TaKaRa Virall RNA 抽提说明书9766
- 格式:pdf
- 大小:463.04 KB
- 文档页数:8

总RNA的提取(Trizol法提取)在收集到生物材料之后,最好能即刻进行RNA制备工作。
若需暂时储存,则应以液氮将生物材料急速冷冻后,储存于-80℃冷冻柜。
在制备RNA时,将储存于冷冻柜的材料取出,立即以加入液氮研磨的方式打破细胞,不可以先行解冻,以避免RNase的作用。
1.提取组织RNA时,每50~100mg组织用1ml Trizol试剂对组织进行裂解;提取细胞RNA时,先离心沉淀细胞,每5-10╳106个细胞加1ml Trizol后,反复用枪吹打或剧烈振荡以裂解细胞;2.将上述组织或细胞的Trizol裂解液转入EP管中,在室温15~30C下放置5分钟;3.在上述EP管中,按照每1ml TRIZOL加0.2ml氯仿的量加入氯仿,盖上EP管盖子,在手中用力震荡15秒,在室温下(15℃~30℃)放置2~3分钟后,12000g(2℃~8℃)离心15分钟;4.取上层水相置于新EP管中,按照每1ml TRIZOL加0.5ml异丙醇的量加入异丙醇,在室温下(15℃~30℃)放置10分钟,12000g(2℃~8℃)离心10分钟;5.弃上清,按照每1ml TRIZOL加1ml75%乙醇进行洗涤,涡旋混合,7500g(2℃~8℃)离心5分钟,弃上清;6.让沉淀的RNA在室温下自然干燥;7.用Rnase-free water溶解RNA沉淀。
PCR实验室常用DNA聚合酶有三种:TaKaRa Taq TM,TaKaRa E X Taq TM和Pyrobest TM DNA Polymerase。
TaKaRa Taq TM是一般的DNA聚合酶,保真性较差,但价钱便宜,一般用于基因表达的检测等。
TaKaRa E X Taq TM是具有Proof reading 活性的耐热性DNA聚合酶,具有一定的保真性,而且其扩增得到的PCR产物3’端附有一个“A”碱基,如果希望直接将产物克隆到T-vector可以用此酶。
Pyrobest TM DNA Polymerase也是具有Proof reading活性的耐热性DNA聚合酶,其特点是保真性极高,扩增得到的PCR产物为平滑末端。

September 2010RNeasy® Plus Mini HandbookFor purification of total RNA from animalcells and easy-to-lyse animal tissues usinggDNA Eliminator columnsQIAGEN Sample and Assay TechnologiesQIAGEN is the leading provider of innovative sample and assay technologies, enabling the isolation and detection of contents of any biological sample. Our advanced, high-quality products and services ensure success from sample to result.QIAGEN sets standards in:Purification of DNA, RNA, and proteinsNucleic acid and protein assaysmicroRNA research and RNAiAutomation of sample and assay technologiesOur mission is to enable you to achieve outstanding success and breakthroughs. For more information, visit .ContentsKit Contents 4 Storage 4 Product Use Limitations 4 Product Warranty and Satisfaction Guarantee 5 Technical Assistance 5 Safety Information 6 Quality Control 7 Introduction 8 Principle and procedure 8 Equipment and Reagents to Be Supplied by User 11 Important Notes 12 Determining the amount of starting material 12 Handling and storing starting material 15 Disrupting and homogenizing starting material 15 ProtocolsPurification of Total RNA from Animal Cells 17 Purification of Total RNA from Animal Tissues 23 Troubleshooting Guide 30 Appendix A: General Remarks on Handling RNA 34 Appendix B: Storage, Quantification, and Determination of Qualityof RNA 36 Appendix C: Protocol for Formaldehyde Agarose Gel Electrophoresis 39 Appendix D: Purification of Total RNA Containing Small RNAsfrom Cells 41 Appendix E: Separate Purification of Small RNA (Containing miRNA)and Larger RNA from Animal Cells Using the RNeasy Plus Mini Kitand RNeasy MinElute Cleanup Kit 43 References 46 Ordering Information 47Kit ContentsRNeasy Plus Mini Kit (50) (250)Catalog no. 74134 74136Number of preps 50 250gDNA Eliminator Mini Spin Columns (uncolored)50 250(each in a 2 ml Collection Tube)50 250RNeasy Mini Spin Columns (pink)(each in a 2 ml Collection Tube)Collection Tubes (1.5 ml) 50 250Collection Tubes (2 ml) 50 250Buffer RLT Plus* 45 ml 220 mlBuffer RW1* 45 ml 220 mlBuffer RPE† (concentrate) 11 ml 55 mlRNase-Free Water 10 ml 50 mlHandbook 11* Contains a guanidine salt. Not compatible with disinfectants containing bleach. See page 6for safety information.† Before using for the first time, add 4 volumes of ethanol (96–100%) as indicated on thebottle to obtain a working solution.StorageThe RNeasy Plus Mini Kit should be stored dry at room temperature (15–25ºC)and is stable for at least 9 months under these conditions.Product Use LimitationsThe RNeasy Plus Mini Kit is intended for molecular biology applications. Thisproduct is not intended for the diagnosis, prevention, or treatment of a disease.All due care and attention should be exercised in the handling of the product.We recommend all users of QIAGEN® products to adhere to the NIH guidelinesthat have been developed for recombinant DNA experiments, or to otherapplicable guidelines.Product Warranty and Satisfaction Guarantee QIAGEN guarantees the performance of all products in the manner describedin our product