Long Taq DNA Polymerase
- 格式:pdf
- 大小:216.37 KB
- 文档页数:2

竭诚为您提供优质文档/双击可除pcr反应体系中模板dna推荐使用量篇一:pcR常见问题分析pcR常见问题分析*假阳性*出现非特异性扩增现象:pcR扩增后出现的条带与预计的大小不一致,或大或小,或者有时同时出现特异性扩增带*如何进行长片段的扩增首先需选择正确的耐热聚合酶。
longtaqdnapolymerase 是具有3′-5′外切核酸酶活性的耐热dna聚合酶,扩增效率高并且错配率低,对简单模板可扩增长达1kb的片段,对复杂二级结构(gcrich等)和具有重复序列的模板可良好扩增长达15kb的片段。
此外,较长产物的扩增还需要相应调整引物设计、延伸时间、变性时间和缓冲液ph等。
引物使用标准方法设计,通常18~24mers会得到较好的产量。
按1kb/分钟的速率增加延伸时间使dna聚合酶完成聚合反应。
为了保证有效的长片段扩增,可以将延伸温度设置为68℃左右。
同时为防止模板损伤,在每个循环将94℃变性时间减少到30秒或更短,扩增前升温至94℃的时间应低于1分钟。
*如何提高pcR扩增的保真性?下游应用为基因筛选、测序、突变检测和分子诊断的用户,对pcR保真性要求很高。
降低pcR扩增的错误率可以通过使用各种具有高保真性的dna 聚合酶来实现。
dna聚合酶的保真度用错误率来表示,包括碱基错配率、pcR产物突变百分数等。
在目前已发现的所有耐高温dna聚合酶中,pfu酶的出错率最低,保真度最高(见附表)。
除对酶进行选择,研究者也可通过优化反应条件来进一步减少pcR突变率,包括优化缓冲液组成、耐热聚合酶的浓度及对pcR循环条件进行优化等。
聚合酶的错误率。
*应用/acl分析,通过对/acl目标基因进行扩增和克隆,检测pcR产物突变百分数,从而测定dna篇二:pcR相关问题q-1:设计pcR用引物时的注意事项a-1:可以从以下几个方面考虑。
1.引物长度通常为20~25mer。
但进行lapcR时,引物长度应增长为30~35mer。


qiagen pcr各个温度设定原理Qiagen PCR是一种用于聚合酶链式反应(Polymerase Chain Reaction,PCR)的设备和试剂盒。
在PCR过程中,温度的设定对反应的成功与否起着至关重要的作用。
本文将从不同温度的设定原理来介绍Qiagen PCR的工作原理。
PCR反应需要进行三个不同温度的循环:变性、退火和延伸。
这三个温度的设定是由PCR仪器自动控制的,确保反应的进行顺利。
第一个温度是变性温度,通常设定为94-98°C。
在这个温度下,PCR反应管中的DNA双链结构会被解开,使得两个DNA链分离。
这个步骤是为了使DNA的两个链能够与引物结合,并为后续的延伸反应提供单链的DNA模板。
第二个温度是退火温度,通常设定为50-65°C。
在这个温度下,引物与DNA单链模板结合,形成引物-模板复合物。
引物是PCR反应中的两个短链DNA片段,能够特异性地与待扩增的DNA序列结合。
退火温度的设定要根据引物的序列特性来确定,以确保引物能够与目标序列特异性结合,而不与其他非特异性序列结合。
第三个温度是延伸温度,通常设定为72°C。
在这个温度下,DNA 聚合酶(Taq DNA Polymerase)会结合在引物-模板复合物上,并通过加入新的核苷酸将DNA链延伸。
Taq DNA Polymerase是一种热稳定的DNA聚合酶,可以在高温下工作,因此延伸温度通常设定为较高的温度。
通过这样的三个温度循环,反复进行变性、退火和延伸,可以使待扩增的DNA序列不断复制,最终产生大量的目标DNA。
PCR反应一般会进行30-40个循环,以确保足够的扩增产物。
除了这三个基本的温度设定外,Qiagen PCR还可以根据实验需要进行一些特殊的温度控制。
例如,在某些特定的PCR反应中,可能需要设定一个较低的退火温度,以增加引物与模板的结合特异性。
此外,PCR反应中的温度控制还可以根据反应体系的组成来进行调整,以优化反应的效果。
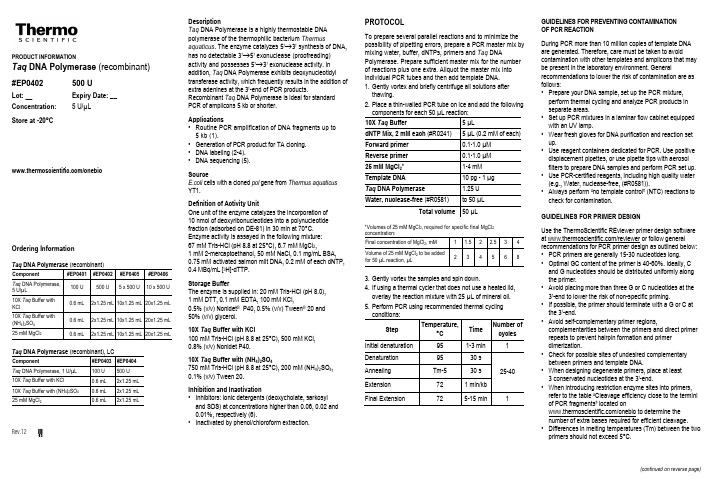
PRODUCT INFORMATIONTaq DNA Polymerase (recombinant)#EP0402 500 ULot: __ Expiry Date: __ Concentration: 5 U/µLStore at -20°C/onebioOrdering InformationTaq DNA Polymerase (recombinant)Component #EP0401 #EP0402 #EP0405 #EP0406 Taq DNA Polymerase,5 U/µL100 U 500 U 5 x 500 U 10 x 500 U 10X Taq Buffer withKCl0.6 mL 2x1.25 mL 10x1.25 mL 20x1.25 mL 10X Taq Buffer with(NH4)2SO40.6 mL 2x1.25 mL 10x1.25 mL 20x1.25 mL 25 mM MgCl2 0.6 mL 2x1.25 mL 10x1.25 mL 20x1.25 mL Taq DNA Polymerase (recombinant), LCComponent #EP0403 #EP0404Taq DNA Polymerase, 1 U/µL 100 U 500 U10X Taq Buffer with KCl 0.6 mL 2x1.25 mL10X Taq Buffer with (NH4)2SO4 0.6 mL 2x1.25 mL25 mM MgCl2 0.6 mL 2x1.25 mL Rev.12 V DescriptionTaq DNA Polymerase is a highly thermostable DNApolymerase of the thermophilic bacterium Thermusaquaticus. The enzyme catalyzes 5’→3’ synthesis of DNA,has no detectable 3’→5’ exonuclease (proofreading)activity and possesses 5’→3’ exonuclease activity. Inaddition, Taq DNA Polymerase exhibits deoxynucleotidyltransferase activity, which frequently results in the addition ofextra adenines at the 3’-end of PCR products.Recombinant Taq DNA Polymerase is ideal for standardPCR of amplicons 5 kb or shorter.Applications•Routine PCR amplification of DNA fragments up to5 kb (1).•Generation of PCR product for TA cloning.•DNA labeling (2-4).•DNA sequencing (5).SourceE.coli cells with a cloned pol gene from Thermus aquaticusYT1.Definition of Activity UnitOne unit of the enzyme catalyzes the incorporation of10 nmol of deoxyribonucleotides into a polynucleotidefraction (adsorbed on DE-81) in 30 min at 70°C.Enzyme activity is assayed in the following mixture:67 mM Tris-HCl (pH 8.8 at 25°C), 6.7 mM MgCl2,1 mM 2-mercaptoethanol, 50 mM NaCl, 0.1 mg/mL BSA,0.75 mM activated salmon milt DNA, 0.2 mM of each dNTP,0.4 MBq/mL [3H]-dTTP.Storage BufferThe enzyme is supplied in: 20 mM Tris-HCl (pH 8.0),1 mM DTT, 0.1 mM EDTA, 100 mM KCl,0.5% (v/v) Nonidet® P40, 0.5% (v/v) Tween® 20 and50% (v/v) glycerol.10X Taq Buffer with KCl100 mM Tris-HCl (pH 8.8 at 25°C), 500 mM KCl,0.8% (v/v) Nonidet P40.10X Taq Buffer with (NH4)2SO4750 mM Tris-HCl (pH 8.8 at 25°C), 200 mM (NH4)2SO4,0.1% (v/v) Tween 20.Inhibition and Inactivation•Inhibitors: ionic detergents (deoxycholate, sarkosyland SDS) at concentrations higher than 0.06, 0.02 and0.01%, respectively (6).•Inactivated by phenol/chloroform extraction.PROTOCOLTo prepare several parallel reactions and to minimize thepossibility of pipetting errors, prepare a PCR master mix bymixing water, buffer, dNTPs, primers and Taq DNAPolymerase. Prepare sufficient master mix for the numberof reactions plus one extra. Aliquot the master mix intoindividual PCR tubes and then add template DNA.1.Gently vortex and briefly centrifuge all solutions afterthawing.2.Place a thin-walled PCR tube on ice and add the followingcomponents for each 50 µL reaction:10X Taq Buffer 5 µLdNTP Mix, 2 mM each (#R0241) 5 µL (0.2 mM of each)Forward primer 0.1-1.0 µMReverse primer 0.1-1.0 µM25 mM MgCl2* 1-4 mMTemplate DNA 10 pg - 1 µgTaq DNA Polymerase 1.25 UWater, nuclease-free (#R0581) to 50 µLTotal volume 50 µL*Volumes of 25 mM MgCl2, required for specific final MgCl2concentration:Final concentration of MgCl2, mM 1 1.5 2 2.5 3 4Volume of 25 mM MgCl2 to be addedfor 50 µL reaction, µL2 3 4 5 6 83.Gently vortex the samples and spin down.4.If using a thermal cycler that does not use a heated lid,overlay the reaction mixture with 25 µL of mineral oil.5.Perform PCR using recommended thermal