化学信息学——第八讲hyperchem 软件学习
- 格式:pptx
- 大小:5.34 MB
- 文档页数:60


HyperChem软件中Script的常用语句注解1. 计算方法及参数设定calculation-method item计算方法设定item: MolecularMechanics, SemiEmpirical, AbInitio, DFT molecular-mechanics-method item分子力学方法设定item: mm+, amber, bio+, opls, amber94, charmm22, tndo optim-max-cycles x优化最大迭代次数x设定optim-convergence goal 优化收敛目标goal设定optim-converged返回是否收敛(true/false)optim-algorithm item 优化算法设定item: PolakRibiere, NewtonRaphson, etc.assign-basisset item abinitio计算基组设定item: 3-21G, STO-3G, etc.semi-empirical-method item半经验计算方法设定item: am1, pm3, extendedhuckel, cndo, indo, zindos, mndod, tndo, etc.accelerate-scf-convergence true/false 是否加速SCF收敛scf-convergence goal SCF收敛目标goal设定max-iterations n SCF计算的最大迭代次数n设定2. 计算结果输出omsgs-to-file filename 将信息输出到filename文件append-omsgs-to-file filename 将信息追加到filename文件omsgs-not-to-file 关闭输出文件query-response-has-tag yes/no 提取的信息回显开关query-value HSV提取系统数据结构中的信息并单行输出HSV: current-file-name, coordinates,dipole-moment, dipole-moment-componentstotal-energy, heat-of-formation,scf-binding-energy, scf-core-energy, scf-electronic-energy start-logging HyperChem运行记录开启stop-logging HyperChem运行记录关闭export-property-file filename将分子轨道信息写入filename文件3. 分子结构及其文件操作file-format item定义分子结构文件格式item: hin, mol, zmt, ent, skc, xyz, ml2open-file filename 读入filename分子结构文件write-file filename 将分子结构写入filename文件menu-build-add-hydrogens 给分子加氢menu-build-model-build 给分子加氢并规格化do-optimization 进行优化操作do-single-point 进行单点计算do-molecular-dynamics进行分子动力学操作4. 屏幕显示设置screen-refresh-period n屏幕刷新周期atom-labels item原子标识设定item: none, number, symbol, name, charge, etc.show-hydrogens yes/no 分子中氢显示开关show-multiple-bonds yes/no分子中重键显示开关request string弹出窗口并显示由双引号表达的字符串string 5. 其它read-script filename读取名为filename的script文件exit-script 退出script实例: example.scrcalculation-method molecular-mechanicsmolecular-mechanics-method mm+optim-max-cycles 2048optim-convergence 0.1optim-algorithm PolakRibiereomsgs-to-file results.txtquery-response-has-tag nofile-format molopen-file 000A-0001.molmenu-build-model-builddo-optimizationquery-value total-energyfile-format zmtwrite-file 000A-0001.zmtomsgs-not-to-fileexit-script附: HyperChem State Variables (HSV)abinitio-buffer-size: Variable, Read/Write.Type: integer in range (1 .. 2147483647).Two electron integral buffer size.abinitio-calculate-gradient: Variable, Read/Write.Type:boolean.Enable Ab Initio gradient calculation (Single Point only).abinitio-cutoff: Variable, Read/Write.Type: float in range (0 .. 1e+010).Two electron integral cutoff.abinitio-d-orbitals: Variable, Read/Write.boolean.Type:Either five (False) or six (True).abinitio-direct-scf: Variable, Read/Write.boolean.Type:Enable Ab Initio Direct SCF calculation.abinitio-f-orbitals: Variable, Read/Write.boolean.Type:Either seven (False) or ten (True).abinitio-integral-format: Variable, Read/Write.Type: enum(raffenetti, regular).Either regular or raffenetti.abinitio-integral-path: Variable, Read/Write.string.Type:Path for storing integrals.abinitio-mo-initial-guess: Variable, Read/Write.Type: enum(core-hamiltonian, projected-huckel, projected-cndo, projected-indo).Either core-hamiltonian, projected-huckel, projected-cndo, projected-indo. abinitio-mp2-correlation-energy: Variable, Read/Write.boolean.Type:Enable Ab Initio MP2 correlation energy.abinitio-mp2-frozen-core: Variable, Read/Write.boolean.Type:Enable Ab Initio MP2 frozen core.abinitio-scf-convergence: Variable, Read/Write.Type: float in range (0 .. 100).SCF Convergence for Ab Initio.abinitio-use-ghost-atoms: Variable, Read/Write.boolean.Type:Include or ignore ghost atoms.accelerate-scf-convergence: Variable, Read/Write.Type:boolean.Whether to use DIIS procedure.add-amino-acid: Command.Arg list: string.String-1 gives the name of an amino acid residue to add to the system.add-nucleic-acid: Command.Arg list: string.String-1 names the nucleotide to add to the current system.align-molecule: Command.Arg list: .Align the inertial axes of the molecular system.align-viewer: Command.Arg list: enum(x, y, z, line).Align the viewer's line-of-sight with the indicated axis or LINE.allow-ions: Variable, Read/Write.Type:boolean.Whether to allow excess valence on atoms.alpha-orbital-occupancy: Variable, Read/Write.Type: vector of float.(i) Number of electrons in the i-th MO.alpha-scf-eigenvector: Variable, Read/Write.Type: vector of float-list.(i) Coefficients for the i-th MO.amino-alpha-helix: Command.Arg list: (void).Subsequent additions of amino acid residues are to use alpha-helix torsions. amino-beta-sheet: Command.Arg list: (void).Subsequent additions of amino acid residues are to use beta-sheet torsions. amino-isomer: Variable, Read/Write.Type: enum(l, d).Whether amino acids are l or d.amino-omega: Variable, Read/Write.Type: float angle in range (-360 .. 360).The Omega amino acid backbone angle.amino-phi: Variable, Read/Write.Type: float angle in range (-360 .. 360).The Phi amino acid backbone angle.amino-psi: Variable, Read/Write.Type: float angle in range (-360 .. 360).The Psi amino acid backbone angle.animate-vibrations: Variable, Read/Write.boolean.Type:Whether or not to animate vibrations.annotation-color: Variable, Read/Write.string.Type:Default color for annotations.annotation-filled: Variable, Read/Write.boolean.Type:The circle and rectangle annotations are filledannotation-layer-hidden: Variable, Read/Write.boolean.Type:The annotation layer is hidden andd only molecule layer shows. annotation-layer-in-front: Variable, Read/Write.boolean.Type:The annotation layer is in front of molecule layer.append-dynamics-average: Command.Arg list: string.Add a named selection to dynamics average gathering.append-dynamics-graph: Command.Arg list: string.Add a named selection to dynamics graph display.append-omsgs-to-file: Command.Arg list: string.String-1 gives the name of a file to which o-msgs are to be appended. assign-basisset: Command.Arg list: string.Assign a basis set to a selection or system.atom-basisset: Variable, Read/Write.Type: array of string.(iat, imol) The basis set of atom iat in molecule imol.atom-charge: Variable, Read/Write.(iat, imol) The charge of atom iat in molecule imol.atom-color: Variable, Read/Write.Type: array of enum(ByElement, Black, Blue, Green, Cyan, Red, Violet, Yellow, White).(iat, imol) The current color of the atom.atom-count: Variable, Readonly.Type: vector of integer.(imol) The number of atoms in molecule imol.atom-extra-basisset: Variable, Read/Write.Type: array of string, float.(iat, imol) The basis set of atom iat in molecule imol.atom-info: Variable, Readonly.(unknown).Type:Funny composite to support backends.atom-label-text: Variable, Readonly.Type: array of string.(iat, imol) RO. The text of the current atom label.atom-labels: Variable, Read/Write.Type: enum(None, Symbol, Name, Number, Type, Charge, Spin, Mass, BasisSet, Chirality, RMSGradient, Custom).Label for atoms.atom-mass: Variable, Read/Write.Type: array of float.(iat, imol) The mass of atom iat in molecule imol.atom-name: Variable, Read/Write.Type: array of string.(iat, imol) The name of atom iat in molecule imol.atom-spin-density-at-nucleus: Variable, Read/Write.Type: array of float.(iat, imol) The electron density of nucleus of atom iat in molecule imol.atom-spin-population: Variable, Read/Write.Type: array of float.(iat, imol) The spin density of atom iat in molecule imol.atom-type: Variable, Read/Write.Type: array of string.(iat, imol) The type of atom iat in molecule imol.atomic-number: Variable, Read/Write.(iat, imol) The atomic number of atom iat in molecule imol.atomic-symbol: Variable, Readonly.Type: array of string.(iat, imol) The element symbol of the atom.auxilliary-basis: Variable, Read/Write.Type: integer in range (0 .. 3).1=A1, 2=A2, 3=P1back-clip: Variable, Read/Write.float.Type:Set back clipping plane.backend-active: Variable, Read/Write.boolean.Type:Whether current channel is an active backend.backend-communications: Variable, Read/Write.Type: enum(Local, Remote).Whether to compute on local or remote host.backend-host-name: Variable, Read/Write.string.Type:The name of remote host for backend communications.backend-process-count: Variable, Read/Write.Type: integer in range (1 .. 