WEKA完整中文教程
- 格式:pdf
- 大小:2.82 MB
- 文档页数:52
Weka--ARFF数据格式介绍和转换简介Weka 是⼀个由Java编写的开源免费的数据挖掘⼯具,全称怀卡托智能分析环境(Waikato Environment for Knowledge Analysis),它是基于JAVA环境下开源的机器学习(machine learning)以及数据挖掘(data mining)的软件,运⾏该⼯具需要安装Java环境。
Weka同时提供了命令⾏和GUI两种使⽤⽅式⽅式,前者效率更⾼,后者使⽤更简单。
软件安装1. 下载并安装Java环境参见教程:2.安装WekaWindows上下载.exe安装后直接双击运⾏安装官⽅⽹址:数据集介绍在Weka中,⼀个数据集由 weka.core.Instances 实现。
数据集中每个样例是由weka.core.Instance实现。
每个样例由多个属性组成,其中简单的属性类型见表1。
表1:Weka数据集的简单属性属性类型描述样例列表型(nominal)⼀组值得预定义列表{1,2,3}, {good, bad}数值型(numeric)⼀个实数或者整数12, 2.3, 50字符串(string)⼀个任意长的字符序列,包含在双引号内"better", "worse"除了简单属性,Weka还有附加类型的属性date和relational,将会在之后介绍。
Weka的数据集存储在ARFF格式的⽂件中,下⾯是⼀个ARFF⽂件的格式说明:% This is a toy example, the UCI weather dataset.% Any relation to real weather is purely coincidental.@relation golfWeatherMichigan_1988/02/10_14days@attribute outlook {sunny, overcast, rainy}@attribute windy {TRUE, FALSE}@attribute temperature real@attribute humidity real@attribute play {yes, no}@datasunny,FALSE,85,85,nosunny,TRUE,80,90,noovercast,FALSE,83,86,yesrainy,FALSE,70,96,yesrainy,FALSE,68,80,yes以%开头的两⾏是注释,主要介绍该数据集的来源,内容和意义等;@relation 是该数据集的关系名称;@attribute 是该数据集每个实例的属性说明,上例中共有5个属性,其中3个列表型属性,2个数值型属性,没有字符串型属性;@data 下⾯⾏就是数据集内容,每⾏代表⼀个实例,每个实例由5个之前定义过的属性。

数据挖掘-WEKA 实验报告一一、实验内容1、Weka 工具初步认识(掌握weka程序运行环境)2、实验数据预处理。
(掌握weka中数据预处理的使用)对weka自带测试用例数据集weather.nominal.arrf文件,进行一下操作。
1)、加载数据,熟悉各按钮的功能。
2)、熟悉各过滤器的功能,使用过滤器Remove、Add对数据集进行操作。
3)、使用weka.unsupervised.instance.RemoveWithValue 过滤器去除humidity属性值为high的全部实例。
4)、使用离散化技术对数据集glass.arrf中的属性RI和Ba 进行离散化(分别用等宽,等频进行离散化)。
(1)打开已经安装好的weka,界面如下,点击openfile即可打开weka自带测试用例数据集weather.nominal.arrf文件(2)打开文件之后界面如下:(3)可对数据进行选择,可以全选,不选,反选等,还可以链接数据库,对数据进行编辑,保存等。
还可以对所有的属性进行可视化。
如下图:(4)使用过滤器Remove、Add对数据集进行操作。
(5)点击此处可以增加属性。
如上图,增加了一个未命名的属性unnamed.再点击下方的remove按钮即可删除该属性.(5)使用weka.unsupervised.instance.RemoveWithValue过滤器去除humidity属性值为high的全部实例。
没有去掉之前:(6)去掉其中一个属性之后:(7)选择choose里的removewithvalue:(8)选择huminity属性:(9)使用离散化技术对数据集glass.arrf中的属性RI和Ba进行离散化(分别用等宽,等频进行离散化)。
RI等宽:(10)Ba等频:二、思考与分析.1.使用数据集编辑器打开weather.nominal.arrf文件,实例编号为2的分类属性值是多少?如图所示:实例编号为2的分类值属性为no加载weather.nomina.arrf文件后,temperature属性可以有哪些合法值?Temperature可以取值为:hot、mild、coolWord 资料。

Weka总结引言Weka是一个免费、开源的数据挖掘和机器学习软件,于1997年首次发布。
它由新西兰怀卡托大学的机器学习小组开发,提供了一系列数据预处理、分类、回归、聚类和关联规则挖掘等功能。
本文将对Weka进行总结,并讨论其主要功能和优点。
主要功能1. 数据预处理Weka提供了各种数据预处理技术,用于数据的清洗、转换和集成。
最常用的预处理技术包括缺失值处理、离散化、属性选择和特征缩放等。
通过这些预处理技术,用户可以减少数据中的噪声和冗余信息,提高机器学习模型的性能。
2. 分类Weka支持多种分类算法,包括决策树、贝叶斯分类器、神经网络和支持向量机等。
用户可以根据自己的需求选择适当的算法进行分类任务。
Weka还提供了交叉验证和自动参数调整等功能,帮助用户评估和优化分类器的性能。
3. 回归除了分类,Weka还支持回归问题的解决。
用户可以使用线性回归、多项式回归和局部回归等算法,对给定的数据集进行回归分析。
Weka提供了模型评估和可视化工具,帮助用户理解回归模型和评估其预测性能。
4. 聚类Weka的聚类算法可用于将数据集中相似的样本归类到一起。
Weka支持K-means、DBSCAN、谱聚类和层次聚类等常用的聚类算法。
用户可以根据数据的特点选择适当的算法并解释聚类结果。
5. 关联规则挖掘关联规则挖掘是一种常见的数据挖掘任务,用于发现数据集中的频繁项集和关联规则。
通过Weka,用户可以使用Apriori和FP-growth等算法来挖掘数据中的关联规则。
Weka还提供了支持多种评估指标的工具,用于评估关联规则的质量和可信度。
优点1. 易于使用Weka的用户界面友好且易于使用。
它提供了直观的图形界面,使用户可以快速上手并进行各种数据挖掘任务。
此外,Weka还支持命令行操作,方便用户在脚本中使用和集成Weka的功能。
2. 强大的功能Weka提供了丰富的数据挖掘和机器学习功能,涵盖了数据预处理、分类、回归、聚类和关联规则挖掘等领域。

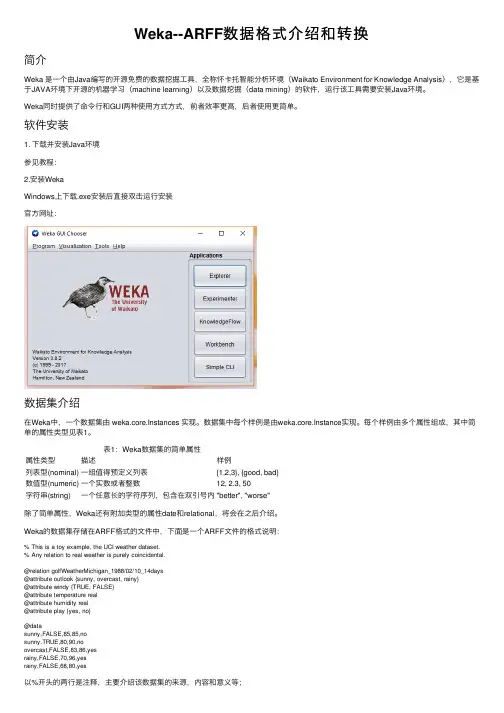
Weka数据挖掘软件使用指南Weka 数据挖掘软件使用指南1. Weka简介该软件是WEKA的全名是怀卡托智能分析环境(Waikato Environment for Knowledge Analysis),它的源代码可通过得到。
Weka作为一个公开的数据挖掘工作平台,集合了大量能承担数据挖掘任务的机器学习算法,包括对数据进行预处理,分类,回归、聚类、关联规则以及在新的交互式界面上的可视化。
如果想自己实现数据挖掘算法的话,可以看一看Weka的接口文档。
在Weka中集成自己的算法甚至借鉴它的方法自己实现可视化工具并不是件很困难的事情。
2. Weka启动打开Weka主界面后会出现一个对话框,如图:主要使用右方的四个模块,说明如下:Explorer使用Weka探索数据的环境,包括获取关联项,分类预测,聚簇等;(本文主要总结这个部分的使用)Experimenter运行算法试验、管理算法方案之间的统计检验的环境;KnowledgeFlow这个环境本质上和Explorer所支持的功能是一样的,但是它有一个可以拖放的界面。
它有一个优势,就是支持增量学习;SimpleCLI提供了一个简单的命令行界面,从而可以在没有自带命令行的操作系统中直接执行Weka命令;(某些情况下使用命令行功能更好一些)3.主要操作说明点击进入Explorer模块开始数据探索环境:3.1主界面进入Explorer模式后的主界面如下:3.1.1标签栏主界面最左上角(标题栏下方)的是标签栏,分为五个部分,功能依次是:1. Preprocess. 选择和修改要处理的数据;2. Classify. 训练和测试关于分类或回归的学习方案;3. Cluster. 从数据中学习聚类;4. Associate.从数据中学习关联规则;5. Select attributes. 选择数据中最相关的属性;6. Visualize.查看数据的交互式二维图像。
3.1.2载入、编辑数据标签栏下方是载入数据栏,功能如下:1.Open file.打开一个对话框,允许你浏览本地文件系统上的数据文件(.dat);2.Open URL.请求一个存有数据的URL 地址;3.Open DB.从数据库中读取数据;4.Generate.从一些数据生成器中生成人造数据。

1)Explorer用来进行数据实验、挖掘的环境,它提供了分类,聚类,关联规则,特征选择,数据可视化的功能。
(An environment for exploring data with WEKA)2)Experimentor用来进行实验,对不同学习方案进行数据测试的环境。
(An environment for performing experiments and conducting statistical tests between learning schemes.)3)KnowledgeFlow功能和Explorer差不多,不过提供的接口不同,用户可以使用拖拽的方式去建立实验方案。
另外,它支持增量学习。
(This environment supports essentially the same functions as the Explorer but with a drag-and-drop interface. One advantage is that it supports incremental learning.)4)SimpleCLI简单的命令行界面。
(Provides a simple command-line interface that allows direct execution of WEKA commands for operating systems that do not provide their own command line interface.)二、实验内容1.选用数据文件为:2.在WEKA中点击explorer 打开文件3.对数据整理分析4.将数据分类:单机classify ——在test options 中 选择第一项(Use training set )——点击classifier 下面的choose 按钮 选择trees 中的J48由上图可知该树有5个叶子是否出去游玩由天气晴朗(sunny)、天气预报(overcast)以及阴雨天(rainy)因素决定5.关联规则我们打算对前面的“bank-data”数据作关联规则的分析。