literature. The purchaser must determine the suitability of theproduct for its particular use. Should any product fail to perform satisfactorily due to any reason other than misuse, QIAGEN will replace it free of charge orrefund the purchase price. We reserve the right to change, alter, or modify anyproduct to enhance its performance and design. If a QIAGEN product does notmeet your expectations, simply call your local Technical Service Department or distributor. We will credit your account or exchange the product — as you wish. Separate conditions apply to QIAGEN scientific instruments, service products, and to products shipped on dry ice. Please inquire for more information.A copy of QIAGEN terms and conditions can be obtained on request, and is also provided on the back of our invoices. If you have questions about product specifications or performance, please call QIAGEN Technical Services or your local distributor (see back cover or visit ).Technical AssistanceAt QIAGEN, we pride ourselves on the quality and availability of our technical support. Our Technical Service Departments are staffed by experienced scientists with extensive practical and theoretical expertise in sample and assay technologies and the use of QIAGEN products. If you have any questions or experience any difficulties regarding the RNeasy Plus Mini Kit or QIAGEN products in general, please do not hesitate to contact us.QIAGEN customers are a major source of information regarding advanced or specialized uses of our products. This information is helpful to other scientists as well as to the researchers at QIAGEN. We therefore encourage you to contact us if you have any suggestions about product performance or new applications and techniques.For technical assistance and more information, please see our Technical Support Center at /Support or call one of the QIAGEN Technical Service Departments or local distributors (see back cover or visit ).Safety InformationWhen working with chemicals, always wear a suitable lab coat, disposablegloves, and protective goggles. For more information, please consult theappropriate material safety data sheets (MSDSs). These are available online in convenient and compact PDF format at /ts/msds.asp whereyou can find, view, and print the MSDS for each QIAGEN kit and kitcomponent.CAUTION: DO NOT add bleach or acidic solutions directly to the sample-preparation waste.Buffer RLT Plus contains guanidine thiocyanate and Buffer RW1 contains a small amount of guanidine thiocyanate. This chemical can form highly reactive compounds when combined with bleach. If liquid containing these buffers is spilt, clean with suitable laboratory detergent and water. If the spilt liquid contains potentially infectious agents, clean the affected area first with laboratory detergent and water, and then with 1% (v/v) sodium hypochlorite. The following risk and safety phrases apply to the components of the RNeasy Plus Mini Kit.Buffer RLT PlusContains guanidine thiocyanate: harmful. Risk and safety phrases:*R20/21/22-32, S13-26-36-46Buffer RW1Contains ethanol: flammable. Risk phrase:* R1024-hour emergency informationEmergency medical information in English, French, and German can be obtained 24 hours a day from:Poison Information Center Mainz, GermanyTel: +49-6131-19240* R10: Flammable; R20/21/22: Harmful by inhalation, in contact with skin, and if swallowed; R32: Contact with acids liberates very toxic gas; S13: Keep away from food, drink and animal feeding stuffs; S26: In case of contact with eyes, rinse immediately with plenty of water and seek medical advice; S36: Wear suitable protective clothing; S46: If swallowed, seek medical advice immediately and show the container or label.Quality ControlIn accordance with QIAGEN’s ISO-certified Quality Management System, each lot of RNeasy Plus Mini Kit is tested against predetermined specifications to ensure consistent product quality.IntroductionThe RNeasy Plus Mini Kit is designed to purify RNA from small amounts ofanimal cells or tissues. The kit is compatible with a wide range of cultured cellsand with easy-to-lyse tissues. Genomic DNA contamination is effectively removed using a specially designed gDNA Eliminator spin column. The purifiedRNA is ready to use and is ideally suited for downstream applications that aresensitive to low amounts of DNA contamination, such as quantitative, real-timeRT-PCR.