32).The number of processes to run.backend-user-id: Variable, Read/Write.string.Type:The user id to use on the remote host for backend communications.backend-user-password: Variable, Read/Write.string.Type:The password for user id to use on the remote host for backend communications. balls-highlighted: Variable, Read/Write.boolean.Type:Balls and Balls-and-Cylinders should be highlighted when shaded.balls-radius-ratio: Variable, Read/Write.Type: float in range (0.001 .. 1).Size of the Balls relative to the maximum value.balls-shaded: Variable, Read/Write.boolean.Type:Balls and Balls-and-Cylinders should be shaded.basisset-count: Variable, Readonly.integer.Type:Number of coefficients required to describe a molecular orbital.bend-energy: Variable, Readonly.Type: float in range (-1e+010 .. 1e+010).Results from backend computation.beta-orbital-occupancy: Variable, Read/Write.Type: vector of float.(i) Number of electrons in the i-th MO.beta-scf-eigenvector: Variable, Read/Write.Type: vector of float-list.(i) Coefficients for the i-th MO.bond-color: Variable, Read/Write.Type: enum(ByElement, Black, Blue, Green, Cyan, Red, Violet, Yellow, White).The color used for drawing atoms and bonds.bond-spacing-display-ratio: Variable, Read/Write.Type: float in range (0 .. 1).Bond spacing display ratio.builder-enforces-stereo: Variable, Read/Write.boolean.Type:Whether the model builder implicitly enforces any existing stereochemistry. calculation-method: Variable, Read/Write.Type: enum(MolecularMechanics, SemiEmpirical, AbInitio, DFT).Whether molecular mechanics, semi-empirical, or ab initio.cancel-menu: Variable, Read/Write.Type:boolean.Whether the cancel menu is up, or the normal one.cancel-notify: Command.Arg list: string.String-1 names a variable to stop watching.change-stereochem: Command.Arg list: integer, integer.Immediately change the stereochemistry about (iat, imol).change-user-menuitem: Command.Arg list: integer, string, string.Change the text and procedure associated with the specified user MenuItem. chirality: Variable, Read/Write.Type: array of string.(iat, imol) A, R, S, or ?, for achiral, R, S, or unknown chirality. ci-criterion: Variable, Read/Write.Type: enum(Energy, Orbital).One of: energy, orbital.ci-excitation-energy: Variable, Read/Write.Type: float in range (0 .. 10000).When ci-criterion=energy, maximum excitation energy.ci-occupied-orbitals: Variable, Read/Write.Type: integer in range (0 .. 32767).When ci-criterion=orbital, count of occupied orbitals included. ci-state-to-optimize: Variable, Read/Write.Type: integer in range (0 .. 32767).Which CI state to optimize with conjugate directionsci-unoccupied-orbitals: Variable, Read/Write.Type: integer in range (0 .. 32767).When ci-criterion=orbital, count of unoccupied orbitals included. clip-cursor: Variable, Read/Write.Type: float in range (0 .. 1000).Select Z axis clip cursor tool.clip-icon-step: Variable, Read/Write.Type: float in range (0 .. 1000).Select clip step.color-element: Command.Arg list: integer, enum().Element Int-1 gets color String-2 as its default color.color-selection: Command.Arg list: string.String-1 names a color for the current selection.compile-script-file: Command.Arg list: string, string.Compile file string-1, writing result to string-2 configuration: Variable, Read/Write.integer.Type:The current UV configuration of the system.configuration-interaction: Variable, Read/Write.Type: enum(NoCI, SinglyExcited, Microstate).One of: no-ci, singly-excited, microstate.connectivity-in-pdb-file: Variable, Read/Write.Type:boolean.Whether connectivity information is to be included in a PDB file.constrain-bond-angle: Command.Arg list: float angle in range (-360 .. 360).Float-1 gives the angle constraint for the three currently selected atoms. constrain-bond-down: Command.Arg list: integer, integer, integer, integer.Constrain the bond from (iat1, imol1) to (iat2, imo2) to be down.constrain-bond-length: Command.Arg list: float in range (0 .. 100).Float-1 gives the length constraint for the two currently selected atoms. constrain-bond-torsion: Command.Arg list: float angle in range (-360 .. 360).Float-1 gives the torsion constraint for the four currently selected atoms. constrain-bond-up: Command.Arg list: integer, integer, integer, integer.Constrain the bond from (iat1, imol1) to (iat2, imo2) to be up.constrain-change-stereo: Command.Arg list: integer, integer.Constrain atom (iat, imol) to change the current stereochemistry.constrain-drawing: Variable, Read/Write.boolean.Type:Whether to constrain bond lengths and angles to canonicalize drawing of moleculeconstrain-fix-stereo: Command.Arg list: integer, integer.Constrain atom (iat, imol) to enforce the current stereochemistry.constrain-geometry: Command.Arg list: string.String-1 describes the geometry constraint around the currently selected atom. coordinates: Variable, Read/Write.Type: array of float, float, float.(iat, imol) The x, y, and z coordinates of atom iat in molecule imol. coordination: Variable, Readonly.Type: array of integer.(iat, imol) The coordination number for the specified atom.correlation-functional: Variable, Read/Write.Type: enum(None, Perdew86, VWN, LYP, PZ81, PW91, PBE96, HCTH98).Perdew, LYP, etc.cpk-max-double-buffer-atoms: Variable, Read/Write.Type: integer in range (0 .. 2147483647).Maximum number of double buffered atoms in cpk rendering mode. create-atom: Command.Arg list: integer in range (0 .. 103).Create a new atom at the origin with atomic number nAtno.current-file-name: Variable, Readonly.string.Type:The name of the current file.custom-title: Variable, Read/Write.string.Type:Custom Title string, append string to title.cutoff-inner-radius: Variable, Read/Write.Type: float in range (0 .. 1e+010).The distance (in Angstroms) to begin a switched cutoff.cutoff-outer-radius: Variable, Read/Write.Type: float in range (0 .. 1e+010).The distance (in Angstroms) at which nonbonded interactions become zero. cutoff-type: Variable, Read/Write.Type: enum(None, Switched, Shifted).Electrostatic cutoff to apply to molecular mechanics calculations.cycle-atom-stereo: Command.Arg list: integer, integer.Advance the stereo constraint about atom (iat, imol).cycle-bond-stereo: Command.Arg list: integer, integer, integer, integer.Advance the stereo constraint along the bond (iat1, imol1)--(iat2, imol2). cylinders-color-by-element: Variable, Read/Write.boolean.Type:Color Cylinders using element colors.cylinders-width-ratio: Variable, Read/Write.Type: float in range (0 .. 1).Width of the Cylinders relative to the maximum value.d-orbitals-on-second-row: Variable, Read/Write.Type:boolean.Include D orbitals on second row.declare-float: Command.Arg list: string.Declare a new floating-point variable.declare-integer: Command.Arg list: string.Declare a new integer variable.declare-string: Command.Arg list: string.Declare a new string variable.default-element: Variable, Read/Write.Type: integer in range (0 .. 103).The atomic number of the default element for drawing operations. delete-atom: Command.Arg list: integer, integer.Delete the specified atom.delete-file: Command.Arg list: string.filename to be deleted.delete-named-selection: Command.Arg list: string.Remove the named selection String-1 from the list of named selections. delete-selected-atoms: Command.Arg list: (void).Delete the currently selected atoms.dipole-moment: Variable, Read/Write.Type: float in range (-1e+010 .. 1e+010).Dipolemoment.dipole-moment-components: Variable, Read/Write.Type: float, float, float.Dipole moment components.do-langevin-dynamics: Command.Arg list: (void).Perform a Langevin dynamics computation on the system.do-molecular-dynamics: Command.Arg list: (void).Perform a molecular dynamics computation on the system.do-monte-carlo: Command.Arg list: (void).Perform a Monte Carlo computation on the system.do-optimization: Command.Arg list: (void).Perform a structure optimization on the system.do-qm-calculation: Variable, Read/Write.boolean.Type:For single-point QM calculations, whether to re-compute wave function. do-qm-graph: Variable, Read/Write.boolean.Type:For single-point QM calculations, to graph some data.do-qm-isosurface: Variable, Read/Write.boolean.Type:For single-point QM calculations, to generate iso-surface of results.do-single-point: Command.Arg list: (void).Perform a single-point computation on the system.do-vibrational-analysis: Command.Arg list: (void).Perform a vibrational analysis computation on the system.dot-surface-angle: Variable, Read/Write.Type: float angle in range (-90 .. 