下面是一个简单的Java Weka使用案例。
在这个例子中,我们将使用Weka库对一个简单的数据集进行分类。
首先,你需要确保你已经安装了Weka库并将其添加到你的Java项目中。
你可以从Weka官网下载最新版本的Weka库。
然后,你可以使用以下代码加载数据集并使用J48决策树算法进行分类:
在这个例子中,我们首先使用DataSource类加载数据集。
然后,我们创建一个
J48对象,设置其未剪枝和随机种子属性,并使用数据集构建分类器。
最后,我们输出分类器的内容。
这只是一个简单的Weka使用案例,你可以根据自己的需求使用不同的算法和数据集进行更复杂的分类任务。




Weka入门教程3. 分类与回归背景知识WEKA把分类(Classification)和回归(Regression)都放在“Classify”选项卡中,这是有原因的。
在这两个任务中,都有一个目标属性(输出变量)。
我们希望根据一个样本(WEKA 中称作实例)的一组特征(输入变量),对目标进行预测。
为了实现这一目的,我们需要有一个训练数据集,这个数据集中每个实例的输入和输出都是已知的。
观察训练集中的实例,可以建立起预测的模型。
有了这个模型,我们就可以新的输出未知的实例进行预测了。
衡量模型的好坏就在于预测的准确程度。
在WEKA中,待预测的目标(输出)被称作Class属性,这应该是来自分类任务的“类”。
一般的,若Class属性是分类型时我们的任务才叫分类,Class属性是数值型时我们的任务叫回归。
选择算法这一节中,我们使用C4.5决策树算法对bank-data建立起分类模型。
我们来看原来的“bank-data.csv”文件。
“ID”属性肯定是不需要的。
由于C4.5算法可以处理数值型的属性,我们不用像前面用关联规则那样把每个变量都离散化成分类型。
尽管如此,我们还是把“Children”属性转换成分类型的两个值“YES”和“NO”。
另外,我们的训练集仅取原来数据集实例的一半;而从另外一半中抽出若干条作为待预测的实例,它们的“pep”属性都设为缺失值。
经过了这些处理的训练集数据在这里下载;待预测集数据在这里下载。
我们用“Explorer”打开训练集“bank.arff”,观察一下它是不是按照前面的要求处理好了。
切换到“Classify”选项卡,点击“Choose”按钮后可以看到很多分类或者回归的算法分门别类的列在一个树型框里。
3.5版的WEKA中,树型框下方有一个“Filter...”按钮,点击可以根据数据集的特性过滤掉不合适的算法。
我们数据集的输入属性中有“Binary”型(即只有两个类的分类型)和数值型的属性,而Class变量是“Binary”的;于是我们勾选“Binary attributes”“Numeric attributes”和“Binary class”。
拉/压力传感器(轮辐式),F2822型示例:型号F28222A D P R 1X 914113.01 07/2021 C N威卡(WIKA )操作说明,F2822型© 07/2021 WIKA Alexander Wiegand SE & Co. KG 保留所有权利。
WIKA ®是威卡(WIKA )在各个国家的注册商标。
在开始任何工作之前,请仔细阅读操作说明!请妥善保管以备后用!型号F2822操作说明页码 3 - 203威卡(WIKA )操作说明,F2822型A D P R 1X 914113.01 07/2021 C N目录1. 一般信息 42. 设计与功能 52.1 型号F2822概述 . . . . . . . . . . . . . . . . . . . . .52.2 描述 . . . . . . . . . . . . . . . . . . . . . . . . .52.3 供货范围........................53. 安全 63.1 符号说明........................63.2 预期用途........................63.3 不当使用........................73.4 操作人员责任 . . . . . . . . . . . . . . . . . . . . . .73.5 人员资质........................83.6 个人防护设备 . . . . . . . . . . . . . . . . . . . . . .83.7 标签/安全标识......................94. 运输、包装和储存 104.1 运输 . . . . . . . . . . . . . . . . . . . . . . . . 104.2 包装和储存 . . . . . . . . . . . . . . . . . . . . . . 105. 调试、运行 115.1 调试前注意事项 . . . . . . . . . . . . . . . . . . . . 115.2 安装说明.......................115.3 拉/压力传感器安装 . . . . . . . . . . . . . . . . . . . 125.4 电气连接.......................145.5 连接放大器 . . . . . . . . . . . . . . . . . . . . . . 145.6 引脚分配 型号F2822. . . . . . . . . . . . . . . . . . . 146. 故障 157. 维护和清洁 167.1 维护 . . . . . . . . . . . . . . . . . . . . . . . . 167.2 清洁 . . . . . . . . . . . . . . . . . . . . . . . . 167.3 再校准 . . . . . . . . . . . . . . . . . . . . . . . 168. 拆卸、返修和处置 178.1 拆卸 . . . . . . . . . . . . . . . . . . . . . . . . 178.2 返修 . . . . . . . . . . . . . . . . . . . . . . . . 178.3 处置 . . . . . . . . . . . . . . . . . . . . . . . . 179. 规格 189.1 认证 . . . . . . . . . . . . . . . . . . . . . . . . 1910. 附件 19附件:欧盟符合性声明 204威卡(WIKA )操作说明,F2822型A D P R 1X 914113.01 07/2021 C N1. 一般信息■操作说明中描述的拉/压力传感器均采用先进的技术进行设计和制造。
PrimerPROTECTED guest · Join · Help · Sign In ·PAGE DISCUSSION (2)HISTORY NOTIFY ME Join this WikiRecent Changes Manage WikiSearch Home All pages All tags All files FAQ Not So FAQTroubleshooting Introduction WEKA is a comprehensive toolbench for machine learning and data mining. Its main strengths lie in theclassification area, where all current ML approaches --and quite a few older ones --have been implementedwithin a clean, object-oriented Java class hierarchy. Regression, Association Rules and clustering algorithmshave also been implemented.However, WEKA is also quite complex to handle --amply demonstrated by many questions on the WEKAmailing list . Concerning the graphical user interface, the WEKA development group offers documentation forthe Explorer and the Experimenter. However, there is little documentation on using the command line interfaceto WEKA, although it is essential for realistic learning tasks.This document serves as a practical introduction to the command line interface. Since there has been a recentreorganization in class hierarchies for WEKA, all examples may only work with versions 3.4.4 and above only(until the next reorganization, that is ;-) Basic concepts and issues can more easily be transferred to earlierversions, but the specific examples may need to be slightly adapted (mostly removing the third class hierarchylevel and renaming some classes).While for initial experiments the included graphical user interface is quite sufficient, for in-depth usage thecommand line interface is recommended, because it offers some functionality which is not available via the GUI-and uses far less memory. Should you get Out of Memory errors, increase the maximum heap size for yourjava engine, usually via -Xmx1024M or -Xmx1024m for 1GB. Windows users should modify RunWeka.bat toadd the parameter -Xmx1024M before the -jar option, yielding java -Xmx1024M -jar weka.jar -thedefault setting of 16 to 64MB is usually