cyclingconditions:StepTemperature,°CTimeNumber ofcyclesInitial denaturation 95 1-3 min 1Denaturation 95 30 s25-40Annealing Tm-5 30 sExtension72 1 min/kbFinal Extension 72 5-15 min 1 GUIDELINES FOR PREVENTING CONTAMINATIONOF PCR REACTIONDuring PCR more than 10 million copies of template DNA are generated. Therefore, care must be taken to avoid contamination with other templates and amplicons that may be present in the laboratory environment. General recommendations to lower the risk of contamination are as follows:•Prepare your DNA sample, set up the PCR mixture, perform thermal cycling and analyze PCR products in separate areas.•Set up PCR mixtures in a laminar flow cabinet equipped with an UV lamp.•Wear fresh gloves for DNA purification and reaction set up.•Use reagent containers dedicated for PCR. Use positive displacement pipettes, or use pipette tips with aerosol filters to prepare DNA samples and perform PCR set up. •Use PCR-certified reagents, including high quality water (e.g., Water, nuclease-free, (#R0581)).•Always perform “no template control” (NTC) reactions to check for contamination.GUIDELINES FOR PRIMER DESIGNUse the ThermoScientific REviewer primer design software at /reviewer or follow general recommendations for PCR primer design as outlined below: •PCR primers are generally 15-30 nucleotides long. •Optimal GC content of the primer is 40-60%. Ideally, C and G nucleotides should be distributed uniformly along the primer.•Avoid placing more than three G or C nucleotides at the 3’-end to lower the risk of non-specific priming.•If possible, the primer should terminate with a G or C at the 3’-end.•Avoid self-complementary primer regions, complementarities between the primers and direct primer repeats to prevent hairpin formation and primer dimerization.•Check for possible sites of undesired complementary between primers and template DNA.•When designing degenerate primers, place at least3 conservated nucleotides at the 3’-end.•When introducing restriction enzyme sites into primers, refer to the table “Cleavage efficiency close to the termini of PCR fragments” located on/onebio to determine the number of extra bases required for efficient cleavage. •Differences in melting temperatures (Tm) between the two primers should not exceed 5°C.