90).Dot surface angle.double-buffered-display: Variable, Read/Write.boolean.Type:Whether display operations are double-buffered.dynamics-average-period: Variable, Read/Write.Type: integer in range (1 .. 32767).Computation results from dynamics run.dynamics-bath-relaxation-time: Variable, Read/Write.Type: float in range (0 .. 1e+010).Bath relaxation time for dynamics.dynamics-collection-period: Variable, Read/Write.Type: integer in range (1 .. 32767).Dynamics data collection interval.dynamics-constant-temp: Variable, Read/Write.boolean.Type:Whether to keep temperature fixed at dynamics-simulation-temp. dynamics-cool-time: Variable, Read/Write.Type: float in range (0 .. 1e+010).Time taken to change from dynamics-simulation-temp to dynamics-final-temp. dynamics-final-temp: Variable, Read/Write.Type: float in range (0 .. 1e+010).Temperature to cool back to when annealing.dynamics-friction-coefficient: Variable, Read/Write.Type: float in range (0 .. 1000000).Friction coefficient for Langevin dynamics.dynamics-heat-time: Variable, Read/Write.Type: float in range (0 .. 1e+010).Time taken to change from dynamics-starting-temp ->dynamics-simulation-temp.dynamics-info-elapsed-time: Variable, Readonly.Type: float in range (0 .. 1e+010).Elapsed time in dynamics run.dynamics-info-kinetic-energy: Variable, Readonly.Type: float in range (-1e+010 .. 1e+010).Computation results from dynamics run.dynamics-info-last-update: Variable, Readonly.boolean.Type:Last update from dynamics run.dynamics-info-potential-energy: Variable, Readonly.Type: float in range (-1e+010 .. 1e+010).Computation results from dynamics run.dynamics-info-temperature: Variable, Readonly.Type: float in range (0 .. 1e+010).Computation results from dynamics run.dynamics-info-total-energy: Variable, Readonly.Type: float in range (-1e+010 .. 1e+010).Computation results from dynamics run.dynamics-playback: Variable, Read/Write.Type: enum(none, playback, record).Playback a recorded dynamics run.dynamics-playback-end: Variable, Read/Write.Type: integer in range (0 .. 32767).End playback of recorded dynamics run.dynamics-playback-period: Variable, Read/Write.Type: integer in range (1 .. 32767).Dynamics playback interval.dynamics-playback-start: Variable, Read/Write.Type: integer in range (0 .. 32767).Start playback of recorded dynamics run.dynamics-restart: Variable, Read/Write.boolean.Type:Use saved velocities.dynamics-run-time: Variable, Read/Write.Type: float in range (0 .. 1e+010).Total integration time at dynamics-simulation-temp. dynamics-seed: Variable, Read/Write.Type: integer in range (-32768 .. 32767).Seed for dynamics initialization random number generator. dynamics-simulation-temp: Variable, Read/Write.Type: float in range (0 .. 1e+010).High temperature for the dynamics run.dynamics-snapshot-filename: Variable, Read/Write.string.Type:Name file of to store dynamics run.dynamics-snapshot-period: Variable, Read/Write.Type: integer in range (1 .. 32767).Set recording interval of dynamics run.dynamics-starting-temp: Variable, Read/Write.Type: float in range (0 .. 1e+010).Starting temperature for the dynamics run.dynamics-temp-step: Variable, Read/Write.Type: float in range (0 .. 1e+010).Step size (K) by which temperature is changed.error: Variable, Read/Write.string.Type:The current error.errors-are-not-omsgs: Command.Arg list: (void).Specifies that error messages are to appear in message boxes. errors-are-omsgs: Command.Arg list: (void).Specifies that error messages should be treated like o-msgs. estatic-energy: Variable, Readonly.Type: float in range (-1e+010 .. 1e+010).Results from backend computation.exchange-functional: Variable, Read/Write.Type: enum(None, Hartree-Fock, Slater, Becke88, PW91, Gill96, PBE96, HCTH98, B3-LYP, B3-PW91, EDF1, Becke97).Slater, Becke88, etc.excited-state: Variable, Read/Write.boolean.Type:False for lowest state, true for next-lowest state.execute-client: Command.Arg list: string.Run a client application.execute-hyperchem-client: Command.Arg list: string.Run a client application. App can reliably connect to instance of HyperChem. execute-string: Command.Arg list: string.Execute the string variable as a script.exit-script: Command.Arg list: (void).Exit the current script.explicit-hydrogens: Variable, Read/Write.boolean.Type:Whether hydrogens are to be drawn explicitly.export-dipole: Variable, Read/Write.boolean.Type:Whether or not to export dipole moment data to .EXT file.export-ir: Variable, Read/Write.boolean.Type:Whether or not to export IR data to .EXT file.export-orbitals: Variable, Read/Write.boolean.Type:Whether or not to export orbital data to .EXT file.export-property-file: Command.Arg list: string.Writes properties to the named file.export-uv: Variable, Read/Write.boolean.Type:Whether or not to export UV data to .EXT file.factory-settings: Command.Arg list: (void).Reset chem to its out-of-the-box state.field-direction: Variable, Read/Write.Type: integer in range (1 .. 3).direction (X,Y or Z) of the static electric field applied to the systemfield-strength: Variable, Read/Write.Type: float in range (-1000 .. 1000).strength (a.u.) of the static electric field applied to the systemfile-diff-message: Command.Arg list: string, string, string, string.Compare file1 to file2; if they are the same say string3, else say string4.file-format: Variable, Read/Write.string.Type:The molecule file format.file-needs-saved: Variable, Read/Write.Type:boolean.Whether the current system needs to be saved.formal-charge: Variable, Read/Write.Type: array of integer.(iat, imol) Positive or negative formal charge on atom used by model builder. front-clip: Variable, Read/Write.float.Type:Set front clipping plane.global-inhibit-redisplay: Variable, Readonly.Type:boolean.Whether redisplay of the system is inhibited (readonly)gradient-x: Variable, Readonly.Type: float in range (-1e+010 .. 1e+010).Molecular gradient in the X directiongradient-y: Variable, Readonly.Type: float in range (-1e+010 .. 1e+010).Molecular gradient in the Y directiongradient-z: Variable, Readonly.Type: float in range (-1e+010 .. 1e+010).Molecular gradient in the Z directiongradients: Variable, Read/Write.Type: array of float, float, float.(iat, imol) The x, y, and z gradients of atom iat in molecule imol.graph-beta: Variable, Read/Write.boolean.Type:If true and UHF, graph beta-spin orbitals instead of alpha.graph-contour-increment: Variable, Read/Write.Type: float in range (-1e+010 .. 1e+010).Increment between contour lines.graph-contour-increment-other: Variable, Read/Write.boolean.Type:Whether to use graph-increment-other (true) or use defaults (false).graph-contour-levels: Variable, Read/Write.Type: integer in range (1 .. 32767).The number of contour levels to plot.graph-contour-start: Variable, Read/Write.Type: float in range (-1e+010 .. 1e+010).Value for first contour line.graph-contour-start-other: Variable, Read/Write.boolean.Type:Whether to use graph-contour-start (true) or use defaults (false).graph-data-row: Variable, Readonly.Type: vector of float-list.(i) The values on the i-th row of graph data.graph-data-type: Variable, Read/Write.Type: enum(electrostatic, charge-density, orbital, orbital-squared, spin-density).The type of wavefunction data to plot.graph-horizontal-grid-size: Variable, Read/Write.Type: integer in range (2 .. 8192).Number of data grid points for plotting in the horizontal direction.graph-orbital-offset: Variable, Read/Write.Type: integer in range (0 .. +Inf).Display orbital offset.graph-orbital-selection-type: Variable, Read/Write.Type: enum(lumo-plus, homo-minus, orbital-number).Display orbital type.graph-plane-offset: Variable, Read/Write.Type: float in range (-1e+010 .. 