too small. If you get errors that classes are not found, check yourCLASSPATH : does it include weka.jar ? You can explicitly set CLASSPATH via the -cp command line optionas well.We will begin by describing basic concepts and ideas. Then, we will describe the weka.filters package, which isused to transform input data, e.g. for preprocessing, transformation, feature generation and so on.Then we will focus on the machine learning algorithms themselves. These are called Classifiers in WEKA. Wewill restrict ourselves to common settings for all classifiers and shortly note representatives for all mainapproaches in machine learning.Afterwards, practical examples are given. In Appendix A you find an example java program which utilizesvarious WEKA classes in order to give some functionality which is not yet integrated in WEKA --namely tooutput predictions for test instances within a cross-validation. It also outputs the complete class probabilitydistribution.Finally, in the doc directory of WEKA you find a documentation of all java classes within WEKA. Prepare touse it since this overview is not intended to be complete. If you want to know exactly what is going on, take alook at the mostly well-documented source code, which can be found in weka-src.jar and can be extracted viathe jar utility from the Java Development Kit.If you find any bugs, less comprehensible statements, have comments or want to offer suggestions, pleasecontact me .Basic conceptsDatasetA set of data items, the dataset, is a very basic concept of machine learning. A dataset is roughly equivalent to a two-dimensional spreadsheet or database table. In WEKA, it is implemented by the Instances class. A dataset is a collection of examples, each one of class Instance . Each Instance consists of a number ofattributes, any of which can be nominal (= one of a predefined list of values), numeric (= a real or integernumber) or a string (= an arbitrary long list of characters, enclosed in "double quotes"). The externalrepresentation of an Instances class is an ARFF file, which consists of a header describing the attribute typesand the data as comma-separated list. Here is a short, commented example. A complete description of theARFF file format can be found here .% This is a toy example, the UCI weather dataset.% Any relation to real weather is purely coincidental.}}Comment lines at the beginning of the dataset should give an indication of its source, context and meaning.@relation golfWeatherMichigan_1988/02/10_14daysHere we state the internal name of the dataset. Try to be as comprehensive as possible.@attribute outlook {sunny, overcast rainy}@attribute windy {TRUE, FALSEHere we define two nominal attributes, outlook and windy. The former has three values: sunny, overcast and rainy; the latter two: TRUE and FALSE. Nominal values with special characters, commas or spaces are enclosed in 'single quotes'.@attribute temperature real@attribute humidity realThese lines define two numeric attributes. Instead of real, integer or numeric can also be used. While double floating point values are stored internally, only seven decimal digits are usually processed.@attribute play {yes, no}The last attribute is the default target or class variable used for prediction. In our case it is a nominal attribute with two values, making this a binary classification problem.@datasunny,FALSE,85,85,nosunny,TRUE,80,90,noovercast,FALSE,83,86,yesrainy,FALSE,70,96,yesrainy,FALSE,68,80,yesThe rest of the dataset consists of the token @data, followed by comma-separated values for the attributes --one line per example. In our case there are five examples.In our example, we have not mentioned the attribute type string, which defines "double quoted" string attributes for text mining. In recent WEKA versions, date/time attribute types are also supported.By default, the last attribute is considered the class/target variable, i.e. the attribute which should be predicted as a function of all other attributes. If this is not the case, specify the target variable via -c. The attribute numbers are one-based indices, i.e. -c 1specifies the first attribute.Some basic statistics and validation of given ARFF files can be obtained via the main() routine ofweka.core.Instances:java weka.core.Instances data/soybean.arffweka.core offers some other useful routines, e.g. converters.C45Loader and converters.CSVLoader, which can be used to import C45 datasets and comma/tab-separated datasets respectively, e.g.:java weka.core.converters.CSVLoader data.csv >data.arffjava weka.core.converters.C45Loader c45_filestem >data.arffClassifierAny learning algorithm in WEKA is derived from the abstract Classifier class. Surprisingly little is needed for a basic classifier: a routine which generates a classifier model from a training dataset (= buildClassifier) and another routine which evaluates the