(continued on reverse page)Estimation of primer melting temperatureFor primers containing less than 25 nucleotides, the approx. melting temperature (Tm) can be calculated using the following equation:Tm= 4 (G + C) + 2 (A + T),where G, C, A, T represent the number of respective nucleotides in the primer.If the primer contains more than 25 nucleotides specialized computer programs e.g., REviewer™ (/reviewer), are recommended to account for interactions of adjacent bases, effect of salt concentration, etc.COMPONENTS OF THE REACTION MIXTURE Template DNAOptimal amounts of template DNA in the 50 µL reaction volume are 0.01-1 ng for both plasmid and phage DNA, and 0.1-1 µg for genomic DNA. Higher amount of template increases the risk of generation of non-specific PCR products. Lower amount of template reduces the accuracy of the amplification.All routine DNA purification methods are suitable for template preparation e.g., Thermo Scientific GeneJET Genomic DNA Purification Kit (#K0721) or GeneJET™ Plasmid Miniprep Kit (#K0502). Trace amounts of certain agents used for DNA purification, such as phenol, EDTA and proteinase K, can inhibit DNA polymerases. Ethanol precipitation and repeated washes of the DNA pellet with 70% ethanol normally removes trace contaminants from DNA samples.MgCl2 concentrationDue to the binding of Mg2+ to dNTPs, primers and DNA templates, Mg2+ concentration needs to be optimized for maximal PCR yield. The recommended concentration range is 1-4 mM. If the Mg2+ concentration is too low, the yield of PCR product could be reduced. On the contrary, non-specific PCR products may appear and the PCR fidelity may be reduced if the Mg2+ concentration is too high.If the DNA samples contain EDTA or other metal chelators, the Mg2+ ion concentration in the PCR mixture should be increased accordingly (1 molecule of EDTA binds one Mg2+). dNTPsThe recommended final concentration of each dNTP is0.2 mM. In certain PCR applications, higher dNTP concentrations may be necessary. Due to the binding ofMg2+ to dNTPs, the MgCl2 concentration needs to be adjusted accordingly. It is essential to have equal concentrations of all four nucleotides (dATP, dCTP, dGTP and dTTP) present in the reaction mixture. To achieve 0.2 mM concentration of each dNTP in the PCRmixture, use the following volumes of dNTP mixes:Volume ofPCR mixturedNTP Mix,2 mM each(#R0241)dNTP Mix,10 mM each(#R0191)dNTP Mix,25 mM each(#R1121)50 µL 5 µL 1 µL 0.4 µL25 µL 2.5 µL 0.5 µL 0.2 µL20 µL 2 µL 0.4 µL 0.16 µLPrimersThe recommended concentration range of the PCR primersis 0.1-1 µM. Excessive primer concentrations increase theprobability of mispriming and generation of non-specific PCRproducts.For degenerate primers higher primer concentrations in therange of 0.3-1 µM are often favorable.CYCLING PARAMETERSInitial DNA denaturationIt is essential to completely denature the template DNA atthe beginning of PCR to ensure efficient utilization of thetemplate during the first amplification