1e+010).Offset along viewer's Z axis of the plane of the data to plot.graph-vertical-grid-size: Variable, Read/Write.Type: integer in range (2 .. 8192).Number of data grid points for plotting in the vertical direction.grid-max-value: Variable, Readonly.Type: float in range (-1e+010 .. 1e+010).The isosurface maximum grid value.grid-min-value: Variable, Readonly.Type: float in range (-1e+010 .. 1e+010).The isosurface minimum grid value.hbond-energy: Variable, Readonly.Type: float in range (-1e+010 .. 1e+010).Results from backend computation.heat-of-formation: Variable, Read/Write.Type: float in range (-1e+010 .. 1e+010).Heat of formation.help: Command.Arg list: string.Give help on topic String-1.hide-errors: Variable, Read/Write.boolean.Type:Whether to display error messages on the screen (channel specific). hide-messages: Variable, Read/Write.boolean.Type:Whether to display MESSAGE value on the screen.hide-toolbar: Variable, Read/Write.boolean.Type:Command to toggle the toolbar.hide-warnings: Variable, Read/Write.boolean.Type:Whether to display warning messages on the screen (channel specific). huckel-constant: Variable, Read/Write.Type: float in range (0 .. 10).Extended Huckel constant.huckel-scaling-factor: Variable, Read/Write.Type: float in range (0 .. 100000).Extended Huckel scaling factor.huckel-weighted: Variable, Read/Write.boolean.Type:Extended Huckel weighting factor.。

分子图形与分子模型设计——HyperChem使用简介厦门大学化学系2005年3月HyperChem使用简介HyperChem是HyperCube Inc.的产品,它具有非常强大的综合计算与分析功能,是优秀的分子图形和分子设计的工具软件之一。
HyperChem是运行在Windows系统的分子计算与建模软件,具有量子化学(半经验和从头算)、分子力学、分子动力学、随机动力学、Monte Carlo模拟等计算功能,计算结果可以用三维图形显示。
它还提供用户VB、C/C++和FORTRAN等语言的应用程序接口。
HyperChem 7.5版本已经推出。
图1是HyperChem的工作窗口,最下部是工作状态档。
在菜单下面是常用工具档。
图1 HyperChem的工作窗口HyperChem的操作可以使用鼠标和键盘两种。
在工具档从左开始有8个工具图标,当鼠标点图标之后,鼠标在工作区的形状也改变为该图标的形状:1.绘图工具。
鼠标双击该图标可直接进入缺省元素周期表,选择所要绘制元素。
2.选择工具。
3.xy轴方向旋转工具,也可使用键盘的上下左右光标键进行相同的操作。
4.z轴方向旋转工具,也可使用键盘的Home和End键进行相同的操作。
5.xy轴平移工具,也可使用键盘的Shift+上下左右光标键进行相同的操作。
6.z轴平移工具。
7.缩放工具,也可使用键盘的PgUp和PgDn键进行相同的操作。
8.z轴截片工具。
鼠标操作有较为多样:左点击、右点击、左拖拉、右拖拉、左右拖拉、Shift+左点击、Shift+右点击、双击等。
一般的旋转和平移操作是使用鼠标的左键进行,当完成了某个基团、分子的选择之后,可以使用右键对所选部分进行旋转和平移操作。
HyperChem的详细操作将结合具体的实例进行讲解。
以下通过对HyperChem 5.1的菜单命令的介绍,说明它的主要功能和使用方法。
一、File1.New (Ctrl+N):新建一个沿尚未命名文件。
HyperChem 程序及其应用1、绘制丙二烯分子骨架模型,并测量有关分子构型的几何信息2、指定输出文件File---Start Log。
(1)先用半经验方法进行分子优化,从Setup中选择Semi-empirical…设定参数如下所示(2)选择Options…可设置收敛限和迭代次数,如下所示:(3)从Compute中选择Geometry Optimzation…进行集合构型优化:(4)优化完成之后,在Compute选择Single Point可进行单点计算。
3、采用从头算的方法:(1)Setup中选择Ab Initio…设定参数如下:(2)从Compute中选择Geometry Optimzation…进行集合构型优化:(3)完成集合构型优化后,从Compute选择Single Point可进行单点计算。
4、计算结束后,停止数据输出,从File---Stop Log。
5、分析有关分子的性质并简单分析讨论分子性质(1)采用从头算方法后,分析振动光谱:(该图显示谱线的位置、强度和振动模式)虚振动频率-185.84意味着,此结构不是一个稳定结构,而是一个过渡态。
(2)计算电子光谱最低能量跃迁π-π*在373.90,是禁阻跃迁允许的跃迁是116.84单态π-π*跃迁。
(3)分子偶极矩(4)轨道特征1、最高占据轨道2、最低空轨道(5)绘分子图,测电子光谱从Comput选择Plot Molecular Graphs1、2D图像2、3D图像6、结论与经验1、丙烯分子为一平面型分子,并且其振动频率存在虚频-185.84,意味着此平面结构不是一个稳定结构,而是一个过渡态。
2、半经验算法计算分子总能量为-16180.6852898 (kcal/mol),从头算方法计算分子总能量为-72576.4084722 (kcal/mol),所以计算方法的选择很重要。
3、计算分子的电子光谱能够得到该分子最低能量跃迁π-π*在373.90,是禁阻跃迁;允许的跃迁是116.84单态π-π*跃迁。

分子图形与分子模型设计——HyperChem使用简介厦门大学化学系2005年3月HyperChem使用简介HyperChem是HyperCube Inc.的产品,它具有非常强大的综合计算与分析功能,是优秀的分子图形和分子设计的工具软件之一。
HyperChem是运行在Windows系统的分子计算与建模软件,具有量子化学(半经验和从头算)、分子力学、分子动力学、随机动力学、Monte Carlo模拟等计算功能,计算结果可以用三维图形显示。
它还提供用户VB、C/C++和FORTRAN等语言的应用程序接口。
HyperChem 7.5版本已经推出。
图1是HyperChem的工作窗口,最下部是工作状态档。
在菜单下面是常用工具档。
图1 HyperChem的工作窗口HyperChem的操作可以使用鼠标和键盘两种。
在工具档从左开始有8个工具图标,当鼠标点图标之后,鼠标在工作区的形状也改变为该图标的形状:1.绘图工具。
鼠标双击该图标可直接进入缺省元素周期表,选择所要绘制元素。
2.选择工具。
3.xy轴方向旋转工具,也可使用键盘的上下左右光标键进行相同的操作。
4.z轴方向旋转工具,也可使用键盘的Home和End键进行相同的操作。
5.xy轴平移工具,也可使用键盘的Shift+上下左右光标键进行相同的操作。
6.z轴平移工具。
7.缩放工具,也可使用键盘的PgUp和PgDn键进行相同的操作。
8.z轴截片工具。
鼠标操作有较为多样:左点击、右点击、左拖拉、右拖拉、左右拖拉、Shift+左点击、Shift+右点击、双击等。
一般的旋转和平移操作是使用鼠标的左键进行,当完成了某个基团、分子的选择之后,可以使用右键对所选部分进行旋转和平移操作。
HyperChem的详细操作将结合具体的实例进行讲解。
以下通过对HyperChem 5.1的菜单命令的介绍,说明它的主要功能和使用方法。
一、File1.New (Ctrl+N):新建一个沿尚未命名文件。
河北师范大学计算量子化学研究所蔡新华教授量子化学在线教学.100/qc/lzhx-0.htmHyperChem 程序及其应用一、HyperChem 程序的运行环境HyperChem 程序包,用C++写源程序,具有工作站、微机等不同的版本,作为教学示例,我们向大家介绍适用于微机运行的程序版本。
可以通过网络选购HyperChem程序包,也可以免费下载演示版本以供学习只用,现在该公司提供的最新版本为V6.0。
该公司的网址为:该公司与98年诺贝尔奖金得主Pople的关系可以参看其网页有关介绍。
1、软件环境Windows95、Windows98或Windows2000系统。
2、硬件环境486以上的微机,内存应在8M以上,硬盘至少有32M以上自由空间。
为了能够以最佳方式显示分子图像,最好有VGA以上显示器。
3、程序安装使用该程序应注意程序版权(注册)。
安装程序默认子目录为:C:\hyper6安装完成后,该目录可以看到如下文件,其中,绿色烧杯为执行程序图标。
有关该程序的使用说明、参考手册等全套文档均可免费获得。
二、程序基本使用方法我们以演示版本为例,说明该程序的基本使用方法。
1、启动程序在屏幕上,双击绿色烧杯可以得到如下画面:点击 Try进入工作区窗口窗口各部分功能简介标题名称:最大、最小化、退出按钮菜单条:FILE、EDIT、BUILD、SELECT、DISPLAY、DATABASE、SETUP 、COMPUTE、CANCEL、SCRIPT、HELP工具条:工作区:状态行:2、打开已存在的数据文件File-Open选择分子图形的显示方式Display- Labels可以选择原子、化学键等标记方式:Dispay-RenderingRenderings-BallsRendering—Balls and Cylinders3、建立计算分子的数据文件以丁二烯为例:选择Build-Default Element可以显示指定元素的基本性质:选择绘图工具后,得到碳碳骨架。


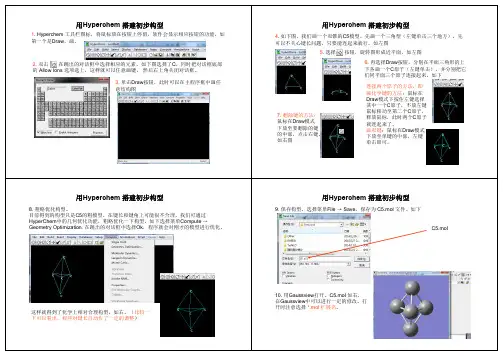
双击在跳出的对话框中选择相应的元素,如下图选择了C,同时把对话框底部Allow Ions 选项选上,这样就可以任意画键。
然后右上角关闭对话框。
意结构图选择按钮,旋转图形成近平面,如左图们同平面三个原子连接起来。
如下7. 删除键的方法:鼠标在Draw模式下放至要删除的键的中部,点击右键。
Hyperchem搭建初步构型的粗模型,在键长和键角上可能很不合理,我们可通过中的几何优化功能,粗略优化一下构型。
如下选择菜单Compute →在跳出的对话框中选择Ok,程序就会对刚才的模型进行优化。
这样就得到了化学上相对合理构型,如右。
(比较一下可以看出,程序对键长自动作了一定的调整)用Hyperchem9. 保存构型。
选择菜单File →Save,保存为10. 用Gaussview打开,C5.mol 如右,在Gaussview中可以进行一定的修改。
打开时注意选择*.mol 扩展名。
中选择File →Save 保存为C5.gjf,文件用Scite编辑器打开,内容如下第1行%开头定义chk文件,类似还可以定义内存%mem,%rwf等。
第2行#开头定义计算关键字第3行空第4行说明性文字第5行空第6行0 指体系电荷,如负1价离子则写-1. 1 为体系多重度= 2s+1 s为体系总自旋第7-11行为原子坐标。
第12空行用Gaussian优化构型下面以优化C5为例进行说明:1. 编辑C5.gjf文件,内容如右%mem定义内存为4GB%nprocshared=8定义使用CPU数目#P B3LYP/6-31+G*定义了优化Freq。
文件传到计算服务器上。
转换文件格式(注意Linux上区分大小写)5. 运行ps x 查看,如果正常,则显示如下:Gaussian优化构型传回文件。
查看工作是否计算完可以通过类似5步中的ps 录下运行tail C5.out 查看文件末尾信息,如果最后一行显示如下:Normal termination …说明任务正常结束,结果文件可用。

HyperChem应用乙烯最低电子激发态的从头计算优化乙烯的基态通过前面几个例子的学习,你已经对从头计算有了初步的了解,所以下面的一些指令将省略。
用STO-3G基组构造乙烯:1. 在File菜单中选择New,刷新工作区。
2. 确保Explicit Hydrogens没有选择。
3. 从Default Element中选择碳,并画一条C-C单键。
单击碳键中部使它变成双键。
4. 从Build菜单选择Add H和Model Build构造乙烯。
5. 从Setup菜单选择Ab Initio,并选基组为Minimal (STO-3G)。
同时令Total charge = 0,Spin multiplicity = 1,Spin pairing = RHF,Accelerate convergence =Yes,SCF Convergence limit = 0.0001. 按下CI按钮选择CI Method 为None。
优化乙烯基态:1. 选择Compute菜单的Geometry Optimization。
选择Polak-Ribiere方法,RMS gr adient 为0.01。
选择OK关闭对话框。
得到的结果为:C-C bond length:1.31埃,C-H bond length:1.08埃,H-C-H angle:115.7度。
计算相关能:1. 在Setup菜单选择Ab Initio,按下Options按钮并选择MP2 correlation energy。
单击OK回到工作区。
2. 在Compute菜单选择Single Point。
得到结果:SCF能量:-48364.64 kcal/mol ,MP2总能量:-48438.61 kcal/mol,包括-74.97kc al/mol相关能。
具体运算结果可能会与这个值有微小的差别。
乙烯基态轨道观察乙烯的轨道和轨道能量图:1. 从Compute菜单选择Orbitals。

HyperChem基本操作画原子1. 打开Element Table对话框。
这里有两种方法:在Build菜单中选择Default Element,或者双击Drawing工具。
Default Element对话框允许从周期表中选择缺省元素。
2. 如果单击Properties...按钮,将显示当前选择元素的物理属性。
也可以按下Shift键同时单击元素按钮,结果是一样的。
单击OK键,物理属性框消失。
3. 如果Allow Ions或者Explicit Hydrogens打开(用对勾选择),左键单击这些选项使其关闭。
4. 在缺省元素列表中选择Carbon,接着关闭元素对话框。
缺省元素将设置为碳。
当然也可以把打开的Default Element对话框移走,这样可以看到HyperChem工作区。
当画原子非常多的分子时,这是非常有效的。
5. 左键单击Drawing工具,把指针移到工作区。
6. 左键单击工作区左下角,将出现一个小圈,代表未成键的碳原子。
7. 在工作区不同位置画更多的原子。
画价键1. 把指针移到刚才画的第一个碳上。
2. 按下鼠标左键。
这是价键在第一个原子的位置。
3. 保持鼠标按钮按下的同时拖向工作区的顶端。
4. 放开鼠标按钮。
这是价键在第二个原子的位置。
一条线代表两个碳原子之间的价键。
5. 用仍旧停留在价键末端的指针, 用左键拖向工作区右下角。
6. 放开鼠标按钮。
这是第三个原子的位置。
7. 在空白工作区画六个价键,形成一个环。
现在你清楚了如何画原子和分子,并且学会了一些基本技巧。
选择原子在这个练习中,通过选择原子,你可以学到基本的选择技巧。
首先必须设置选择的级别[原子(atoms),基(residues),或分子(molecules)]。
这里设置为原子(atoms)。
1. 左键单击Select菜单。
2. 左键单击选择Atoms。
接下来,关闭Multiple Selections:1. 左键单击Select菜单。
化学软件——HyperChem(分子模拟)介绍HyperChem是一款以高质量,灵活易操作而闻名的分子模拟软件。
通过利用3D对量子化学计算,分子力学及动力学进行模拟动画,HyperChem 为您提供比其它 Windows 软件更多的模拟工具。
图形界面:图形界面,有半经验方法( AM1 , PM3 等), UHF , RHF 和 CI 和7.0 版新增加的密度泛函。
可进行单点能,几何优化,分子轨道分析,预测可见- 紫外光谱,蒙特卡罗和分子力学计算。
主要功能:1. 结构输入和对分子操作。
2. 显示分子。
3. 化学计算。
用量子化学或经典势能曲面方法,进行单点、几何优化和过渡态寻找计算。
可以进行的计算类型有:单点能,几何优化,计算振动频率得到简正模式,过渡态寻找,分子动力学模拟 Langevin 动力学模拟, Metropolis Monte Carlo 模拟。
支持的计算方法有:从头计算,半经验方法,分子力学,混合计算。
4. 可以用来研究的分子特性有:同位素的相对稳定性;生成热;活化能;原子电荷; HOMO-LUMO能量间隔;电离势;电子亲和力;偶极矩;电子能级; MP2 电子相关能; CI 激发态能量;过渡态结构和能量;非键相互作用能; UV-VIS 吸收谱; IR 吸收谱;同位素对振动的影响;对结构特性的碰撞影响;团簇的稳定性。
5. 支持用户定制的外部程序。
6. 其它模块: RAYTRACE 模块, RMS Fit , SEQUENCE 编辑器,晶体构造器;糖类构造器,构像搜寻,QSAR 特性,脚本编辑器。