generated model on an unseen test dataset (= classifyInstance), or generates a probability distribution for all classes (= distributionForInstance).A classifier model is an arbitrary complex mapping from all-but-one dataset attributes to the class attribute. The specific form and creation of this mapping, or model, differs from classifier to classifier. For example, ZeroR's model just consists of a single value: the most common class, or the median of all numeric values in case of predicting a numeric value (= regression learning). ZeroR is a trivial classifier, but it gives a lower bound on the performance of a given dataset which should be significantly improved by more complex classifiers. As such it is a reasonable test on how well the class can be predicted without considering the other attributes.Later, we will explain how to interpret the output from classifiers in detail --for now just focus on the Correctly Classified Instances in the section Stratified cross-validation and notice how it improves from ZeroR to J48: java weka.classifiers.rules.ZeroR -t weather.arffjava weka.classifiers.trees.J48 -t weather.arffThere are various approaches to determine the performance of classifiers. The performance can most simply be measured by counting the proportion of correctly predicted examples in an unseen test dataset. This value is the accuracy, which is also 1-ErrorRate. Both terms are used in literature.The simplest case is using a training set and a test set which are mutually independent. This is referred to as hold-out estimate. To estimate variance in these performance estimates, hold-out estimates may be computed by repeatedly resampling the same dataset --i.e. randomly reordering it and then splitting it into training and test sets with a specific proportion of the examples, collecting all estimates on test data and computing average and standard deviation of accuracy.A more elaborate method is cross-validation. Here, a number of folds n is specified. The dataset is randomly reordered and then split into n folds of equal size. In each iteration, one fold is used for testing and the other n-1folds are used for training the classifier. The test results are collected and averaged over all folds. This gives the cross-validation estimate of the accuracy. The folds can be purely random or slightly modified to create the same class distributions in each fold as in the complete dataset. In the latter case the cross-validation is called stratified. Leave-one-out (loo) cross-validation signifies that n is equal to the number of examples. Out of necessity, loo cv has to be non-stratified, i.e. the class distributions in the test set are not related to those in the training data. Therefore loo cv tends to give less reliable results. However it is still quite useful in dealing with small datasets since it utilizes the greatest amount of training data from the dataset.weka.filtersThe weka.filters package is concerned with classes that transforms datasets --by removing or adding attributes, resampling the dataset, removing examples and so on. This package offers useful support for data preprocessing, which is an important step in machine learning.All filters offer the options -i for specifying the input dataset, and -o for specifying the output dataset. If any of these parameters is not given, this specifies standard input resp. output for use within pipes. Other parameters are specific to each filter and can be found out via -h, as with any other class. The weka.filters package is organized into supervised and unsupervised filtering, both of which are again subdivided into instance and attribute filtering. We will discuss each of the four subsection separately.weka.filters.supervisedClasses below weka.filters.supervised in the class hierarchy are for supervised filtering, i.e. taking advantage of the class information. A class must be assigned via -c, for WEKA default behaviour use -c last.attributeDiscretize is used to discretize numeric attributes into nominal ones, based on the class information, via Fayyad & Irani's MDL method, or optionally with Kononeko's MDL method. At least some learning schemes or classifiers can only process nominal data, e.g. rules.Prism; in some cases discretization may also reduce learning time.java weka.filters.supervised.attribute.Discretize -i data/iris.arff -o iris-nom.java weka.filters.supervised.attribute.Discretize -i data/cpu.arff -o cpu-classvNominalToBinary encodes all nominal attributes into binary (two-valued) attributes, which can be used to transform the dataset into a purely numeric representation, e.g. for visualization via multi-dimensional scaling.java weka.filters.supervised.attribute.NominalToBinary -i data/contact-lenses.arKeep in mind that most classifiers in WEKA utilize transformation filters internally, e.g. Logistic and SMO, so you will