* The purified RNA can also be used in other applications, including:RT-PCRDifferential displaycDNA synthesisNorthern, dot, and slot blot analysesPrimer extensionPoly A+ RNA selectionRNase/S1 nuclease protectionMicroarraysThe RNeasy Plus Mini Kit allows the parallel processing of multiple samples inless than 25 minutes. Time-consuming and tedious methods such as CsCl step-gradient ultracentrifugation and alcohol precipitation steps, or methods involving the use of toxic substances such as phenol and/or chloroform, are replaced by the RNeasy Plus procedure.Principle and procedureThe RNeasy Plus procedure integrates QIAGEN’s patented technology for selective removal of double-stranded DNA with well-established RNeasy technology. Efficient purification of high-quality RNA is guaranteed, without the need for additional DNase digestion.Biological samples are first lysed and homogenized in a highly denaturing guanidine-isothiocyanate–containing buffer, which immediately inactivates RNases to ensure isolation of intact RNA. The lysate is then passed through a gDNA Eliminator spin column. This column, in combination with the optimized high-salt buffer, allows efficient removal of genomic DNA.Ethanol is added to the flow-through to provide appropriate binding conditions for RNA, and the sample is then applied to an RNeasy spin column, where total* Visit /geneXpression for information on standardized solutions for gene expression analysis, including QuantiTect® Kits and Assays for quantitative, real-time RT-PCR.RNA binds to the membrane and contaminants are efficiently washed away. High-quality RNA is then eluted in 30 μl, or more, of water.With the RNeasy Plus procedure, all RNA molecules longer than 200 nucleotides are isolated. The procedure provides an enrichment for mRNA, since most RNAs <200 nucleotides (such as 5.8S rRNA, 5S rRNA, and tRNAs, which together comprise 15–20% of total RNA) are selectively excluded. The size distribution of the purified RNA is comparable to that obtained by centrifugation through a CsCl cushion, where small RNAs do not sediment efficiently.In this handbook, different protocols are provided for different starting materials. The protocols differ primarily in the lysis and homogenization of the sample. Once the sample is applied to the gDNA Eliminator spin column, the protocols are similar (see flowchart, next page).RNeasy Plus Procedure CellsortissueGenomic DNATotal RNAEluted RNA Lyse and homogenize Remove genomic DNA Add ethanolBind total RNAWashEluteEquipment and Reagents to Be Supplied by User When working with chemicals, always wear a suitable lab coat, disposable gloves, and protective goggles. For more information, consult the appropriate material safety data sheets (MSDSs), available from the product supplier.For all protocols14.3 M β-mercaptoethanol (β-ME) (commercially available solutions areusually 14.3 M)Sterile, RNase-free pipet tipsMicrocentrifuge (with rotor for 2 ml tubes)96–100% ethanol*70% ethanol* in waterDisposable glovesFor tissue samples: RNA later® RNA Stabilization Reagent (see ordering information, page 47) or liquid nitrogenEquipment for sample disruption and homogenization (see pages 15–16).Depending on the method chosen, one or more of the following arerequired:Trypsin and PBSQIAshredder homogenizer (see ordering information, page 47)Blunt-ended needle and syringeMortar and pestleTissueLyser II or TissueLyser LT (see ordering information, page 48) TissueRuptor® homogenizer (see ordering information, page 48)* Do not use denatured alcohol, which contains other substances such as methanol or methylethylketone.Important