7. 新的力场方法: Amber 2 , Amber 3 ,用于糖类的 Amber , Amber 94 ,Amber 96 。
8. ESR 谱。
9. 电极化率。
10. 二维和三维势能图。
11. 蛋白质设计。
12. 电场。
13. 梯度的图形显示。
14. 新增功能:密度泛函理论 (DFT) 计算; NMR 模拟;数据库; Charmm 蛋白质模拟;半经验方法TNDO ;磁场中分子计算;激发态几何优化; MP2 相关结构优化;新的芳香环图;交互式参数控制;增强的聚合物构造功能;新增基组。
利用Hyperchem软件进行分子结构构建及性质计算实验目的1.初步了解分子模型方法的原理和应用。
2.学习使用Hyperchem软件构建简单的分子并使用适当方法优化结构。
3.学习使用Hyperchem软件计算简单分子的几何和电子性质。
实验原理化学的学习使我们认识了许多分子的分子式及二维结构,如何得到分子的三维结构,以及分子在空间的几何特征和电子特征,则可以借助于理论计算的工具和方法去模拟计算。
HyperChem软件是HyperCube公司开发的Windows界面程序。
是常用的分子设计和模拟软件。
它可以应用于构建简单及复杂的分子模型并进行综合计算与分析。
分子构建过程可以通过熟练各个菜单及工具栏的操作来实现,计算和分析需要我们了解常用的计算方法。
在本实验室中我们需要了解一下计算方法,这些方法位于HyperChem的Setup菜单下。
(1)分子力学(Molecular Mechanics)方法:分子力学又叫力场方法,目前广泛地用于计算分子的构象和能量。
适用于超大规模体系,超低精度计算。
分子力学的基本假设:玻恩-奥本海默近似,原子核的运动与电子的运动可以看成是独立的;分子是一组靠各种作用力维系在一起的原子集合。
这些原子在空间上若过于靠近,便相互排斥;但又不能远离,否则连接它们的化学键以及由这些键构成的键角等会发生变化,即出现键的拉伸或压缩、键角的扭变等,会引起分子内部应力的增加。
每个真实的分子结构,都是在上述几种作用达到平衡状态的表现。
分子力学从几个主要的典型结构参数和作用力出发来讨论分子结构,即用位能函数来表示当键长、键角、二面角等结构参数以及非键作用等偏离“理想”值时分子能量的变化。
不同的分子力场方法采用不同的势能函数。
MM+:适用于有机分子的计算。
Amber:适用于有机分子、蛋白质和核酸等大分子的计算。
(2)半经验计算(Semi-empirical)方法:是求解HF(Hartree-Fock)方程时采用各种近似,或者直接使用拟合的经验参数来近似求解自洽场。
HyperChem 程序及其应用一、HyperChem 程序的运行环境HyperChem 程序包,用C++写源程序,具有工作站、微机等不同的版本,作为教学示例,我们向大家介绍适用于微机运行的程序版本。
可以通过网络选购HyperChem程序包,也可以免费下载演示版本以供学习只用,现在该公司提供的最新版本为V6.0。
该公司的网址为:该公司与98年诺贝尔奖金得主Pople的关系可以参看其网页有关介绍。
1、软件环境Windows95、Windows98或Windows2000系统。
2、硬件环境486以上的微机,内存应在8M以上,硬盘至少有32M以上自由空间。
为了能够以最佳方式显示分子图像,最好有VGA以上显示器。
3、程序安装使用该程序应注意程序版权(注册)。
安装程序默认子目录为:C:\hyper6安装完成后,该目录可以看到如下文件,其中,绿色烧杯为执行程序图标。
有关该程序的使用说明、参考手册等全套文档均可免费获得。
二、程序基本使用方法我们以演示版本为例,说明该程序的基本使用方法。
1、启动程序在屏幕上,双击绿色烧杯可以得到如下画面:点击 Try进入工作区窗口窗口各部分功能简介标题名称:最大、最小化、退出按钮菜单条:FILE、EDIT、BUILD、SELECT、DISPLAY、DATABASE、SETUP 、COMPUTE、CANCEL、SCRIPT、HELP工具条:工作区:状态行:2、打开已存在的数据文件File-OpenDisplay- Labels可以选择原子、化学键等标记方式:Dispay-RenderingRenderings-BallsRendering—Balls and Cylinders3、建立计算分子的数据文件以丁二烯为例:选择Build-Default Element可以显示指定元素的基本性质:选择绘图工具后,得到碳碳骨架。
左键、右键增减键序把2D骨架转换为3D图像Select选定原子后,状态行提示原子标号、坐标等结构信息。
《化学信息学》课程教学大纲课程名称:化学信息学英文译名:Chemical Informatics课程编号:开课单位:生物工程学院预修课程:适用专业:化学工程与工艺、生物工程、制药工程开课学期:课内总学时:32学分:2 考核方式:考查一、教学目的与要求1.掌握Internet 的基本知识,服务功能。
着重掌握IE 浏览Internet来获得科学信息的方法。
掌握国际与国内一些典型的搜索引擎的使用方法与技术,并灵活运用以获得所需的科学信息。
2.掌握现代化学文献的基础知识、化学文献的检索系统和四大检索方法,即网络检索、光盘检索、联机检索和手工检索等。
牢固掌握计算机网络检索化学文献的方法与技术,特别是结合毕业论文的课题利用网络数据库查阅国内外有关的化学文献。
3.牢固掌握ChemWin、HyperChem和Origin等现代化学图文处理软件的使用技术,能灵活绘制各种类型的化学分子式、分子图、反应式、立体模型图、表面图、化学实验装置图、化学工艺流程图。
初步掌握一些基本的量子力学和量子化学计算方法,进行化学建模、分子设计、分子的电子结构及相关性质的计算等。
二、课程内容及学时分配第一章化学信息学与Internet(一) 化学信息的构成及化学信息学(二) 因特网(Internet)概述第二章化学化工网站与网上数据库(一) 通用信息检索引擎:yahoo,google,baidu(二) 重要的化学化工综合同站:,(三) 网上化学结构数据库、反应数据库和初性数据库:CambridgeSoft,PubMed(四) 网上化学化工分类信息与信息资源:第三章化学化工文献检索(一) 文献慨述(二) 美国<化学文摘)(印刷出版物)(三) CA索引和手工检索方法(四) CAS的电子出版物(五) CA on CD的使用方法(六) 美国科学信息研究所ISI和《科学引文索引》SCI简介(七) 其他文献数据库:Dialog,GenBank,EI,Elsevier第四章网上专利检索(一) 中国专利文摘数据库:(二) 美国专利数据库:,(三) 其他专利数据库与知识产权保护组织:IBM,PCM,IPDL第五章化学实验数据统计与谱分析(一) 实验数据的统计分布:实验误差,统计分布(二) 分布的数字特征计算:位置、散布、分布、相关特征参数(三) 化学分析的采样(四) 比较实验:总体标准差、均值比较(五) 回归分析:一元、多元回归,相关系数、显著性检验(六) 数据的最小二乘平滑处理:(七) 曲线拟合和谱峰分辨(八) 谱的微分与积分第六章科学数据处理与绘图工具软件Origin(一) Origin的窗口组成(二) 数据的输入、编辑和保存(三) 数据的统计处理:排序、求和、标准差、t检验、多元线性回归(四) 数据关系的图形表示:分布图、线图、点线图、直方图、扇形图(五) 谱与曲线的处理:曲线平滑、背景校正、基线、谱的微积分第七章化学中的多元分析方法.(一) 化学与多元分析:多元分析的问题与特征、主要方法、变量类型,数据预处理(二) 相关分析与变量分组:相关系数矩阵、复相关系数、偏相关系数矩阵(三) 降维、信息压缩与显示技术:主成分分析、非线性映射(四) 分类与判别:距离判别法、K-最临近法、线性判别法(五) 聚类分析(六) 因子分析:偏最小二乘(七) 其他的信息挖掘技术:最大熵、模拟退火、神经网络、遗传算法第八章化学结构的表示与可视化(一) 分子拓扑结构的矩阵表示与矩阵指数:邻接矩阵、距离矩阵、关联矩阵、环矩阵(二) 分子拓扑结构的线性标记方法:WLN(三) 有机分子结构式绘图软件ChemWindow(四) 分子几何结构的表示:坐标系、常见文件格式(五) 二维分子结构显示软件WebLab ViewPro第九章分子结构信息与分子设计(一) 分子及其聚集体的计算与模拟方法评述:量子力学、分子力学、分子动力学、随机模拟、蒙特卡罗(二) 定量构效关系方法:Free-Wilson、Hansch(三) 分子设计中的重要参数:Hammett、Taft、Es、VDW体积与半径、logP、连接性指数、MR、溶解度参数、量子化学参数(四) 分子计算与模拟软件举例:Sybyl、InsightII、Alchemy、Charmm(五) 分子建模与分析软件HyperChem功能简介(六) 分子与材料设计简介三、教材[1]缪强. 面向21世课程教材《化学信息学导论》,高等教育出版社,2001年7月四、教材与主要参考书[2]沈勇,张大经,郑康成编著.《现代化学信息基础教程》,广州:中山大学出院版社,2000年11月[3]吴功宜,吴英编著. 《Internet 基础》,北京:清华大学出版社,2000 年[4]林卓然编著.《计算机应用基础教程WINDOWS98版》,广州:中山大学出版社,2000 年[5]王源编著. 《现代化学文献检索》,上海:上海科学技术文献出版社,1999 年[6]杨均辉等. 《四大文献索引及联机检索》. 广州:中山大学出版社,1996 年[7]胡立江等编著. 《精通ChemDraw》,北京:清华大学出版社,2002年[8]俞庆森,朱龙观编著. 面向21世课程教材《分子设计导论》,高等教育出版社,2000年7月[9]赵文元,王亦军编著.《计算机在化学化工中的应用技术》,科学出版社2001年[10]邵学广,蔡文生编著.中国科学院研究生教学丛书《化学信息学》,科学出版社,2001年6月。
Hyperchem程序应用-环戊烷画分子结构模型图:加氢、模型化:用半经验方法CNDO进行优化:用从头算计算方法进行单点计算:显示原子电荷:测键长、键角、二面角:分子性质:戊烷分子2D、3D静电势图:分子总电荷密度图(2D、3D):等值面图:分子轨道图-最高占据轨道2D、3D图:分子轨道图-最低空轨道2D 、3D 图:分子结构模型表示:输出结果:HyperChem log start -- Sun Dec 12 21:25:30 2010.Geometry optimization, SemiEmpirical, molecule = (untitled).CNDOFletcherReeves optimizerConvergence limit = 0.0001000 Iteration limit = 50Accelerate convergence = YESOptimization algorithm = Fletcher-ReevesCriterion of RMS gradient = 0.1000 kcal/(A mol) Maximum cycles = 225RHF Calculation:Singlet state calculationNumber of electrons = 30Number of Double Occupied Levels = 15Charge on the System = 0Total Orbitals = 30Starting CNDO calculation with 30 orbitalsE=0.0000 Grad=0.000 Conv=NO(0 cycles 0 points) [Iter=1 Diff=9872.30361]E=0.0000 Grad=0.000 Conv=NO(0 cycles 0 points) [Iter=2 Diff=6.19972]E=0.0000 Grad=0.000 Conv=NO(0 cycles 0 points) [Iter=3 Diff=0.35983]E=0.0000 Grad=0.000 Conv=NO(0 cycles 0 points) [Iter=4 Diff=0.02932]E=0.0000 Grad=0.000 Conv=NO(0 cycles 0 points) [Iter=5 Diff=0.00064]E=0.0000 Grad=0.000 Conv=NO(0 cycles 0 points) [Iter=6 Diff=0.00003]E=-3902.0813 Grad=75.208 Conv=NO(0 cycles 1 points) [Iter=1 Diff=50.66003] E=-3902.0813 Grad=75.208 Conv=NO(0 cycles 1 points) [Iter=2 Diff=3.70425] E=-3902.0813 Grad=75.208 Conv=NO(0 cycles 1 points) [Iter=3 Diff=0.31058] E=-3902.0813 Grad=75.208 Conv=NO(0 cycles 1 points) [Iter=4 Diff=0.03342] E=-3902.0813 Grad=75.208 Conv=NO(0 cycles 1 points) [Iter=5 Diff=0.00075] E=-3902.0813 Grad=75.208 Conv=NO(0 cycles 1 points) [Iter=6 Diff=0.00012] E=-3902.0813 Grad=75.208 Conv=NO(0 cycles 1 points) [Iter=7 Diff=0.00000] E=-3854.4941 Grad=131.923 Conv=NO(0 cycles 2 points) [Iter=1 Diff=20.67949] E=-3854.4941 Grad=131.923 Conv=NO(0 cycles 2 points) [Iter=2 Diff=1.47203] E=-3854.4941 Grad=131.923 Conv=NO(0 cycles 2 points) [Iter=3 Diff=0.11775] E=-3854.4941 Grad=131.923 Conv=NO(0 cycles 2 points) [Iter=4 Diff=0.01208] E=-3854.4941 Grad=131.923 Conv=NO(0 cycles 2 points) [Iter=5 Diff=0.00021] E=-3854.4941 Grad=131.923 Conv=NO(0 cycles 2 points) [Iter=6 Diff=0.00001] E=-3929.7410 Grad=5.160 Conv=NO(1 cycles 3 points) [Iter=1 Diff=0.02148]E=-3929.7410 Grad=5.160 Conv=NO(1 cycles 3 points) [Iter=2 Diff=0.00163]E=-3929.7410 Grad=5.160 Conv=NO(1 cycles 3 points) [Iter=3 Diff=0.00015]E=-3929.7410 Grad=5.160 Conv=NO(1 cycles 3 points) [Iter=4 Diff=0.00002]E=-3930.0144 Grad=3.723 Conv=NO(1 cycles 4 points) [Iter=1 Diff=0.02156]E=-3930.0144 Grad=3.723 Conv=NO(1 cycles 4 points) [Iter=2 Diff=0.00164]E=-3930.0144 Grad=3.723 Conv=NO(1 cycles 4 points) [Iter=3 Diff=0.00016]E=-3930.0144 Grad=3.723 Conv=NO(1 cycles 4 points) [Iter=4 Diff=0.00002]E=-3930.1650 Grad=3.794 Conv=NO(1 cycles 5 points) [Iter=1 Diff=0.08681]E=-3930.1650 Grad=3.794 Conv=NO(1 cycles 5 points) [Iter=2 Diff=0.00661]E=-3930.1650 Grad=3.794 Conv=NO(1 cycles 5 points) [Iter=3 Diff=0.00063]E=-3930.1650 Grad=3.794 Conv=NO(1 cycles 5 points) [Iter=4 Diff=0.00008]E=-3930.0955 Grad=7.526 Conv=NO(1 cycles 6 points) [Iter=1 Diff=0.03555]E=-3930.0955 Grad=7.526 Conv=NO(1 cycles 6 points) [Iter=2 Diff=0.00271]E=-3930.0955 Grad=7.526 Conv=NO(1 cycles 6 points) [Iter=3 Diff=0.00026]E=-3930.0955 Grad=7.526 Conv=NO(1 cycles 6 points) [Iter=4 Diff=0.00003]E=-3930.1970 Grad=4.816 Conv=NO(2 cycles 7 points) [Iter=1 Diff=0.31929]E=-3930.1970 Grad=4.816 Conv=NO(2 cycles 7 points) [Iter=2 Diff=0.02495]E=-3930.1970 Grad=4.816 Conv=NO(2 cycles 7 points) [Iter=3 Diff=0.00236]E=-3930.1970 Grad=4.816 Conv=NO(2 cycles 7 points) [Iter=5 Diff=0.00001] E=-3930.0500 Grad=8.453 Conv=NO(2 cycles 8 points) [Iter=1 Diff=0.10464] E=-3930.0500 Grad=8.453 Conv=NO(2 cycles 8 points) [Iter=2 Diff=0.00818] E=-3930.0500 Grad=8.453 Conv=NO(2 cycles 8 points) [Iter=3 Diff=0.00078] E=-3930.0500 Grad=8.453 Conv=NO(2 cycles 8 points) [Iter=4 Diff=0.00010] E=-3930.0500 Grad=8.453 Conv=NO(2 cycles 8 points) [Iter=5 