usually not have to use these filters explicity. However, if you plan to run a lot of experiments, pre-applying the filters yourself may improve runtime performance.instanceResample creates a stratified subsample of the given dataset. This means that overall class distributions are approximately retained within the sample. A bias towards uniform class distribution can be specified via -B.java weka.filters.supervised.instance.Resample -i data/soybean.arff -o soybean-5 java weka.filters.supervised.instance.Resample -i data/soybean.arff -o soybean-uStratifiedRemoveFolds creates stratified cross-validation folds of the given dataset. This means that per default the class distributions are approximately retained within each fold. The following example splits soybean.arff into stratified training and test datasets, the latter consisting of 25% (=1/4) of the data.java weka.filters.supervised.instance.StratifiedRemoveFolds -i data/soybean.arff -c last-N4-F1-Vjava weka.filters.supervised.instance.StratifiedRemoveFolds -i data/soybean.arff -c last-N4-F1weka.filters.unsupervisedClasses below weka.filters.unsupervised in the class hierarchy are for unsupervised filtering, e.g. the non-stratified version of Resample. A class should not be assigned here.attributeStringToWordVector transforms string attributes into a word vectors, i.e. creating one attribute for each word which either encodes presence or word count (-C) within the string. -W can be used to set an approximate limit on the number of words. When a class is assigned, the limit applies to each class separately. This filter is useful for text mining.Obfuscate renames the dataset name, all attribute names and nominal attribute values. This is intended for exchanging sensitive datasets without giving away restricted information.Remove is intended for explicit deletion of attributes from a dataset, e.g. for removing attributes of the iris dataset:java weka.filters.unsupervised.attribute.Remove -R1-2-i data/iris.arff -o iris java weka.filters.unsupervised.attribute.Remove -V-R3-last -i data/iris.arff -instanceResample creates a non-stratified subsample of the given dataset, i.e. random sampling without regard to the class information. Otherwise it is equivalent to its supervised variant.java weka.filters.unsupervised.instance.Resample -i data/soybean.arff -o soybeanRemoveFolds creates cross-validation folds of the given dataset. The class distributions are not retained. The following example splits soybean.arff into training and test datasets, the latter consisting of 25% (=1/4) of the data.java weka.filters.unsupervised.instance.RemoveFolds -i data/soybean.arff -o soyb java weka.filters.unsupervised.instance.RemoveFolds -i data/soybean.arff -o soybRemoveWithValues filters instances according to the value of an attribute.java weka.filters.unsupervised.instance.RemoveWithValues -i data/soybean.arff \ -o soybean-without_herbicide_injury.arff -V-C last-L19weka.classifiersClassifiers are at the core of WEKA. There are a lot of common options for classifiers, most of which are related to evaluation purposes. We will focus on the most important ones. All others including classifier-specificparameters can be found via -h, as usual.-t specifies the training file (ARFF format)-T specifies the test file in (ARFF format). If this parameter is missing, a crossvalidation will be performed (default: 10-fold cv)-x This parameter determines the number of folds for the cross-validation. A cv will only be performed if -T is missing.-c As we already know from the weka.filters section, this parameter sets the class variable with a one-based index.-d The model after training can be saved via this parameter. Each classifier has a different binary format for the model, so it can only be read back by the exact same classifier on a compatible dataset. Only the model on the training set is saved, not the multiple models generated via cross-validation.-l Loads a previously saved model, usually for testing on new, previously unseen data. In that case, a compatible test file should be specified, i.e. the same attributes in the same order.-p #If a test file is specified, this parameter shows you the predictions and one attribute (0 for none) for all test instances. If no test file is specified, this outputs nothing. In that case, you will have to use callClassifier from Appendix A.-i A more detailed performance description via precision, recall, true-and false positive rate is additionally output with this parameter. All these values can also be computed from the confusion matrix.-o This parameter switches the human-readable output of the model description off. In case of support vector machines or NaiveBayes, this makes some sense unless you want to parse and visualize a lot ofinformation.We now give a short list of selected classifiers in WEKA. Other classifiers below weka.classifiers in package overview may also be used. This is more easy to see in the Explorer GUI.