NotesDetermining the amount of starting materialIt is essential to use the correct amount of starting material in order to obtain optimal RNA yield and purity. The maximum amount that can be used is limited by:The type of sample and its DNA and RNA contentThe volume of Buffer RLT Plus required for efficient lysis and the maximum loading volume of the RNeasy spin columnThe DNA removal capacity of the gDNA Eliminator spin columnThe RNA binding capacity of the RNeasy spin columnWhen processing samples containing high amounts of DNA or RNA, less than the maximum amount of starting material shown in Table 1 should be used, so that the DNA removal capacity of the gDNA Eliminator spin column and the RNA binding capacity of the RNeasy spin column are not exceeded.When processing samples containing average or low amounts of RNA, the maximum amount of starting material shown in Table 1 can be used. However, even though the RNA binding capacity of the RNeasy spin column is not reached, the maximum amount of starting material must not be exceeded. Otherwise, lysis will be incomplete and cellular debris may interfere with the binding of RNA to the RNeasy spin column membrane, resulting in lower RNA yield and purity.More information on using the correct amount of starting material is given in each protocol. Table 2 shows expected RNA yields from various cells and tissues.Note: If the DNA removal capacity of the gDNA Eliminator spin column is exceeded, the purified RNA will be contaminated with DNA. Although the gDNA Eliminator spin column can bind more than 100 μg DNA, we recommend using samples containing less than 20 μg DNA to ensure removal of virtually all genomic DNA. If the binding capacity of the RNeasy spin column is exceeded, RNA yields will not be consistent and may be reduced. If lysis of the starting material is incomplete, RNA yields will be lower than expected, even if the binding capacity of the RNeasy spin column is not exceeded.Table 1. RNeasy Mini spin column specificationsMaximum binding capacity 100 μg RNA Maximum loading volume 700 μlRNA size distribution RNA >200 nucleotides Minimum elution volume 30 μl Maximum amount of starting materialAnimalcells 1 x 107 cellsAnimal tissues 30 mgTable 2. Yields of total RNA with the RNeasy Plus Mini KitCell cultures (1 x 106 cells) Average yield oftotal RNA* (μg)Mouse/rattissues (10 mg)Average yield oftotal RNA* (μg)NIH/3T3 10 Embryo (13 day) 25HeLa 15 Brain 5–10COS-7 35Heart 4–8 LMH 12 Kidney 20–30Huh 15Liver 40–60Lung 10–20* Amounts can vary due to factors such as species, developmental stage, and growthconditions. Since the RNeasy procedure enriches for mRNA and other RNA species >200nucleotides, the total RNA yield does not include 5S rRNA, tRNA, and other low-molecular-weight RNAs, which make up 15–20% of total cellular RNA.Counting cells or weighing tissue is the most accurate way to quantitate theamount of starting material. However, the following may be used as a guide.Animal cellsThe number of HeLa cells obtained in various culture vessels after confluentgrowth is given in Table 3.Table 3. Growth area and number of HeLa cells in various culturevesselsCell-culture vessel Growth area (cm3)* Number of cells†Multiwell plates96-well 0.32–0.6 4–5104x48-well 1 1 x 10524-well 2 2.5 x 10512-well 4 5 x 1056-well 9.5 1 x 106Dishes 35 mm 8 1 x 10660mm 21 2.5 x 106100 mm 56 7 x 106145–150mm 145 2 x 107Flasks 40–50 ml 25 3 x 106250–300ml 75 1 x 107650–750 ml 162–175 2 x 107* Per well, if multiwell plates are used; varies slightly depending on the supplier.