Diff=0.00000] E=-3930.3799 Grad=2.276 Conv=NO(3 cycles 9 points) [Iter=1 Diff=0.00575] E=-3930.3799 Grad=2.276 Conv=NO(3 cycles 9 points) [Iter=2 Diff=0.00059] E=-3930.3799 Grad=2.276 Conv=NO(3 cycles 9 points) [Iter=3 Diff=0.00007] E=-3930.4749 Grad=1.731 Conv=NO(3 cycles 10 points) [Iter=1 Diff=0.00574] E=-3930.4749 Grad=1.731 Conv=NO(3 cycles 10 points) [Iter=2 Diff=0.00059] E=-3930.4749 Grad=1.731 Conv=NO(3 cycles 10 points) [Iter=3 Diff=0.00007] E=-3930.5400 Grad=1.433 Conv=NO(3 cycles 11 points) [Iter=1 Diff=0.02291] E=-3930.5400 Grad=1.433 Conv=NO(3 cycles 11 points) [Iter=2 Diff=0.00237] E=-3930.5400 Grad=1.433 Conv=NO(3 cycles 11 points) [Iter=3 Diff=0.00030] E=-3930.5400 Grad=1.433 Conv=NO(3 cycles 11 points) [Iter=4 Diff=0.00005] E=-3930.5815 Grad=1.970 Conv=NO(3 cycles 12 points) [Iter=1 Diff=0.00052] E=-3930.5815 Grad=1.970 Conv=NO(3 cycles 12 points) [Iter=2 Diff=0.00005] E=-3930.5830 Grad=1.813 Conv=NO(4 cycles 13 points) [Iter=1 Diff=0.13831] E=-3930.5830 Grad=1.813 Conv=NO(4 cycles 13 points) [Iter=2 Diff=0.01226] E=-3930.5830 Grad=1.813 Conv=NO(4 cycles 13 points) [Iter=3 Diff=0.00133] E=-3930.5830 Grad=1.813 Conv=NO(4 cycles 13 points) [Iter=4 Diff=0.00019] E=-3930.5830 Grad=1.813 Conv=NO(4 cycles 13 points) [Iter=5 Diff=0.00000] E=-3930.4128 Grad=6.041 Conv=NO(4 cycles 14 points) [Iter=1 Diff=0.06925] E=-3930.4128 Grad=6.041 Conv=NO(4 cycles 14 points) [Iter=2 Diff=0.00614] E=-3930.4128 Grad=6.041 Conv=NO(4 cycles 14 points) [Iter=3 Diff=0.00066] E=-3930.4128 Grad=6.041 Conv=NO(4 cycles 14 points) [Iter=4 Diff=0.00010] E=-3930.6182 Grad=1.216 Conv=NO(5 cycles 15 points) [Iter=1 Diff=0.00777] E=-3930.6182 Grad=1.216 Conv=NO(5 cycles 15 points) [Iter=2 Diff=0.00071] E=-3930.6182 Grad=1.216 Conv=NO(5 cycles 15 points) [Iter=3 Diff=0.00008] E=-3930.6323 Grad=1.558 Conv=NO(5 cycles 16 points) [Iter=1 Diff=0.00082] E=-3930.6323 Grad=1.558 Conv=NO(5 cycles 16 points) [Iter=2 Diff=0.00007] E=-3930.6365 Grad=0.975 Conv=NO(6 cycles 17 points) [Iter=1 Diff=0.00493] E=-3930.6365 Grad=0.975 Conv=NO(6 cycles 17 points) [Iter=2 Diff=0.00053] E=-3930.6365 Grad=0.975 Conv=NO(6 cycles 17 points) [Iter=3 Diff=0.00007] E=-3930.6555 Grad=0.993 Conv=NO(6 cycles 18 points) [Iter=1 Diff=0.00493] E=-3930.6555 Grad=0.993 Conv=NO(6 cycles 18 points) [Iter=2 Diff=0.00053] E=-3930.6555 Grad=0.993 Conv=NO(6 cycles 18 points) [Iter=3 Diff=0.00007] E=-3930.6479 Grad=2.055 Conv=NO(6 cycles 19 points) [Iter=1 Diff=0.00305] E=-3930.6479 Grad=2.055 Conv=NO(6 cycles 19 points) [Iter=2 Diff=0.00033] E=-3930.6479 Grad=2.055 Conv=NO(6 cycles 19 points) [Iter=3 Diff=0.00004] E=-3930.6563 Grad=1.187 Conv=NO(7 cycles 20 points) [Iter=1 Diff=0.02653] E=-3930.6563 Grad=1.187 Conv=NO(7 cycles 20 points) [Iter=2 Diff=0.00282]E=-3930.6563 Grad=1.187 Conv=NO(7 cycles 20 points) [Iter=4 Diff=0.00008] E=-3930.6553 Grad=1.447 Conv=NO(7 cycles 21 points) [Iter=1 Diff=0.00680] E=-3930.6553 Grad=1.447 Conv=NO(7 cycles 21 points) [Iter=2 Diff=0.00072] E=-3930.6553 Grad=1.447 Conv=NO(7 cycles 21 points) [Iter=3 Diff=0.00010] E=-3930.6553 Grad=1.447 Conv=NO(7 cycles 21 points) [Iter=4 Diff=0.00002] E=-3930.6733 Grad=0.377 Conv=NO(8 cycles 22 points) [Iter=1 Diff=0.00126] E=-3930.6733 Grad=0.377 Conv=NO(8 cycles 22 points) [Iter=2 Diff=0.00013] E=-3930.6733 Grad=0.377 Conv=NO(8 cycles 22 points) [Iter=3 Diff=0.00002] E=-3930.6770 Grad=0.591 Conv=NO(8 cycles 23 points) [Iter=1 Diff=0.00126] E=-3930.6770 Grad=0.591 Conv=NO(8 cycles 23 points) [Iter=2 Diff=0.00013] E=-3930.6770 Grad=0.591 Conv=NO(8 cycles 23 points) [Iter=3 Diff=0.00002] E=-3930.6760 Grad=1.185 Conv=NO(8 cycles 24 points) [Iter=1 Diff=0.00060] E=-3930.6760 Grad=1.185 Conv=NO(8 cycles 24 points) [Iter=2 Diff=0.00006] E=-3930.6772 Grad=0.765 Conv=NO(9 cycles 25 points) [Iter=1 Diff=0.01277] E=-3930.6772 Grad=0.765 Conv=NO(9 cycles 25 points) [Iter=2 Diff=0.00156] E=-3930.6772 Grad=0.765 Conv=NO(9 cycles 25 points) [Iter=3 Diff=0.00022] E=-3930.6772 Grad=0.765 Conv=NO(9 cycles 25 points) [Iter=4 Diff=0.00004] E=-3930.6943 Grad=0.537 Conv=NO(9 cycles 26 points) [Iter=1 Diff=0.01278] E=-3930.6943 Grad=0.537 Conv=NO(9 cycles 26 points) [Iter=2 Diff=0.00156] E=-3930.6943 Grad=0.537 Conv=NO(9 cycles 26 points) [Iter=3 Diff=0.00022] E=-3930.6943 Grad=0.537 Conv=NO(9 cycles 26 points) [Iter=4 Diff=0.00004] E=-3930.7007 Grad=0.713 Conv=NO(9 cycles 27 points) [Iter=1 Diff=0.05107] E=-3930.7007 Grad=0.713 Conv=NO(9 cycles 27 points) [Iter=2 Diff=0.00624] E=-3930.7007 Grad=0.713 Conv=NO(9 cycles 27 points) [Iter=3 Diff=0.00088] E=-3930.7007 Grad=0.713 Conv=NO(9 cycles 27 points) [Iter=4 Diff=0.00017] E=-3930.7007 Grad=0.713 Conv=NO(9 cycles 27 points) [Iter=5 Diff=0.00000] E=-3930.6814 Grad=1.564 Conv=NO(9 cycles 28 points) [Iter=1 Diff=0.04619] E=-3930.6814 Grad=1.564 Conv=NO(9 cycles 28 points) [Iter=2 Diff=0.00565] E=-3930.6814 Grad=1.564 Conv=NO(9 cycles 28 points) [Iter=3 Diff=0.00080] E=-3930.6814 Grad=1.564 Conv=NO(9 cycles 28 points) [Iter=4 Diff=0.00015] E=-3930.6814 Grad=1.564 Conv=NO(9 cycles 28 points) [Iter=5 Diff=0.00000] E=-3930.7007 Grad=0.746 Conv=NO(10 cycles 29 points) [Iter=1 Diff=0.30872] E=-3930.7007 Grad=0.746 Conv=NO(10 cycles 29 points) [Iter=2 Diff=0.03123] E=-3930.7007 Grad=0.746 Conv=NO(10 cycles 29 points) [Iter=3 Diff=0.00379] E=-3930.7007 Grad=0.746 Conv=NO(10 cycles 29 points) [Iter=4 Diff=0.00060] E=-3930.7007 Grad=0.746 Conv=NO(10 cycles 29 points) [Iter=5 Diff=0.00001] E=-3930.4949 Grad=5.995 Conv=NO(10 cycles 30 points) [Iter=1 Diff=0.22743] E=-3930.4949 Grad=5.995 Conv=NO(10 cycles 30 points) [Iter=2 Diff=0.02299] E=-3930.4949 Grad=5.995 Conv=NO(10 cycles 30 points) [Iter=3 Diff=0.00279] E=-3930.4949 Grad=5.995 Conv=NO(10 cycles 30 points) [Iter=4 Diff=0.00044] E=-3930.4949 Grad=5.995 Conv=NO(10 cycles 30 points) [Iter=5 Diff=0.00001] E=-3930.7065 Grad=0.567 Conv=NO(11 cycles 31 points) [Iter=1 Diff=0.00702] E=-3930.7065 Grad=0.567 Conv=NO(11 cycles 31 points) [Iter=2 Diff=0.00087]E=-3930.7065 Grad=0.567 Conv=NO(11 cycles 31 points) [Iter=4 Diff=0.00002] E=-3930.7183 Grad=0.492 Conv=NO(11 cycles 32 points) [Iter=1 Diff=0.00702] E=-3930.7183 Grad=0.492 Conv=NO(11 cycles 32 points) [Iter=2 Diff=0.00087] E=-3930.7183 Grad=0.492 Conv=NO(11 cycles 32 points) [Iter=3 Diff=0.00013] E=-3930.7183 Grad=0.492 Conv=NO(11 cycles 32 points) [Iter=4 Diff=0.00002] E=-3930.7261 Grad=0.615 Conv=NO(11 cycles 33 points) [Iter=1 Diff=0.02813] E=-3930.7261 Grad=0.615 Conv=NO(11 cycles 33 points) [Iter=2 Diff=0.00348] E=-3930.7261 Grad=0.615 Conv=NO(11 cycles 33 points) [Iter=3 Diff=0.00050] E=-3930.7261 Grad=0.615 Conv=NO(11 cycles 33 points) [Iter=4 Diff=0.00010] E=-3930.7292 Grad=1.133 Conv=NO(11 cycles 34 points) [Iter=1 Diff=0.00269] E=-3930.7292 Grad=1.133 Conv=NO(11 cycles 34 points) [Iter=2 Diff=0.00033] E=-3930.7292 Grad=1.133 Conv=NO(11 cycles 34 points) [Iter=3 Diff=0.00005] E=-3930.7300 Grad=0.957 Conv=NO(12 cycles 35 points) [Iter=1 Diff=0.34365] E=-3930.7300 Grad=0.957 Conv=NO(12 cycles 35 points) [Iter=2 Diff=0.04173] E=-3930.7300 Grad=0.957 Conv=NO(12 cycles 35 points) [Iter=3 Diff=0.00599] E=-3930.7300 Grad=0.957 Conv=NO(12 cycles 35 points) [Iter=4 Diff=0.00114] E=-3930.7300 Grad=0.957 Conv=NO(12 cycles 35 points) [Iter=5 Diff=0.00002] E=-3930.5986 Grad=5.283 Conv=NO(12 cycles 36 points) [Iter=1 Diff=0.21604] E=-3930.5986 Grad=5.283 Conv=NO(12 cycles 36 points) [Iter=2 Diff=0.02624] E=-3930.5986 Grad=5.283 Conv=NO(12 cycles 36 points) [Iter=3 Diff=0.00377] E=-3930.5986 Grad=5.283 Conv=NO(12 cycles 36 points) [Iter=4 Diff=0.00071] E=-3930.5986 Grad=5.283 Conv=NO(12 cycles 36 points) [Iter=5 Diff=0.00001] E=-3930.7395 Grad=0.780 Conv=NO(13 cycles 37 points) [Iter=1 Diff=0.01219] E=-3930.7395 Grad=0.780 Conv=NO(13 cycles 37 points) [Iter=2 Diff=0.00152] E=-3930.7395 Grad=0.780 Conv=NO(13 cycles 37 points) [Iter=3 Diff=0.00022] E=-3930.7395 Grad=0.780 Conv=NO(13 cycles 37 points) [Iter=4 Diff=0.00004] E=-3930.7498 Grad=0.615 Conv=NO(13 cycles 38 points) [Iter=1 Diff=0.01218] E=-3930.7498 Grad=0.615 Conv=NO(13 cycles 38 points) [Iter=2 Diff=0.00152] E=-3930.7498 Grad=0.615 Conv=NO(13 cycles 38 points) [Iter=3 