•trees.J48 A clone of the C4.5 decision tree learner•bayes.NaiveBayes A Naive Bayesian learner. -K switches on kernel density estimation for numerical attributes which often improves performance.•meta.ClassificationViaRegression-W functions.LinearRegression Multi-response linear regression.•functions.Logistic Logistic Regression.•functions.SMO Support Vector Machine (linear, polynomial and RBF kernel) with Sequential Minimal Optimization Algorithm due to [Platt, 1998]. Defaults to SVM with linear kernel, -E 5 -C 10gives an SVM with polynomial kernel of degree 5 and lambda=10.•lazy.KStar Instance-Based learner. -E sets the blend entropy automatically, which is usuallypreferable.•lazy.IBk Instance-Based learner with fixed neighborhood. -K sets the number of neighbors to use.IB1is equivalent to IBk -K 1•rules.JRip A clone of the RIPPER rule learner.Based on a simple example, we will now explain the output of a typical classifier, weka.classifiers.trees.J48. Consider the following call from the command line, or start the WEKA explorer and train J48 on weather.arff:java weka.classifiers.trees.J48 -t data/weather.arff -iJ48 pruned tree------------------outlook = sunny| humidity <= 75: yes (2.0)| humidity > 75: no (3.0)outlook = overcast: yes (4.0)outlook = rainy| windy = TRUE: no (2.0)| windy = FALSE: yes (3.0)Number of Leaves : 5Size of the tree : 8The first part, unless you specify -o, is a human-readable form of the training set model. In this case, it is a decision tree. outlook is at the root of the tree and determines the first decision. In case it is overcast, we'll always play golf. The numbers in (parentheses) at the end of each leaf tell us the number of examples in this leaf. If one or more leaves were not pure (= all of the same class), the number of misclassified examples wouldalso be given, after a /slash/Time taken to build model: 0.05 secondsTime taken to test model on training data: 0 secondsAs you can see, a decision tree learns quite fast and is evaluated even faster. E.g. for a lazy learner, testing would take far longer than training.= Error on training data ==Correctly Classified Instance 14 100 %Incorrectly Classified Instances 0 0 %Kappa statistic 1Mean absolute error 0Root mean squared error 0Relative absolute error 0 %Root relative squared error 0 %Total Number of Instances 14== Detailed Accuracy By Class ==TP Rate FP Rate Precision Recall F-Measure Class1 0 1 1 1 yes1 0 1 1 1 no== Confusion Matrix ==a b <--classified as9 0 | a = yes0 5 | b = noThis is quite boring: our classifier is perfect, at least on the training data --all instances were classified correctly and all errors are zero. As is usually the case, the training set accuracy is too optimistic. The detailed accuracy by class, which is output via -i, and the confusion matrix is similarily trivial.== Stratified cross-validation ==Correctly Classified Instances 9 64.2857 %Incorrectly Classified Instances 5 35.7143 %Kappa statistic 0.186Mean absolute error 0.2857Root mean squared error 0.4818Relative absolute error 60 %Root relative squared error 97.6586 %Total Number of Instances 14== Detailed Accuracy By Class ==TP Rate FP Rate Precision Recall F-Measure Class0.778 0.6 0.7 0.778 0.737 yes0.4 0.222 0.5 0.4 0.444 no== Confusion Matrix ==a b <--classified as7 2 | a = yes3 2 | b = noThe stratified cv paints a more realistic picture. The accuracy is around 64%. The kappa statistic measures the agreement of prediction with the true class --1.0 signifies complete agreement. The following error values are not very meaningful for classification tasks, however for regression tasks e.g. the root of the mean squared error per example would be a reasonable criterion. We will discuss the relation between confusion matrix and other measures in the text.The confusion matrix is more commonly named contingency table. In our case we have two classes, andtherefore a 2x2 confusion matrix, the matrix could be arbitrarily large. The number of correctly classified instances is the sum of diagonals in the matrix; all others are incorrectly classified (class "a" gets misclassified as "b" exactly twice, and class "b" gets misclassified as "a" three times).The True Positive (TP)rate is the proportion of examples which were classified as class x, among all examples which truly have class x, i.e. how much part of the class was captured. It is equivalent to Recall. In the confusion matrix, this is the diagonal element divided by the sum over the relevant row, i.e. 7/(7+2)=0.778 for class yes and 2/(3+2)=0.4 for class no in our example.The False Positive (FP)rate is the proportion of examples which were classified as class x, but belong to a different class, among all examples which are not of class x. In the matrix, this is the column sum of