cycle. If the GC contentof the template is 50% or less, an initial 1-3 min denaturationat 95°C is sufficient. For GC-rich templates this step shouldbe prolonged up to 10 min. If longer initial denaturation stepis required, or the DNA is denatured at a higher temperature,Taq DNA Polymerase should be added after the initialdenaturation step to avoid a decrease in its activity.DenaturationA DNA denaturation time of 30 seconds per cycle at 95°Cis normally sufficient. For GC-rich DNA templates, this stepcan be prolonged to 3-4 min. DNA denaturation can also beenhanced by the addition of either 10-15% glycerol or10% DMSO, 5% formamide or 1-1.5 M betaine. The meltingtemperature of the primer-template complex decreasessignificantly in the presence of these reagents. Therefore,the annealing temperature has to be adjusted accordingly.In addition, 10% DMSO and 5% formamide inhibit DNApolymerases by 50%. Thus, the amount of the enzymeshould be increased if these additives are used.Primer annealingThe annealing temperature should be 5°C lower than themelting temperature (Tm) of the primers. Annealing for30 seconds is normally sufficient. If non-specific PCRproducts appear, the annealing temperature should beoptimized stepwise in 1-2°C increments. When additiveswhich change the melting temperature of the primer-templatecomplex are used (glycerol, DMSO, formamide and betaine),the annealing temperature must also be adjusted.ExtensionThe optimal extension temperature for Taq DNA Polymeraseis 70-75°C. The recommended extension step is 1 min at72°C for PCR products up to 2 kb. For larger products, theextension time should be prolonged by 1 min/kb.Number of cyclesThe number of cycles may vary depending on the amount oftemplate DNA in the PCR mixture and the expected PCRproduct yield.If less than 10 copies of the template are present in thereaction, about 40 cycles are required. For higher templateamounts, 25-35 cycles are sufficient.Final extensionAfter the last cycle, it is recommended to incubate the PCRmixture at 72°C for additional 5-15 min to fill-in any possibleincomplete reaction products. If the PCR product will becloned into TA vectors (for instance, using Thermo ScientificInsTAclone PCR Cloning Kit (#K1213)), the final extensionstep may be prolonged to 30 min to ensure the highestefficiency of 3’-dA tailing of PCR product. If the PCR productwill be used for cloning using Thermo Scientific CloneJETPCR Cloning Kit (#K1231), the final extension step can beomitted.TroubleshootingFor troubleshooting please visit/onebioReferences1. Innis, M.A., et al., PCR Protocols and Applications: A LaboratoryManual, Academic, New