these resuspend cells into labeled tubes (for later use in SDS-PAGE analysis).-Note: Except where noted,keep all samples and tubes on ice.Lyse the cells using the sonicator equipped with an appropriate probe.-Note: Lysis is complete when the cloudy cell suspension becomes translucent.The frequency and intensity of sonication should be adjusted such that complete lysis occurs in 10 sec,without frothing (it may denature proteins).-Note: Crude sonicates can be screened for the relative level of expression of GST fugion proteins using the GST substrates CDNB (1-chloro-2,4-dinitrobenzene).Centrifuge in a microcentrifuge for 5 min to remove insoluble materials.Save a10 μl aliquots of the insoluble material for analysis by SDS-PAGE.Transfer the supernatants to fresh tubes,Add 20 μl of a 50% slurry of Glutathione Sepharose 4B (prepared as described above)to each supernatant and mix gently for 5 min at r.t..Add 100 μl of 1 x PBS,vortex briefly,and centrifuge for 5 sec to sediment the Sepharose beads.Discard the supernatant,repeat this 1 x PBS wash twiceElute the fusion protein by adding 10 μl of Glutathione Elution Buffer.Suspend the Sepharose beads and incubate for 5 min at r.t.Centrifuge in a microcentrifuge for 5 min to sediment the Sepharose beads,then transfer the supernatant to fresh tubes.*Glutathione elution buffer: 10 mM reduced glutathione in 50 mM Tris-HCl (pH8.0).Dispense in 1-10 ml aliquots and store at ?20℃until needed.Avoid more than five freeze/thaw cycles.Preparation of Glutathione Sepharose 4B(for bulk matrix for batch purification)Gently shake the bottle of Glutathione Sepharose 4B to resuspend the matrix.Use a pipet to remove sufficient slurry for use and transfer to an appropriate container/tube.(Glutathione Sepharose 4B as supplies is approximately a 75% slurry.The following procedure results in a 50 % slurry; Based on the bed volumerequirement,dispense 1.33 ml of the original Glutathione Sepharose 4B slurry per ml of bed volume required).Sediment the matrix by centrifugation at 500 xg for 5 min,carefully decant the supernatant.Wash the Glutathione Sepharose 4B by adding 10 mol of cold (4℃)1 x PBS per 1.33 ml of the originally slurry of Glutathione Sepharose 4B dispensed,Invert to mix.-Note: Glutathione Sepharose 4B must be thoroughly washed with 1 x PBS to remove the 20% ethanol storage solution.Residual ethanol may interfere with subsequent procedures.Sediment the matrix by centrifugation at 500 x g for 5 min.Decant the supernatant.For each 1.33 ml of the original slurry of Glutathione Sepharose 4B,and 1 ml of 1 x PBS.This results in a 50% slurry.Mix well prior to subsequent pipetting steps.-Note: Glutathione Sepharose 4B equilibrated with 1 x PBS may be stored at 4℃for up to 1 month.Bulk purification of protein expressed in E.coliPick several colonies of E.coli transformed with the pGEX recombinants into separate tubes containing 2 ml of 2 x YTA medium.Grow o/n at 20~37℃.Add 2 ml culture into 100 ml 2 x YTA medium.Grow liquid cultures to an A600 of 0.6-0.8 (about 2 h)with vigorous agitation at20~37℃.Incubate fusion protein expression by adding 50 μl of 100 mM IPTG (final concentration 0.1 mM).Continue incubation for an additional 1~2 hTransfer the liquid cultures to 50 ml tubes.Centrifuge at 3000 rpm for 10 min at 4℃and discard the supernatant.Resuspend each pellet in 5 ml of ice-cold 1 x PBS.Sonicate on ice with three to five brief (10 sec)pulse.Transfer to new microcentrifuge tubes (1 ml,each)and centrifuge for 10 min at 10000 rpm at 4℃to remove insoluble materials.Transfer the supernatants to fresh tubes.Add 50 μl of a 50% slurry of Glutathione Sepharose 4B to each supernatant and rock gently for 10 min at 4℃.Add 1 ml of 1 x PBS to each tube,vortex briefly,and centrifuge 10000 rpm at for 5 sec to sediment the Sepharose beads.Discard sup,repeat 1 x PBS wash twice.Elute the fusion protein by adding 100 μl of Glutathione Elution Buffer.Suspend the Sepharose beads and incubate for 10 min at 4℃.Centrifuge in a microcentrifuge for 5 min to sediment the Sepharose beads,then transfer the supernatant to fresh tubes.。
GST亲和层析介质使用说明书一、简介GST亲和层析介质(GST Agarose)是专门设计用于纯化谷胱甘肽S—转移酶(GST)融合蛋白、其它谷胱甘肽转移酶以及与谷胱甘肽有亲和作用蛋白的分离介质,一步分离就可得到高纯度的GST融合目标蛋白,纯化条件温和,可以保证蛋白的活性。
本产品是自主设计合成的GST琼脂糖凝胶,具有优良的物理和化学稳定性,使用寿命长,操作方便,批次重复性好,易于放大,是研发与生产的理想选择.二、性能参数三、适用范围分离谷胱甘肽S—转移酶(GST)融合蛋白、其它谷胱甘肽转移酶以及与谷胱甘肽有亲和作用的蛋白。
四、操作说明1。
缓冲液配制缓冲液A(平衡缓冲液):10mM Na2HPO4,1。
8mM KH2PO4,140mM NaCl,2。
7mM KCl,调节pH值至8.0。
缓冲液B(洗脱缓冲液):10mM Glutathione(还原型),50mM Tris-HCl,调节pH值至8.0.因Glutathione易氧化,需现用现配。
(注:各种溶液配制完毕后,最好进行脱气处理,0.45 μm滤膜过滤备用)。
2。
样品预处理:按每克湿重菌体/2~5ml平衡缓冲液的比例充分悬浮离心收集的菌体;600w功率,每循环超声3s,冷却3s,循环99×3次,破碎菌体;4℃、15000rpm离心15m,收集上清液,或用0.45μm滤膜过滤.3。
装柱:聚苯乙烯层析柱1)将层析柱固定在铁架台或层析架上,封闭层析柱下端出口,向柱内充入纯水,排开层析柱内空气,先将垫片完全浸没于水面下方,在保持水平的状态下,小心推向底部,避免垫片下方滞留气泡.2) 打开层析柱下端出口,排出柱中纯水;在液面低至距垫片1~1。
5cm高度时封闭下端出口,用移液枪按需要量吸取介质,或用玻璃棒紧靠柱子内壁引流,将介质加入到层析柱中;静置30min,让介质自然沉降。
3)从上端管口将另一垫片缓慢推至介质沉降平面,使介质表面保持水平状态,注意避免垫片与介质接触面滞留气泡(如对实验结果要求不严,也可不放入上垫片,以提高流速)。