class x minus the diagonal element, divided by the rows sums of all other classes; i.e. 3/5=0.6 for class yes and2/9=0.222 for class no.The Precision is the proportion of the examples which truly have class x among all those which were classified as class x. In the matrix, this is the diagonal element divided by the sum over the relevant column, i.e. 7/(7+3) =0.7 for class yes and 2/(2+2)=0.5 for class no.The F-Measure is simply 2*Precision*Recall/(Precision+Recall), a combined measure for precision and recall.These measures are useful for comparing classifiers. However, if more detailed information about the classifier's predictions are necessary, -p #outputs just the predictions for each test instance, along with a range of one-based attribute ids (0 for none). Let's look at the following example. We shall assume soybean-train.arff and soybean-test.arff have been constructed via weka.filters.supervised.instance.StratifiedRemoveFolds as in a previous example.java weka.classifiers.bayes.NaiveBayes-K -t soybean-train.arff-T soybean-test.0 diaporthe-stem-canker 0.9999672587892333 diaporthe-stem-canker1 diaporthe-stem-canker 0.9999992614503429 diaporthe-stem-canker2 diaporthe-stem-canker 0.999998948559035 diaporthe-stem-canker3 diaporthe-stem-canker 0.9999998441238833 diaporthe-stem-canker4 diaporthe-stem-canker 0.9999989997681132 diaporthe-stem-canker5 rhizoctonia-root-rot 0.9999999395928124 rhizoctonia-root-rot6 rhizoctonia-root-rot 0.999998912860593 rhizoctonia-root-rot7 rhizoctonia-root-rot 0.9999994386283236 rhizoctonia-root-rot...The values in each line are separated by a single space. The fields are the zero-based test instance id, followed by the predicted class value, the confidence for the prediction (estimated probability of predicted class), and the true class. All these are correctly classified, so let's look at a few erroneous ones.32 phyllosticta-leaf-spot 0.7789710144361445 brown-spot...39 alternarialeaf-spot 0.6403333824349896 brown-spot...44 phyllosticta-leaf-spot 0.893568420641914 brown-spot...46 alternarialeaf-spot 0.5788190397739439 brown-spot...73 brown-spot 0.4943768155314637 alternarialeaf-spot...In each of these cases, a misclassification occurred, mostly between classes alternarialeaf-spot and brown-spot. The confidences seem to be lower than for correct classification, so for a real-life application it may make sense to output don't know below a certain threshold. WEKA also outputs a trailing newline.If we had chosen a range of attributes via -p, e.g. -p first-last, the mentioned attributes would have been output afterwards as comma-separated values, in (parantheses). However, the zero-based instance id in the first column offers a safer way to determine the test instances.Regrettably, -p does not work without test set (in versions before 3.5.8), i.e. for the cross-validation. Although patching WEKA is feasible, it is quite messy and has to be repeated for each new version. Another way to achieve this functionality is callClassifier, which calls WEKA functions from Java and implements this functionality, optionally outputting the complete class probablity distribution also. The output format is the same as above, but because of the cross-validation the instance ids are not in order, which can be remedied via|sort -n.If we had saved the output of -p in soybean-test.preds, the following call would compute the number of correctly classified instances:cat soybean-test.preds |awk'$2=$4&&$0!=""'|wc-lDividing by the number of instances in the test set, i.e. wc -l < soybean-test.preds minus one (= trailing newline), we get the training set accuracy.ExamplesUsually, if you evaluate a classifier for a longer experiment, you will do something like this (for csh): java -Xmx1024m weka.classifiers.trees.J48 -t data.arff -i-k-d J48-data.model >The -Xmx1024m parameter for maximum heap size ensures your task will get enough memory. There is no overhead involved, it just leaves more room for the heap to grow. -i and -k gives you some additional information, which may be useful, e.g. precision and recall for all classes. In case your model performs well, it makes sense to save it via -d-you can always delete it later! The implicit cross-validation gives a more reasonable estimate of the expected accuracy on unseen data than the training set accuracy. The output both of standard error and output should be redirected, so you get both errors and the normal output of your classifier. The last & starts the task in the background. Keep an eye on your task via top and if you notice the hard disk works hard all the time (for linux), this probably means your task needs too much memory and will not finish in time for the exam. ;-) In that case, switch to a faster classifier or use filters, e.g. for Resample to reduce the size of your dataset or StratifiedRemoveFolds to create training and test