York, 1989.2. Celeda, D., et al., PCR amplification and simultaneous digoxigeninincorporation of long DNA probes for fluorescence in situ hybridization,BioTechniques, 12, 98-102, 1992.3. Finckh, U., et al., Producing single-stranded DNA probes with theTaq DNA polymerase: a high yield protocol, BioTechniques, 10, 35-39,1991.4. Yu, H. et al., Cyanine dye dUTP analogs for enzymatic labeling ofDNA probes, Nucleic Acids Res., 22, 3226-3232, 1994.5. Innis, M.A., et al., DNA sequencing with Thermus aquaticus DNApolymerase and direct sequencing of polymerase chain reaction-amplified DNA, Proc. Natl. Acad. Sci. USA, 85, 9436-9440, 1988.6. Weyant, R.S., et al., Effect of ionic and nonionic detergents on theTaq polymerase, Biotechniques, 9, 309-308, 1990.7. Lundberg, K.S., et al., High-fidelity amplification using a thermostableDNA polymerase isolated from Pyrococcus furiosus, Gene, 108, 1-6,1991.CERTIFICATE OF ANALYSISEndodeoxyribonuclease AssayNo conversion of covalently closed circular DNA to nickedDNA was detected after incubation of 10 U of Taq DNAPolymerase with 1 µg of pUC19 DNA for 4 hours at 37°C.Exodeoxyribonuclease AssayNo degradation of DNA was observed after incubation of1 µg of lambda DNA/HindIII fragments with 10 U Taq DNAPolymerase for 4 hours at 37°C.Ribonuclease AssayNo contaminating RNase activity was detected afterincubation of 10 U of Taq DNA Polymerase with 1 µg of[3H]-RNA for 4 hours at 37°C.Functional AssayTaq DNA Polymerase was tested for amplification of 950 bpsingle copy gene from human genomic DNA and foramplification of cDNA.Quality authorized by: Jurgita ZilinskieneNOTICE TO PURCHASER:Use of this product is covered by US Patent No. 6,127,155. The purchaseof this product includes a limited, non-transferable immunity from suitunder the foregoing patent claims for using only this amount of product forthe purchaser’s own internal research. No right under any other patentclaim, no right to perform any patented method and no right to performcommercial services of any kind, including without limitation reporting theresults of purchaser's activities for a fee or other commercialconsideration, is conveyed expressly, by implication, or by estoppel. Thisproduct is for research use only. Diagnostic uses under Roche patentsrequire a separate license from Roche. Further information on purchasinglicenses may be obtained by contacting the Director of Licensing, AppliedBiosystems, 850 Lincoln Centre Drive, Foster City, California.PRODUCT USE LIMITATIONThis product is developed, designed and sold exclusively for researchpurposes and in vitro use only. The product was not tested for use indiagnostics or for drug development, nor is it suitable for administration tohumans or animals. Please refer to /onebio forMaterial Safety Data Sheet of the product.