† Cell numbers are given for HeLa cells (approximate length = 15 μm), assuming confluentgrowth. Cell numbers will vary for different kinds of animal cells, which vary in length from10 to 30 μm.Animal tissuesA 3 mm cube (27 mm3) of most animal tissues weighs 30–35 mg.Handling and storing starting materialRNA in harvested tissue is not protected until the sample is treated with RNA laterRNA Stabilization Reagent, flash-frozen, or disrupted and homogenized in thepresence of RNase-inhibiting or denaturing reagents. Otherwise, unwantedchanges in the gene expression profile will occur. It is therefore important that tissue samples are immediately frozen in liquid nitrogen and stored at –70ºC,or immediately immersed in RNA later RNA Stabilization Reagent.The procedures for tissue harvesting and RNA protection should be carried out as quickly as possible. Frozen tissue samples should not be allowed to thaw during handling or weighing. After disruption and homogenization in Buffer RLT Plus (lysis buffer), samples can be stored at –70ºC for months.Disrupting and homogenizing starting materialEfficient disruption and homogenization of the starting material is an absolute requirement for all total RNA purification procedures. Disruption and homogenization are 2 distinct steps:Disruption: Complete disruption of plasma membranes of cells and organelles is absolutely required to release all the RNA contained in thesample. Incomplete disruption results in significantly reduced RNA yields. Homogenization: Homogenization is necessary to reduce the viscosity of the lysates produced by disruption. Homogenization shears high-molecular-weight genomic DNA and other high-molecular-weight cellular components to create a homogeneous lysate. Incomplete homogenization results in inefficient binding of RNA to the RNeasy spin column membrane and therefore significantly reduced RNA yields.Disruption and homogenization of tissue samples can be carried out rapidly and efficiently using either the TissueRuptor (for processing samples individually) or a TissueLyser system (for processing multiple samples simultaneously). Disruption and homogenization with TissueRuptor and TissueLyser systems generally results in higher RNA yields than with other methods.Disruption and homogenization using the TissueRuptorThe TissueRuptor is a rotor–stator homogenizer that thoroughly disrupts and simultaneously homogenizes single tissue samples in the presence of lysis buffer in 15–90 seconds, depending on the toughness and size of the sample. The blade of the TissueRuptor disposable probe rotates at a very high speed, causing the sample to be disrupted and homogenized by a combination of turbulence and mechanical shearing. For guidelines on using the TissueRuptor, refer to the TissueRuptor Handbook. For other rotor–stator homogenizers, refer to suppliers’ guidelines.Disruption and homogenization using TissueLyser systemsIn bead-milling, tissues can be disrupted by rapid agitation in the presence ofbeads and lysis buffer. Disruption and simultaneous homogenization occur bythe shearing and crushing action of the beads as they collide with the cells. Twobead mills are available from QIAGEN: the TissueLyser LT for low- to medium-throughput disruption, and the TissueLyser II for medium- to high-throughputdisruption.The TissueLyser LT disrupts and homogenizes up to 12 samples at the same time. The instrument needs to be used in combination with the TissueLyser LT Adapter, which holds 12 x 2 ml microcentrifuge tubes containing stainless steel beads of 5 mm or 7 mm mean diameter. For guidelines on using the TissueLyser LT, refer to the TissueLyser LT Handbook.The TissueLyser II disrupts and homogenizes up to 48 tissue samplessimultaneously when used in combination with the TissueLyser Adapter Set2 x 24, which holds 48 x 2 ml microcentrifuge tubes containing stainless steelbeads of 5 mm mean diameter. For guidelines on using the TissueLyser II, refer to the TissueLyser Handbook. If using other bead mills for sample disruption and homogenization, refer to suppliers’ guidelines.Note: Tungsten carbide beads react with QIAzol Lysis Reagent and must not be used to disrupt and homogenize tissues.The TissueLyser II can also disrupt and homogenize up to 192 tissue samplessimultaneously when used in combination with the TissueLyser Adapter Set2 x 96, which holds 192 x 1.2 ml microtubes containing stainless steel beads of 5 mm mean diameter. In this case, we recommend using