sets -for most classifiers, training takes more time than testing.So, now you have run a lot of experiments --which classifier is best? Trycat*.out |grep-A3"Stratified"|grep"^Correctly"...this should give you all cross-validated accuracies. If the cross-validated accuracy is roughly the same as the training set accuracy, this indicates that your classifiers is presumably not overfitting the training set.Now you have found the best classifier. To apply it on a new dataset, use e.g.java weka.classifiers.trees.J48 -l J48-data.model -T new-data.arffYou will have to use the same classifier to load the model, but you need not set any options. Just add the new test file via -T. If you want, -p first-last will output all test instances with classifications and confidence, followed by all attribute values, so you can look at each error separately.The following more complex csh script creates datasets for learning curves, i.e. creating a 75% training set and 25% test set from a given dataset, then successively reducing the test set by factor 1.2 (83%), until it is also 25% in size. All this is repeated thirty times, with different random reorderings (-S) and the results are written to different directories. The Experimenter GUI in WEKA can be used to design and run similar experiments.#!/bin/cshforeach f ($*)set run=1while($run<= 30)mkdir$run>&!/dev/nulljava weka.filters.supervised.instance.StratifiedRemoveFolds -N4-F1-S$run java weka.filters.supervised.instance.StratifiedRemoveFolds -N4-F1-S$run foreach nr (012345)set nrp1=$nr@nrp1++java weka.filters.supervised.instance.Resample -S0-Z83-c last-i$run/t endecho Run $run of $f done.@run++endend。
大数据导论实验报告
实验一
姓名abc
学号asadsdsa
报告日期
实验一
一.实验目的
1实验开源工具Weka的安装和熟悉;
2.数据理解,数据预处理的实验;
二.实验内容
1.weka介绍
2.数据理解
3.数据预处理
4.保存处理后的数据
三.实验过程
1.导入数据并修改选项
2.用weka.filters.unsupervised.attribute.ReplaceMissingValues处理缺失值
3.用weka.filters.unsupervised.attribute.Discretize离散化第一列数据
4.用weka.filters.unsupervised.instance.RemoveDuplicates删除重复数据
5.用weka.filters.unsupervised.attribute.Discretize离散化第六列数据
6.用weka.filters.unsupervised.attribute.Normalize归一化数据
7.保存数据
四.实验结果与分析
1.数据清理后的对比图,上面的是处理前的图,下图是处理后的图
分析:通过两图对比可发现图一中缺失的数据在图二中已经添加上。
2.离散化第一行后的对比图,图片为离散化之后的效果图
分析:此次处理目标为第一列,可发现处理后‘age’这一列的数据离散化了。
3.删除重复数据之后的效果图
5.离散化第六列后的效果图
分析:此次处理目标为第六列,可清楚看到发生的变化6.归一化后的效果图
此次处理的目标是10,12,13,14列,即将未离散化的数值列进行归一化处理。
WEKA连接数据库的配置说明(mysql)1.安装weka2.在weka的安装目录下新建一个文件夹,名为‘lib’(备注:文件夹名可以是任意英文名)3.在lib文件中放入mysql的驱动程序,如:“mysql-connector-java-5.1.6-bin.jar”;4.配置环境变量的‘系统变量’:(备注:两种方式,一种是直接配置绝对路径,一种是相对路径,任选一种)4.1)绝对路径:在变量classpath中追加mysql驱动程序的安装路径,如:“;F:\weka package\install_path\Weka-3-6\lib\mysql-connector-java-5.1.6-bin.jar”4.2)相对路径:4.2.1)在系统变量中新增一个变量:名:WEKA_HOME值:F:\weka package\install_path\Weka-3-64.2.2)再在classpath中追加:“;%WEKA_HOME%\lib\mysql-connector-java-5.1.6-bin.jar”(备注:两种方式在本质上是一样的,任选一种)重要说明:如果你的classpath是小写的,请改成大写的‘CLASSPATH’,经过验证,该系统变量名直接影响到是否配置成功;并且修改环境变量之后记得保存并重启计算机,否则可能修改不生效。
5.在weka的安装目录下,新建一个文件夹‘weka’,然后将安装目录下的‘weka.jar’解压到该文件夹下;(解压后会有三个文件:weka、java_cup、META-INF)(备注:可以删除weka.jar文件,同样用户可以根据所需保存到要其他地方,以备不时之需)6.在解压后的文件中找到“weka>experiment>DatabaseUtils.props”,将“DatabaseUtils.props”改成“DatabaseUtils.props.sample”(名字可以任意),再将“DatabaseUtils.props.mysql”改成“DatabaseUtils.props”(weka运行时会使用DatabaseUtils.props),并且打开这个文件,按照如下方式修改:在以上这段内容之后加上以下内容:(备注:由于weka仅支持名词型(nominal),数值型(numeric),字符串(string),日期(date).所以我们要将现在数据库中的数据类型对应到这四种类型上来)7.按照以上修改后保存文件;然后将“F:\weka package\install_path\Weka-3-6\weka\META-INF”文件下的“MANIFEST.MF”复制到和META-INF同级目录下,然后打开控制台,在控制台中进入到安装目录下的weka文件,输入命令“jar cfm weka.jar MANIFEST.MF java_cup weka”在该目录下会生成一个weka.jar 文件,再将该文件复制到weka的安装目录下。
Weka如何连接数据库以SQL Server2000为例,其他的数据库操作⽅法⼀样,具体细节各异。
1 安装驱动程序,SQL Server2000将三个.jar加到环境变量。
2 修改 weka\experiment下的DatabaseUtils.props⽂件。
我们可以看到有DatabaseUtils.props.odbc DatabaseUtils.props.oracle等我们先将DatabaseUtils.props随便改成⼀个其他的名字,然后将DatabaseUtils.props.mssqlserver改成DatabaseUtils.props,打开现在的DatabaseUtils.props可以看到以下部分:(#表⽰注释)2.1驱动加载# JDBC driver (comma-separated list)jdbcDriver=com.microsoft.jdbc.sqlserver.SQLServerDriver2.2数据库连接,如果在本机上可以将server_name改为127.0.0.1或者localhost# database URLjdbcURL=jdbc:sqlserver://127.0.0.1:14332.3数据类型的转换。
由于weka仅⽀持名词型(nominal)、数值型(numeric)、字符串、⽇期(date)。
所以我们要将现在数据库中的数据类型对应到这四种类型上来。
将以下数据类型对应的句⼦前⾯的注释符合去掉。
由于SQL Server2000有其他的数据类型Weka尚不能识别,所以我们在下⾯再添加上smallint=3datetime=8等等string,getString()= 0; -->nominalboolean,getBoolean() = 1; -->nominaldouble,getDouble() = 2; -->numericbyte,getByte() = 3; -->numericshort,getByte()= 4; -->numericint,getInteger() = 5; -->numericlong,getLong() = 6; -->numericgloat,getFloat() = 7; -->numericdate,getDate() = 8; -->datevarchar=0float=2tinyint=3int=53其他说明,我们暂时⽤不到,不⽤去管了# other optionsCREATE_DOUBLE=DOUBLE PRECISIONCREATE_STRING=VARCHAR(8000)CREATE_INT=INTcheckUpperCaseNames=falsecheckLowerCaseNames=falsecheckForTable=true4 OK,下⾯可以操作了!运⾏weka的Explore界⾯后,通过Open DB..打开SQL Viewer⼯作界⾯(3.5.5版本⽐3.4.10在这⾥精细了许多)。