© 2012 Thermo Fisher Scientific, Inc. All rights reserved. Tween is aregistered trademark of ICI America, Inc. Nonidet is a trademark of Shell.All other trademarks are the property of Thermo Fisher Scientific Inc. and itssubsidiaries.。

热启动酶的原理及应用前言在现代生物科学研究中,热启动酶被广泛应用于DNA特异性放大技术,例如聚合酶链式反应(PCR)。
热启动酶是一种酶类蛋白,能够在高温条件下保持活性,具有特异性结合和扩增DNA的能力。
本文将介绍热启动酶的原理及其在科学研究和生物工程中的应用。
1. 热启动酶的原理热启动酶的原理基于一种天然存在的酶类蛋白,称为热稳定DNA多聚核苷酸激酶(Taq DNA polymerase)。
Taq DNA polymerase是从热温泉中的细菌Thermus aquaticus中分离得到的。
这种细菌栖息在温泉中,能够耐受高温环境。
因此,Taq DNA polymerase具备了在高温条件下保持稳定和具有DNA聚合能力的特点。
热启动酶通过添加特定的抑制剂和改变酶的活性形式,使其在低温下失去或降低DNA聚合酶活性,从而避免在PCR反应体系中的非特异性扩增。
当温度升高时,抑制剂被去除,酶活性被迅速激活,从而实现特异性DNA扩增。
2. 热启动酶的应用热启动酶作为PCR技术中常用的酶类蛋白,其应用非常广泛。
以下列举了一些热启动酶的主要应用领域:•分子生物学研究:热启动酶可用于DNA扩增、基因克隆、DNA测序等领域。
它的高温稳定性保证了PCR反应在高温环境下能够顺利进行,同时能够在DNA复制过程中起到高效率和高特异性的作用。
•医学诊断:PCR技术结合热启动酶的应用在医学诊断领域取得了重大突破。
例如,热启动酶可以检测疾病相关基因的突变,帮助诊断遗传性疾病、癌症等疾病。
•遗传学研究:热启动酶可以用于DNA指纹技术,通过扩增DNA重复序列,快速鉴定个体之间的遗传关系,对犯罪侦破、亲子鉴定等领域非常重要。
•生物工程:热启动酶在基因工程中常被用来扩增外源基因,从而用于遗传转化、重组蛋白表达等目的。
它的高特异性和高效率使得基因工程实验更加准确可靠。
•食品安全检测:热启动酶可以用于食品中病原菌和污染物的检测与鉴定,如大肠杆菌、沙门菌等。

Taq DNA Polymerase Taq DNA Polymerase 是耐热的DNA 聚合酶,具有5’-3’ DNA 聚合酶活性和双链DNA 特异的5’-3’外切核酸酶活性,无3’-5’外切酶活性。
使用该产品扩增得到的PCR 产物的3’末端附有一个“A ”碱基,可直接用于TA 克隆。
■产品说明PCR 、TA 克隆。
■应用范围用活性化的大马哈鱼精子DNA 作为模板/引物,在74℃,30分钟内,将10 nmol 脱氧核苷酸摄入为酸不溶物质所需要的酶量定义为1个活性单位(U )。
■活性定义无核酸内切、外切酶活性,也无核酸污染,其酶纯度检测大于99%。
■品质保证产品内容Taq DNA Polymerase (5U/µl )2+10×Taq Reaction Buffer (Mg plus )10 mM dNTP 保存条件:-20 ℃. E coli 重组蛋白。
■来源20mM Tris-Cl (pH 8.0), 100mM KCl, 0.1mM EDTA, 1mM DTT, 0.5%Nonidet P-40, 0.5%Tween-20和50% 甘油。
■储存缓冲液10×Taq Reaction Buffer :100mM Tris-Cl(pH8.3@25℃), 500mM KCl, 15mM MgCl 。
2■反应缓冲液GeneCopoeia Inc.19520 Amaranth DriveGermantown, Maryland 20874USATel: 301-515-6982; 1-866-360-9531Fax: 301-515-6983Web: 产品套装编号: C0101A , 本套装包含以下产品:产品编号C01010A C01011A C10010C包装规格200µl1.25ml ×20.5mlTaq DNA 聚合酶生产经销商:能基因广州复能基因有限公司广州高新技术产业开发区广州科学城掬泉路3号广州国际企业孵化器D 区8楼 (邮编: 510663)技术热线: 020-******** 电子邮箱: support @ 网址:■基本反应条件体系:1、PCR 10× Reaction Buffer Primer1Primer210mM dNTP TemplateTaq DNA Polymerase ddH O22.5µl0.5µl1~100ng(质粒)10~1,000ng(基因组)0.2µl up to 25µl反应物组成体积终浓度l ×0.2~1µM 0.2~1µM0.2mM1U /Reaction条件:2、PCR 94℃94℃Tm -5℃72℃72℃4℃2 min 30 sec 30 sec 1kb/min 7 min hold30 cycles}1. PCR 反应条件应根据模板、目的片段大小、引物结构等具体条件不同,设定最佳反应条件。
2×LongTaq PCR StarMix with Loading Dye2×含染料长片段预混PCR 反应体系【产品概述】本产品为预混的含有优化浓度的GenStar ®高纯度LongTaq DNA Polymerase 、dNTPs 、Mg 2+、反应缓冲液和稳定剂等成分的即用型2倍浓度的PCR 溶液,对10 kb 以上的超长DNA 片段仍然具备较强的扩增能力,可扩增30 kb 以λ DNA 为模板的片段或15 kb 以基因组DNA 为模板的片段,故适用于构建基因图谱。