Diff=0.00022] E=-3930.7498 Grad=0.615 Conv=NO(13 cycles 38 points) [Iter=4 Diff=0.00004] E=-3930.7446 Grad=1.320 Conv=NO(13 cycles 39 points) [Iter=1 Diff=0.00844] E=-3930.7446 Grad=1.320 Conv=NO(13 cycles 39 points) [Iter=2 Diff=0.00105] E=-3930.7446 Grad=1.320 Conv=NO(13 cycles 39 points) [Iter=3 Diff=0.00016] E=-3930.7446 Grad=1.320 Conv=NO(13 cycles 39 points) [Iter=4 Diff=0.00003] E=-3930.7500 Grad=0.705 Conv=NO(14 cycles 40 points) [Iter=1 Diff=0.04089] E=-3930.7500 Grad=0.705 Conv=NO(14 cycles 40 points) [Iter=2 Diff=0.00529] E=-3930.7500 Grad=0.705 Conv=NO(14 cycles 40 points) [Iter=3 Diff=0.00080] E=-3930.7500 Grad=0.705 Conv=NO(14 cycles 40 points) [Iter=4 Diff=0.00016] E=-3930.7500 Grad=0.705 Conv=NO(14 cycles 40 points) [Iter=5 Diff=0.00000] E=-3930.7607 Grad=1.074 Conv=NO(14 cycles 41 points) [Iter=1 Diff=0.00206] E=-3930.7607 Grad=1.074 Conv=NO(14 cycles 41 points) [Iter=2 Diff=0.00027] E=-3930.7607 Grad=1.074 Conv=NO(14 cycles 41 points) [Iter=3 Diff=0.00004] E=-3930.7620 Grad=0.819 Conv=NO(15 cycles 42 points) [Iter=1 Diff=0.05678]E=-3930.7620 Grad=0.819 Conv=NO(15 cycles 42 points) [Iter=3 Diff=0.00116] E=-3930.7620 Grad=0.819 Conv=NO(15 cycles 42 points) [Iter=4 Diff=0.00024] E=-3930.7620 Grad=0.819 Conv=NO(15 cycles 42 points) [Iter=5 Diff=0.00000] E=-3930.7861 Grad=1.072 Conv=NO(15 cycles 43 points) [Iter=1 Diff=0.05676] E=-3930.7861 Grad=1.072 Conv=NO(15 cycles 43 points) [Iter=2 Diff=0.00749] E=-3930.7861 Grad=1.072 Conv=NO(15 cycles 43 points) [Iter=3 Diff=0.00116] E=-3930.7861 Grad=1.072 Conv=NO(15 cycles 43 points) [Iter=4 Diff=0.00024] E=-3930.7861 Grad=1.072 Conv=NO(15 cycles 43 points) [Iter=5 Diff=0.00000] E=-3930.7898 Grad=2.033 Conv=NO(15 cycles 44 points) [Iter=1 Diff=0.00627] E=-3930.7898 Grad=2.033 Conv=NO(15 cycles 44 points) [Iter=2 Diff=0.00083] E=-3930.7898 Grad=2.033 Conv=NO(15 cycles 44 points) [Iter=3 Diff=0.00013] E=-3930.7898 Grad=2.033 Conv=NO(15 cycles 44 points) [Iter=4 Diff=0.00003] E=-3930.7908 Grad=1.690 Conv=NO(16 cycles 45 points) [Iter=1 Diff=0.94010] E=-3930.7908 Grad=1.690 Conv=NO(16 cycles 45 points) [Iter=2 Diff=0.12528] E=-3930.7908 Grad=1.690 Conv=NO(16 cycles 45 points) [Iter=3 Diff=0.01931] E=-3930.7908 Grad=1.690 Conv=NO(16 cycles 45 points) [Iter=4 Diff=0.00389] E=-3930.7908 Grad=1.690 Conv=NO(16 cycles 45 points) [Iter=5 Diff=0.00006] E=-3930.7996 Grad=2.454 Conv=NO(16 cycles 46 points) [Iter=1 Diff=0.20799] E=-3930.7996 Grad=2.454 Conv=NO(16 cycles 46 points) [Iter=2 Diff=0.02769] E=-3930.7996 Grad=2.454 Conv=NO(16 cycles 46 points) [Iter=3 Diff=0.00427] E=-3930.7996 Grad=2.454 Conv=NO(16 cycles 46 points) [Iter=4 Diff=0.00086] E=-3930.7996 Grad=2.454 Conv=NO(16 cycles 46 points) [Iter=5 Diff=0.00001] E=-3930.8279 Grad=1.061 Conv=NO(17 cycles 47 points) [Iter=1 Diff=0.38133] E=-3930.8279 Grad=1.061 Conv=NO(17 cycles 47 points) [Iter=2 Diff=0.04968] E=-3930.8279 Grad=1.061 Conv=NO(17 cycles 47 points) [Iter=3 Diff=0.00754] E=-3930.8279 Grad=1.061 Conv=NO(17 cycles 47 points) [Iter=4 Diff=0.00150] E=-3930.8279 Grad=1.061 Conv=NO(17 cycles 47 points) [Iter=5 Diff=0.00002] E=-3930.7996 Grad=2.058 Conv=NO(17 cycles 48 points) [Iter=1 Diff=0.15114] E=-3930.7996 Grad=2.058 Conv=NO(17 cycles 48 points) [Iter=2 Diff=0.01968] E=-3930.7996 Grad=2.058 Conv=NO(17 cycles 48 points) [Iter=3 Diff=0.00298] E=-3930.7996 Grad=2.058 Conv=NO(17 cycles 48 points) [Iter=4 Diff=0.00060] E=-3930.7996 Grad=2.058 Conv=NO(17 cycles 48 points) [Iter=5 Diff=0.00001] E=-3930.8440 Grad=0.360 Conv=NO(18 cycles 49 points) [Iter=1 Diff=0.00912] E=-3930.8440 Grad=0.360 Conv=NO(18 cycles 49 points) [Iter=2 Diff=0.00133] E=-3930.8440 Grad=0.360 Conv=NO(18 cycles 49 points) [Iter=3 Diff=0.00022] E=-3930.8440 Grad=0.360 Conv=NO(18 cycles 49 points) [Iter=4 Diff=0.00005] E=-3930.8372 Grad=1.580 Conv=NO(18 cycles 50 points) [Iter=1 Diff=0.00413] E=-3930.8372 Grad=1.580 Conv=NO(18 cycles 50 points) [Iter=2 Diff=0.00060] E=-3930.8372 Grad=1.580 Conv=NO(18 cycles 50 points) [Iter=3 Diff=0.00010] E=-3930.8462 Grad=0.452 Conv=NO(19 cycles 51 points) [Iter=1 Diff=0.00463] E=-3930.8462 Grad=0.452 Conv=NO(19 cycles 51 points) [Iter=2 Diff=0.00056] E=-3930.8462 Grad=0.452 Conv=NO(19 cycles 51 points) [Iter=3 Diff=0.00008] E=-3930.8523 Grad=0.372 Conv=NO(19 cycles 52 points) [Iter=1 Diff=0.00463]E=-3930.8523 Grad=0.372 Conv=NO(19 cycles 52 points) [Iter=3 Diff=0.00008] E=-3930.8562 Grad=0.471 Conv=NO(19 cycles 53 points) [Iter=1 Diff=0.01854] E=-3930.8562 Grad=0.471 Conv=NO(19 cycles 53 points) [Iter=2 Diff=0.00225] E=-3930.8562 Grad=0.471 Conv=NO(19 cycles 53 points) [Iter=3 Diff=0.00032] E=-3930.8562 Grad=0.471 Conv=NO(19 cycles 53 points) [Iter=4 Diff=0.00006] E=-3930.8567 Grad=0.916 Conv=NO(19 cycles 54 points) [Iter=1 Diff=0.00397] E=-3930.8567 Grad=0.916 Conv=NO(19 cycles 54 points) [Iter=2 Diff=0.00048] E=-3930.8567 Grad=0.916 Conv=NO(19 cycles 54 points) [Iter=3 Diff=0.00007] E=-3930.8577 Grad=0.692 Conv=NO(20 cycles 55 points) [Iter=1 Diff=0.20944] E=-3930.8577 Grad=0.692 Conv=NO(20 cycles 55 points) [Iter=2 Diff=0.02334] E=-3930.8577 Grad=0.692 Conv=NO(20 cycles 55 points) [Iter=3 Diff=0.00318] E=-3930.8577 Grad=0.692 Conv=NO(20 cycles 55 points) [Iter=4 Diff=0.00059] E=-3930.8577 Grad=0.692 Conv=NO(20 cycles 55 points) [Iter=5 Diff=0.00001] E=-3930.7786 Grad=3.886 Conv=NO(20 cycles 56 points) [Iter=1 Diff=0.14055] E=-3930.7786 Grad=3.886 Conv=NO(20 cycles 56 points) [Iter=2 Diff=0.01566] E=-3930.7786 Grad=3.886 Conv=NO(20 cycles 56 points) [Iter=3 Diff=0.00214] E=-3930.7786 Grad=3.886 Conv=NO(20 cycles 56 points) [Iter=4 Diff=0.00040] E=-3930.7786 Grad=3.886 Conv=NO(20 cycles 56 points) [Iter=5 Diff=0.00001] E=-3930.8616 Grad=0.408 Conv=NO(21 cycles 57 points) [Iter=1 Diff=0.00370] E=-3930.8616 Grad=0.408 Conv=NO(21 cycles 57 points) [Iter=2 Diff=0.00046] E=-3930.8616 Grad=0.408 Conv=NO(21 cycles 57 points) [Iter=3 Diff=0.00007] E=-3930.8633 Grad=0.323 Conv=NO(21 cycles 58 points) [Iter=1 Diff=0.00040] E=-3930.8633 Grad=0.323 Conv=NO(21 cycles 58 points) [Iter=2 Diff=0.00005] E=-3930.8638 Grad=0.167 Conv=NO(22 cycles 59 points) [Iter=1 Diff=0.00047] E=-3930.8638 Grad=0.167 Conv=NO(22 cycles 59 points) [Iter=2 Diff=0.00005] E=-3930.8640 Grad=0.335 Conv=NO(22 cycles 60 points) [Iter=1 Diff=0.00009] E=-3930.8643 Grad=0.175 Conv=NO(23 cycles 61 points) [Iter=1 Diff=0.00088] E=-3930.8643 Grad=0.175 Conv=NO(23 cycles 61 points) [Iter=2 Diff=0.00010] E=-3930.8643 Grad=0.175 Conv=NO(23 cycles 61 points) [Iter=3 Diff=0.00001] E=-3930.8647 Grad=0.288 Conv=NO(23 cycles 62 points) [Iter=1 Diff=0.00010] E=-3930.8647 Grad=0.182 Conv=NO(24 cycles 63 points) [Iter=1 Diff=0.00091] E=-3930.8647 Grad=0.182 Conv=NO(24 cycles 63 points) [Iter=2 Diff=0.00011] E=-3930.8647 Grad=0.182 Conv=NO(24 cycles 63 points) [Iter=3 Diff=0.00002] E=-3930.8652 Grad=0.280 Conv=NO(24 cycles 64 points) [Iter=1 Diff=0.00010] E=-3930.8655 Grad=0.179 Conv=NO(25 cycles 65 points) [Iter=1 Diff=0.00055] E=-3930.8655 Grad=0.179 Conv=NO(25 cycles 65 points) [Iter=2 Diff=0.00007] E=-3930.8655 Grad=0.210 Conv=NO(25 cycles 66 points) [Iter=1 Diff=0.00011] E=-3930.8655 Grad=0.210 Conv=NO(25 cycles 66 points) [Iter=2 Diff=0.00001] E=-3930.8657 Grad=0.087 Conv=YES(26 cycles 67 points) [Iter=1 Diff=0.00000]ENERGIES AND GRADIENTTotal Energy = -27282.0711491 (kcal/mol)Total Energy = -43.475879746 (a.u.)Binding Energy = -3930.8657598 (kcal/mol)Isolated Atomic Energy = -23351.2053892 (kcal/mol)Electronic Energy = -94220.9390162 (kcal/mol)Core-Core Interaction = 66938.8678671 (kcal/mol)Heat of Formation = -2555.3957598 (kcal/mol)Gradient = 0.0868837 (kcal/mol/Ang)MOLECULAR POINT GROUPC1EIGENV ALUES(eV)Symmetry: 1 A 2 A 3 A 4 A 5 A Eigenvalue: -52.653831 -37.028770 -37.023975 -29.824509 -27.272388Symmetry: 6 A 7 A 8 A 9 A 10 A Eigenvalue: -27.244719 -26.948746 -21.098186 -21.075989 -16.563450Symmetry: 11 A 12 A 13 A 14 A 15 A Eigenvalue: -16.429049 -15.312742 -15.311525 -14.240474 -14.069750Symmetry: 16 A 17 A 18 A 19 A 20 A Eigenvalue: 5.680637 7.398578 7.487293 7.505365 7.740531Symmetry: 21 A 22 A 23 A 24 A 25 A Eigenvalue: 7.752286 8.286099 8.289171 9.845991 9.896807Symmetry: 