the RNeasy 96 Universal Tissue Kit, which provides high-throughput RNA purification from all types of tissue in 96-well format. For ordering information, see page 48.Protocol: Purification of Total RNA from Animal CellsDetermining the correct amount of starting materialIt is essential to use the correct amount of starting material in order to obtain optimal RNA yield and purity. The minimum amount is generally 100 cells, while the maximum amount depends on:The RNA content of the cell typeThe DNA removal capacity of the gDNA Eliminator spin columnThe RNA binding capacity of the RNeasy spin column (100 μg RNA)The volume of Buffer RLT Plus required for efficient lysis (the maximum volume of Buffer RLT Plus that can be used limits the maximum amount of starting material to 1 x 107 cells)RNA content can vary greatly between cell types. The following examples illustrate how to determine the maximum amount of starting material:COS cells have high RNA content (approximately 35 μg RNA per 106 cells).Do not use more than 3 x 106 cells, otherwise the RNA binding capacity of the RNeasy spin column will be exceeded.HeLa cells have average RNA content (approximately 15 μg RNA per 106 cells). Do not use more than 7 x 106 cells, otherwise the RNA bindingcapacity of the RNeasy spin column will be exceeded.NIH/3T3 cells have low RNA content (approximately 10 μg RNA per 106 cells). The maximum amount of starting material (1 x 107 cells) can beused.If processing a cell type not listed in Table 2 (page 13) and if there is no information about its RNA content, we recommend starting with no more than 3–4 x 106 cells. Depending on RNA yield and purity, it may be possible to increase the cell number in subsequent preparations.Do not overload the gDNA Eliminator spin column, as this will lead to copurification of DNA with RNA. Do not overload the RNeasy spin column, as this will significantly reduce RNA yield and purity.As a guide, Table 3 (page 14) shows the expected numbers of HeLa cells in different cell-culture vessels.Important points before startingIf using the RNeasy Plus Mini Kit for the first time, read “Important Notes”(page 12).If preparing RNA for the first time, read Appendix A (page 34).Cell pellets can be stored at –70ºC for later use or used directly in the procedure. Determine the number of cells before freezing. Frozen cellpellets should be thawed slightly so that they can be dislodged by flickingthe tube in step 2. Homogenized cell lysates from step 3 can be stored at –70ºC for several months. Frozen lysates should be incubated at 37ºC in awater bath until completely thawed and salts are dissolved. Avoidprolonged incubation, which may compromise RNA integrity. If anyinsoluble material is visible, centrifuge for 5 min at 3000–5000 x g.Transfer supernatant to a new RNase-free glass or polypropylene tube, andcontinue with step 4.β-mercaptoethanol (β-ME) must be added to Buffer RLT Plus before use.Add 10 μl β-ME per 1 ml Buffer RLT Plus. Dispense in a fume hood andwear appropriate protective clothing. Buffer RLT Plus is stable at roomtemperature (15–25ºC) for 1 month after addition of β-ME.Buffer RPE is supplied as a concentrate. Before using for the first time, add4 volumes of ethanol (96–100%), as indicated on the bottle, to obtain aworking solution.Buffer RLT Plus may form a precipitate during storage. If necessary, redissolve by warming, and then place at room temperature.Buffer RLT Plus and Buffer RW1 contain a guanidine salt and are therefore not compatible with disinfecting reagents containing bleach. See page 6 for safety information.Perform all steps of the procedure at room temperature. During the procedure, work quickly.Perform all centrifugation steps at 20–25ºC in a standard microcentrifuge.Ensure that the centrifuge does not cool below 20ºC.Procedure1.Harvest cells according to step 1a or 1b.1a. Cells grown in suspension (do not use more than 1 x 107 cells):Determine the number of cells. Pellet the appropriate number of cells by centrifuging for 