本产品有含染料(红色)和不含染料(无色)两种选择。
使用含染料的产品在PCR 反应完成后,不需添加上样缓冲液即可直接上样进行电泳;也可经过纯化处理,以用于酶切、连接、荧光测序等后续操作。
红色染料可指示电泳进程,其迁移速度在1% TAE 琼脂糖凝胶中与300 bp 双链DNA 片段相近。
【产品组分】2×LongTaq PCR StarMix 0.5 mlddH 2O 0.5 ml【保存条件】-20℃恒温保存一年,避免反复冻融。
经常使用,可置于4℃保存至少三个月。
【使用方法】用户需自备的试剂:DNA 模板、引物 操作示例:以50 μl 反应体系为例 1. PCR 反应体系的建立:2. PCR 反应条件的设置: DNA 模板*1 μl 正向引物 (10 μM) 1 μl 反向引物 (10 μM) 1 μl 2×LongTaq PCR StarMix 25 µlddH 2O22 μl94℃ 2 min 94℃ 30 sec55~65℃30 sec 25~35循环 72℃ 1 min/1 kb 72℃5 min*模板量:10~1000 ng 基因组DNA ,1~30 ng 质粒,或1~2 μl RT-PCR 反应后的cDNA 。
注意:以上举例为常规PCR 反应系统,仅供参考。
实际反应条件因模板、引物等的结构不同而各异,需根据模板、引物、目的片段的特点设定最佳反应条件,并根据比例放大或缩小反应体系。
Taq DNA Polymerase( Buffer+ )使用说明书储存:-20 ℃保存。
浓度:5U/µl制品说明:Taq DNA Polymerase 是从克隆有Thermu aquaticus DNA Polymerase 基因的大肠杆菌经诱导表后分离纯化的,其分子量为94 KD。
Taq DNA Polymerase具有5′-3′DNA聚合酶活性和5′-3′外切核酸酶活性,无3′-5′外切酶活性。
在PCR反应中,Taq DNA Polymerase 延伸速度为1-2kb/分钟,产物3′端带A,可直接用于T/A载体克隆。
活性单位:1 单位(U)Taq DNA Polymerase 活性定义为在74℃、30分钟内,以活性化的大马哈鱼精子DNA 作为模板引物,将10 nmol脱氧核苷酸掺入到酸不溶物质所需的酶量。
质量控制:SDS-PAGE 检测纯度大于99%,经检测无外源核酸酶活性;PCR方法检测无宿主残余DNA,能有效地扩增人基因组中的单拷贝基因;室温存放一周,无明显活性改变。
酶贮存缓冲液:20mM Tris-HCl (pH8.0), 0.1 mM EDTA, 1mM DTT, 100 mM KCl, Stabilizers, 50% glycerol。
10×Taq Buffer (含Mg2+):200 mM Tris-HCl (pH 8.4), 200 mM KCl, 100 mM(NH4)2SO4,15 mM MgCl2,其他成分。
★ 10×Taq Buffer 分为含Mg2+和不含Mg2+ 两种,可自选。
★不含Mg2+ 的Buffer,另外配有25 mM MgCl2。
★如果没有特别指定,通常提供的为含有Mg2+的Buffer。
适用范围:一般用于DNA片断的PCR扩增、DNA标记、引物延伸、序列测定、平末端加A 等,产物可以直接用于T/A载体克隆。
建议的PCR 条件: (以 50 μl 反应体系为例)Template <0.5 μgForward Primer (10 μM) 1 μlReverse Primer (10 μM) 1 μl10×Buffer+ (with mgcl2) 5 μldNTP Mixture(各2.5mM) 4 μlTaq DNA polymerase(5U/μl) 0.5~1 μldH2O up to 50 μlPCR 反应循环的设置:94o C: 2-5 min94o C: 30 sec50-60o C: 30 sec30 cycles72o C: 1 min/1-2 kb72o C: 5-10 minNOT FOR HUMAN OR DRUG USE完。
厦门大学学位论文原创性声明本人呈交的学位论文是本人在导师指导下,独立完成的研究成果。
本人在论文写作中参考其他个人或集体已经发表的研究成果,均在文中以适当方式明确标明,并符合法律规范和《厦门大学研究生学另夕卜,该学位论文为燃跫转徽,获得()课题术活动规范(试行)》。
(组)经费或实验室的资助,在()实验室完成。
(请在以上括号内填写课题或课题组负责人或实验室名称,未有此项声明内容的,可以不作特别声明。
)声明人(签名):唧锺列£f年f月叼日1.2TaqDNA聚合酶的基本性质与结构TaqDNA聚合酶属于DNA聚合酶I家族,是一种耐热的DNA聚合酶。
该酶的编码基因全长有2496个碱基,编码832个氨基酸,酶蛋白的分子量为94kD。
酶比活性为200000U/mg。
该酶的热稳定性很高,在95。
C时酶活性半衰期为1.5h;在700C的高温下反应2h后,还能保持90%以上的活性;在75。
C.80℃时活性最高,延伸效率约为每秒150个核苷酸。
TaqDNA聚合酶全酶共分为三个结构域,如图1.1所示。
a_inl23图1-1[8I:TaqDNA聚合酶的结构域图。
Fig.1-1:DomainsofTaqDNApolymerase.DNA聚合酶蛋白多肽链N端1-291个氨基酸构成一个结构域,该结构Taq域表达5’.3’核酸外切酶活性,可以水解单链或双链DNA分子5’端的单核苷酸,释放出寡核苷酸。
该结构域也是二价金属离子的结合位点。
蛋白质肽链C端的第424.832个氨基酸构成另一个结构域,该结构域表达5’.3’聚合酶活性。
图1.2是TaqDNA聚合酶的聚合酶活性区域与DNA结合时的晶体结构图[9】。
此结构域被形象的比作人类的右手,可分为手掌区域、拇指区域及手指区域。
其中,手掌区域是酶的催化位点,可催化磷酰基转移。
引物末端的OH对插入的dNTP的0【磷酰基进行亲核攻击,三羧酸盐侧链结合的金属离子(如M92+)可促进引物末端的去质子化,使dNTP的u磷酰基处形成过渡状态。