26 A 27 A 28 A 29 A 30 A Eigenvalue: 10.778082 10.850065 11.798207 11.810519 12.510362ATOMIC ORBITAL ELECTRON POPULATIONSAO: 1 S C 1 Px C 1 Py C 1 Pz C 2 S C0.981183 0.980386 1.020323 0.985765 0.981165AO: 2 Px C 2 Py C 2 Pz C 3 S C 3 Px C1.010096 1.020040 0.956455 0.981115 1.023187AO: 3 Py C 3 Pz C 4 S C 4 Px C 4 Py C0.979342 0.984885 0.981080 0.987665 1.025378AO: 4 Pz C 5 S C 5 Px C 5 Py C 5 Pz C0.975304 0.981111 1.027103 0.981545 0.978970AO: 6 S H 7 S H 8 S H 9 S H 10 S H1.011723 1.021239 1.021008 1.011737 1.012302AO: 11 S H 12 S H 13 S H 14 S H 15 S H1.018996 1.014655 1.015266 1.012561 1.018415NET CHARGES AND COORDINATESAtom Z Charge Coordinates(Angstrom) Massx y z1 6 0.032343 -1.39292 -0.40536 -0.29632 12.011002 6 0.032244 -1.34981 1.07050 -0.40720 12.011003 6 0.031471 -0.25570 -0.75893 0.58456 12.011004 6 0.030573 0.74650 0.32324 0.42738 12.011005 6 0.031272 0.08522 1.42651 -0.31184 12.011006 1 -0.011723 -1.26616 -0.87726 -1.31021 1.008007 1 -0.021239 -2.38081 -0.80365 0.06269 1.008008 1 -0.021008 -1.85072 1.46941 -1.33095 1.008009 1 -0.011737 -1.92795 1.54160 0.43564 1.0080010 1 -0.012302 -0.59748 -0.82756 1.65452 1.0080011 1 -0.018996 0.16333 -1.78099 0.37672 1.0080012 1 -0.014655 1.66579 -0.03935 -0.10959 1.0080013 1 -0.015266 1.14721 0.66647 1.42046 1.0080014 1 -0.012561 0.53113 1.53877 -1.33889 1.0080015 1 -0.018415 0.25658 2.43292 0.15858 1.00800ATOMIC GRADIENTSAtom Z Gradients(kcal/mol/Angstrom)x y z1 6 0.18896 -0.01485 -0.037312 6 -0.06992 0.15860 0.124113 6 -0.04983 -0.22042 0.046674 6 -0.23309 0.00363 -0.055545 6 -0.07703 0.10066 0.046146 1 -0.03418 -0.07446 -0.091057 1 0.04834 -0.03494 -0.000568 1 0.08235 0.00353 0.067629 1 0.01803 -0.00056 0.0062910 1 -0.02103 0.03455 0.0719511 1 -0.01904 0.16346 0.0392112 1 0.08853 0.01197 -0.0319613 1 -0.01082 -0.02264 0.0098214 1 0.07787 0.03463 -0.1507815 1 0.01086 -0.14317 -0.04460Dipole (Debyes) x y z TotalPoint-Chg. 0.032 0.003 0.010 0.034sp Hybrid 0.028 0.003 0.010 0.030pd Hybrid 0.000 0.000 0.000 0.000Sum 0.061 0.007 0.020 0.064Geometry optimization, SemiEmpirical, molecule = (untitled).CNDOFletcherReeves optimizerConvergence limit = 0.0001000 Iteration limit = 50Accelerate convergence = YESOptimization algorithm = Fletcher-ReevesCriterion of RMS gradient = 0.1000 kcal/(A mol) Maximum cycles = 225 RHF Calculation:Singlet state calculationNumber of electrons = 30Number of Double Occupied Levels = 15Charge on the System = 0Total Orbitals = 30Starting CNDO calculation with 30 orbitalsE=0.0000 Grad=0.000 Conv=NO(0 cycles 0 points) [Iter=1 Diff=9901.38267] E=0.0000 Grad=0.000 Conv=NO(0 cycles 0 points) [Iter=2 Diff=8.52345]E=0.0000 Grad=0.000 Conv=NO(0 cycles 0 points) [Iter=3 Diff=0.51968]E=0.0000 Grad=0.000 Conv=NO(0 cycles 0 points) [Iter=4 Diff=0.04223]E=0.0000 Grad=0.000 Conv=NO(0 cycles 0 points) [Iter=5 Diff=0.00150]E=0.0000 Grad=0.000 Conv=NO(0 cycles 0 points) [Iter=6 Diff=0.00007]E=-3930.8657 Grad=0.087 Conv=YES(0 cycles 1 points) [Iter=1 Diff=0.00000]ENERGIES AND GRADIENTTotal Energy = -27282.0711491 (kcal/mol) Total Energy = -43.475879746 (a.u.)Binding Energy = -3930.8657598 (kcal/mol) Isolated Atomic Energy = -23351.2053892 (kcal/mol) Electronic Energy = -94220.9363559 (kcal/mol)Core-Core Interaction = 66938.8652068 (kcal/mol)Heat of Formation = -2555.3957598 (kcal/mol) Gradient = 0.0868814 (kcal/mol/Ang)MOLECULAR POINT GROUPC1EIGENV ALUES(eV)Symmetry: 1 A 2 A 3 A 4 A 5 A Eigenvalue: -52.653828 -37.028770 -37.023975 -29.824505 -27.272387Symmetry: 6 A 7 A 8 A 9 A 10 A Eigenvalue: -27.244719 -26.948744 -21.098185 -21.075987 -16.563450Symmetry: 11 A 12 A 13 A 14 A 15 A Eigenvalue: -16.429049 -15.312742 -15.311525 -14.240474 -14.069750Symmetry: 16 A 17 A 18 A 19 A 20 A Eigenvalue: 5.680636 7.398577 7.487292 7.505364 7.740530Symmetry: 21 A 22 A 23 A 24 A 25 A Eigenvalue: 7.752286 8.286098 8.289171 9.845991 9.896806Symmetry: 26 A 27 A 28 A 29 A 30 A Eigenvalue: 10.778081 10.850063 11.798207 11.810519 12.510361ATOMIC ORBITAL ELECTRON POPULATIONSAO: 1 S C 1 Px C 1 Py C 1 Pz C 2 S C0.981183 0.980386 1.020323 0.985765 0.981165AO: 2 Px C 2 Py C 2 Pz C 3 S C 3 Px C1.010096 1.020040 0.956455 0.981115 1.023187AO: 3 Py C 3 Pz C 4 S C 4 Px C 4 Py C0.979342 0.984885 0.981080 0.987665 1.025378AO: 4 Pz C 5 S C 5 Px C 5 Py C 5 Pz C0.975304 0.981111 1.027103 0.981545 0.978970AO: 6 S H 7 S H 8 S H 9 S H 10 S H1.011723 1.021239 1.021008 1.011737 1.012302AO: 11 S H 12 S H 13 S H 14 S H 15 S H1.018996 1.014655 1.015266 1.012561 1.018415NET CHARGES AND COORDINATESAtom Z Charge Coordinates(Angstrom) Massx y z1 6 0.032343 -1.39292 -0.40536 -0.29632 12.011002 6 0.032244 -1.34981 1.07050 -0.40720 12.011003 6 0.031471 -0.25570 -0.75893 0.58456 12.011004 6 0.030573 0.74650 0.32324 0.42738 12.011005 6 0.031272 0.08522 1.42651 -0.31184 12.011006 1 -0.011723 -1.26616 -0.87726 -1.31021 1.008007 1 -0.021239 -2.38081 -0.80365 0.06269 1.008008 1 -0.021008 -1.85072 1.46941 -1.33095 1.008009 1 -0.011737 -1.92795 1.54160 0.43564 1.0080010 1 -0.012302 -0.59748 -0.82756 1.65452 1.0080011 1 -0.018996 0.16333 -1.78099 0.37672 1.0080012 1 -0.014655 1.66579 -0.03935 -0.10959 1.0080013 1 -0.015266 1.14721 0.66647 1.42046 1.0080014 1 -0.012561 0.53113 1.53877 -1.33889 1.0080015 1 -0.018415 0.25658 2.43292 0.15858 1.00800ATOMIC GRADIENTSAtom Z Gradients(kcal/mol/Angstrom)x y z1 6 0.18903 -0.01497 -0.037262 6 -0.06997 0.15884 0.124053 6 -0.04990 -0.22039 0.046734 6 -0.23292 0.00362 -0.055475 6 -0.07709 0.10066 0.046006 1 -0.03420 -0.07453 -0.091207 1 0.04821 -0.03497 -0.000488 1 0.08237 0.00350 0.067629 1 0.01799 -0.00054 0.0063110 1 -0.02104 0.03449 0.0720411 1 -0.01899 0.16335 0.0391512 1 0.08855 0.01194 -0.0319513 1 -0.01082 -0.02262 0.0098114 1 0.07792 0.03468 -0.1508315 1 0.01087 -0.14306 -0.04451Dipole (Debyes) x y z TotalPoint-Chg. 0.032 0.003 0.010 0.034sp Hybrid 0.028 0.003 0.010 0.030pd Hybrid 0.000 0.000 0.000 0.000Sum 0.061 0.007 0.020 0.064Single Point, AbInitio, molecule = (untitled).Convergence limit = 0.0001000 Iteration limit = 50Accelerate convergence = YESFull MP2 correlation energy is requested.The initial guess of the MO coefficients is from eigenvectors of the core Hamiltonian. Shell Types: S, S=P.RHF Calculation:Singlet state calculationNumber of electrons = 40Number of Doubly-Occupied Levels = 20Charge on the System = 0Total Orbitals (Basis Functions) = 65Primitive Gaussians = 105Starting HyperGauss calculation with 65 basis functions and 105 primitive Gaussians.2-electron Integral buffers will be 3200 words (double precision) long.Two electron integrals will use a cutoff of 1.00000e-010Regular integral format is used.Computing the one-electron integrals ...Computing 2e integrals (s and p orbitals only): done 0%.Computing 2e integrals (s and p orbitals only): done 10%.Computing 2e integrals (s and p orbitals only): done 20%.Computing 2e integrals (s and p orbitals only): done 30%.Computing 2e integrals (s and p orbitals only): done 40%.Computing 2e integrals (s and p orbitals only): done 50%.Computing 2e integrals (s and p orbitals only): done 60%.Computing 2e integrals (s and p orbitals only): done 70%.Computing 2e integrals (s and p orbitals only): done 80%.Computing 2e integrals (s and p orbitals only): done 90%.2173889 integrals have been produced.Computing the initial guess of the MO coefficients ...Iteration = 1 Difference = 155.0197070573Iteration = 2 Difference = 660.9952921963Iteration = 3 Difference = 0.9379147588Iteration = 4 Difference = 0.7714439436Iteration = 5 Difference = 0.4010571545Iteration = 6 Difference = 0.0020506989Iteration = 7 Difference = 0.0001176669Iteration = 8 Difference = 0.0000087172Computing MP2 energy with 20 occupied and 45 virtual orbitals ...Transfering the 2e integrals from AO to MO: done 0%.Transfering the 2e integrals from AO to MO: done 10%.Transfering the 2e integrals from AO to MO: done 20%.Transfering the 2e integrals from AO to MO: done 30%.Transfering the 2e integrals from AO to MO: done 40%.Transfering the 2e integrals from AO to MO: done 50%.Transfering the 2e integrals from AO to MO: done 60%.Transfering the 2e integrals from AO to MO: done 70%.。