5 min at 300 x g in a centrifuge tube (notsupplied). Carefully remove all supernatant by aspiration, andproceed to step 2.Note: Incomplete removal of cell-culture medium will inhibit lysis and dilutethe lysate, affecting the conditions for DNA removal and RNA purification.Both effects may reduce RNA quality and yield.1b. Cells grown in a monolayer (do not use more than 1 x 107 cells): Cells grown in a monolayer in cell-culture vessels can be either lyseddirectly in the vessel (up to 10 cm diameter) or trypsinized andcollected as a cell pellet prior to lysis. Cells grown in a monolayer incell-culture flasks should always be trypsinized.To lyse cells directly:Determine the number of cells. Completely aspirate the cell-culturemedium, and proceed immediately to step 2.Note: Incomplete removal of cell-culture medium will inhibit lysis and dilutethe lysate, affecting the conditions for DNA removal and RNA purification.Both effects may reduce RNA quality and yield.To trypsinize and collect cells:Determine the number of cells. Aspirate the medium, and wash thecells with PBS. Aspirate the PBS, and add 0.10–0.25% trypsin in PBS.After the cells detach from the dish or flask, add medium (containingserum to inactivate the trypsin), transfer the cells to an RNase-freeglass or polypropylene centrifuge tube (not supplied), and centrifugeat 300 x g for 5 min. Completely aspirate the supernatant, andproceed to step 2.Note: Incomplete removal of cell-culture medium will inhibit lysis and dilutethe lysate, affecting the conditions for DNA removal and RNA purification.Both effects may reduce RNA quality and yield.2.Disrupt the cells by adding Buffer RLT Plus.For pelleted cells, loosen the cell pellet thoroughly by flicking thetube. Add the appropriate volume of Buffer RLT Plus (see Table 5).Vortex or pipet to mix, and proceed to step 3.Note: Incomplete loosening of the cell pellet may lead to inefficient lysisand reduced RNA yields. Ensure that β-ME is added to Buffer RLT Plusbefore use (see “Important points before starting”).Table 5. Volumes of Buffer RLT Plus for lysing pelleted cellsNumber of pelleted cells Volume of Buffer RLT Plusμl<5 x 106 350 5 x 106 – 1 x 107600 μlFor direct lysis of cells grown in a monolayer, add the appropriate volume of Buffer RLT Plus (see Table 6) to the cell-culture dish.Collect the lysate with a rubber policeman. Pipet the lysate into amicrocentrifuge tube (not supplied). Vortex or pipet to mix, andensure that no cell clumps are visible before proceeding to step 3.Note: Ensure that β-ME is added to Buffer RLT Plus before use (see“Important points before starting”).Table 6. Volumes of Buffer RLT Plus for direct cell lysisDish diameter Volume of Buffer RLT Plus*<6 cm 350 μl6–10 cm 600 μl* Regardless of the cell number, use the buffer volumes indicated to completely cover the surface of the dish.3.Homogenize the lysate according to step 3a, 3b, or 3c.See “Disrupting and homogenizing starting material”, page 15, for more details on homogenization. If processing ≤1 x 105 cells, they can behomogenized by vortexing for 1 min. After homogenization, proceed tostep 4.Note: Incomplete homogenization leads to significantly reduced RNA yields and can cause clogging of the RNeasy spin column. Homogenization with a rotor–stator or QIAshredder homogenizer generally results in higher RNA yields than with a syringe and needle.3a. Pipet the lysate directly into a QIAshredder spin column placed in a2 ml collection tube, and centrifuge for 2 min at maximum speed.Proceed to step 4.3b. Homogenize the lysate for 30 s using the TissueRuptor. Proceed to step 4.3c. Pass the lysate at least 5 times through a 20-gauge needle (0.9 mm diameter) fitted to an RNase-free syringe. Proceed to step 4.4.Transfer the homogenized lysate to a gDNA Eliminator spin columnplaced in a 2 ml collection tube (supplied). Centrifuge for 30 s at≥8000 x g (≥10,000 rpm). Discard the column, and save the flow-through.Note: Make sure that no liquid remains on the column membrane aftercentrifugation. If necessary, repeat the centrifugation until all liquid haspassed through the membrane.。
总RNA提取试剂盒(配合TRIZOL使用)Catalog No. TR205-50 (50次反应含DNaseI)TR205-200 (200次反应含DNaseI)TR205-50N (50次反应无DNaseI)TR205-200N (200次反应无DNaseI)Highlights∙适用于从细胞,组织,酵母菌,植物,细菌或生物液体(任何TRIZOL或类似产品可以裂解的样品)中提取到总RNA(含microRNA)。
∙无需进行相分离,无需使用氯仿,无需沉淀过程。
∙提取到的RNA不含DNA,并可应用于Q-PCR,高通量测序等实验。
∙整个过程仅需10分钟。
Ver.1.0.9产品组成:50200RNA洗涤液1 室温40 ml 160 ml第一次使用前按说明加指定量乙醇RNA洗涤液2室温12 ml 48 ml第一次使用前按说明加指定量乙醇DNase I -20℃ 300μl x 1管(5U/μl) 300μl x 4管(5U/μl)DNA消化液室温 4 ml 16 mlRNase-free H2O 室温 6 ml 30 ml2号柱室温50个200个收集管(2ml)室温50个200个特性:1.样品来源:TRIZOL等类似试剂裂解的样品。
2.样品范围:≥17核苷酸。
3.RNA纯度:高质量的RNA可应用于高通量测序等敏感的下游实验。
4.RNA结合量:每个柱子可最多结合50μg的RNA。
试剂制备RNA洗涤液在使用之前一定要配好,添加好乙醇后在试剂瓶上做好标记!!!添加10ml或40ml的无水乙醇(95-100%)到40ml或160ml的RNA洗涤液1中。
添加48ml100%的乙醇(或52 ml95%的乙醇)到12ml的RNA洗涤液2添加192ml100%的乙醇(或208 ml95%的乙醇)到48ml的RNA洗涤液2操作步骤:整个操作步骤是由2个步骤组成:(I)样品裂解匀浆(I I)样品纯化(I)样品裂解匀浆此步骤主要参考TRIZOL等类似裂解试剂说明书的裂解液用量,以下给出我公司提供的可完美替换TRIZOL的TRIcom试剂用量作为参考。

RT-PCR实验方法大全(抽提RNA,RT,PCR)-6可能问题出在标本的保存:一般四小时之内就应处理,分离出细胞说的是套式PCR,可以在你的第一次PCR两个引物内,再设计一对引物进行第二次PCR就行了如果你的第一次PCR刚好包括目的片段,那只好设计个更长的了第二次的引物设计要求可以低一点以50μl体系为例引物各1μl第一次PCR产物5μl二次PCR和巢式PCR,即设计两对引物进行扩增,不是一个概念,它是拿第一次的PC R产物,稀释100-1000倍做模板,加入底物,从新进行扩增反应,以期增加产物的量我做RT-PCR时,提总RNA时,都是用灭菌DEPC水,按1:100稀释后测 OD260和OD280,后根据公式:RNA浓度=OD260*稀释度/25(ug/ul),后用1mg total RNA分离mRNA.做逆转录及PCR,效果很好.luoyu10 wrote:各位大哥:我有一个问题请教,RT- PCR要求模板RNA的260nm/280nm的比值最低为多少,如果太低是不是会影响结果?最低到1.8,最好2.0,我感觉稍微低一点影响不算太大。
问:我是RT-PCR的新手,想请教引物如何设计?好的引物所具有的令人满意的特点:* 典型的引物18到24个核苷长。
引物需要足够长,保证序列独特性,并降低序列存在于非目的序列位点的可能性。
但是长度大于24核苷的引物并不意味着更高的特异性。
较长的序列可能会与错误配对序列杂交,降低了特异性,而且比短序列杂交慢,从而降低了产量。
* 选择GC含量为40%到60%或GC含量反映模板GC含量的引物。
* 设计5'端和中间区为G或C的引物。
这会增加引物的稳定性和引物同目的序列杂交的稳定性。
* 避免引物对3'末端存在互补序列,这会形成引物二聚体,抑制扩增。
* 避免3'末端富含GC。
设计引物时保证在最后5个核苷中含有3个A或T。
* 避免3'末端的错误配对。

Takara总RNA提取纯化和反转录1.总RNA的提取、纯化和反转录(Takara)1.1 总RNA的提取(1)取新鲜或者-80℃冻存的组织,添加液氮充分研磨,约每100 mg组织加入1mL RNAiso plus裂解,振荡混匀后室温静置5 min;(2)以每毫升RNAiso对应0.2 mL氯仿的比例加入氯仿,振荡15 s,室温静置3 min;(3)12000 g,4℃,离心15 min,取上清至新的Eppendorf管中(上层:无色水相为RNA,中层:白色为DNA,下层:红色为蛋白);(4)在得到的水相中加入等体积异丙醇,混匀后-20℃放置20~30 min;(提取dsRNA时,加入等体积的异丙醇和3M NaCl混合液(异丙醇:3M NaCl=1:1))(5)12000 g,4℃,离心10 min,去除上清;(6)加入1 mL75%乙醇(DEPC水和无水乙醇配置,-20℃预冷)洗涤沉淀;(7)8000 g,4℃,离心5 min,去除上清,干燥RNA沉淀;(8)用20~30 μL DEPC水溶解RNA;(9)通过甲醛琼脂糖凝胶电泳判断RNA完整性,用Nanodrop 紫外分光光度仪检测RNA纯度和浓度。
1.2 总RNA的纯化1) 在微量离心管中配制下列反应液。
10×DNase I Buffer 5 μLTotal RNA 20~50 μgRecombinant DNase I (RNase-free) 2 μL(10 units)RNase Inhibitor 20 units补DEPC处理的ddH2O至50 μL2) 37℃反应20~30 min。
3) 按以下方法失活Recombinant DNase I。
(1)加入2.5 μL0.5 M EDTA,混匀,80℃保持2 min。
(2)用DEPC处理的ddH2O定容至100 μL。
4) 加入10 μL 3 M醋酸钠和250 μL乙醇,混匀后-80℃放置20 min。
TaKaRa Code N0.9769 TaKaRa MiniBEST Plant RNA Extration Kit 说明书
一、制品说明
本试剂盒是小量提取植物组织Total RNA 的试剂盒。
试剂盒采用了独特的裂解系统,可以对各种简单的植物材料(叶片、茎、幼苗等)富含多糖多酚的植物组织材料(果实、种子)、真菌等进行RNA的提取,适用范围广。
按照标准流程使用本试剂盒可以有效提取分子量大于200nt的RNA,也可以按照可选步骤提取得到包含Small RNA(<200nt)的Total RNA。
试剂盒中包含了RNA提取所需的全部是局,无需额外购买其他试剂。
本试剂盒具有高效、快速、方便的特点,组织或自爆裂解后,提取操作仅需要30min 便可完成操作。
提取过程中无需苯酚氯仿抽提等步骤。
利用该试剂盒提取的RNA纯度高,基本不含蛋白质及及基因组DNA污染。
本试剂盒可以从50mg—100mg植物组织中纯化得到多至数十微克的高纯度RNA。
提取得到的RNA可以直接用于Northern杂交、斑点杂交、mRNA纯化、体外翻译、RNA分解酶的保护分析、RT-PCR、Real Time PCR、构建cDNA文库等各种分子生物学实验。
二、制品内容(50次量)
本试剂盒分两部分|Part 1和Part 2。

1.总RNA的提取、纯化和反转录(Takara)1.1 总RNA的提取(1)取新鲜或者-80℃冻存的组织,添加液氮充分研磨,约每100 mg组织加入1mL RNAiso plus裂解,振荡混匀后室温静置5 min;(2)以每毫升RNAiso对应0.2 mL氯仿的比例加入氯仿,振荡15 s,室温静置3 min;(3)12000 g,4℃,离心15 min,取上清至新的Eppendorf管中(上层:无色水相为RNA,中层:白色为DNA,下层:红色为蛋白);(4)在得到的水相中加入等体积异丙醇,混匀后-20℃放置20~30 min;(提取dsRNA时,加入等体积的异丙醇和3M NaCl混合液(异丙醇:3M NaCl=1:1))(5)12000 g,4℃,离心10 min,去除上清;(6)加入1 mL75%乙醇(DEPC水和无水乙醇配置,-20℃预冷)洗涤沉淀;(7)8000 g,4℃,离心5 min,去除上清,干燥RNA沉淀;(8)用20~30 μL DEPC水溶解RNA;(9)通过甲醛琼脂糖凝胶电泳判断RNA完整性,用Nanodrop紫外分光光度仪检测RNA纯度和浓度。
1.2 总RNA的纯化1) 在微量离心管中配制下列反应液。
10×DNase I Buffer 5 μLTotal RNA 20~50 μgRecombinant DNase I (RNase-free) 2 μL(10 units)RNase Inhibitor 20 units补DEPC处理的ddH2O至50 μL2) 37℃反应20~30 min。
3) 按以下方法失活Recombinant DNase I。
(1)加入2.5 μL0.5 M EDTA,混匀,80℃保持2 min。
(2)用DEPC处理的ddH2O定容至100 μL。
4) 加入10 μL 3 M醋酸钠和250 μL乙醇,混匀后-80℃放置20 min。
5) 4℃,12 000 g离心10 min,弃上清。
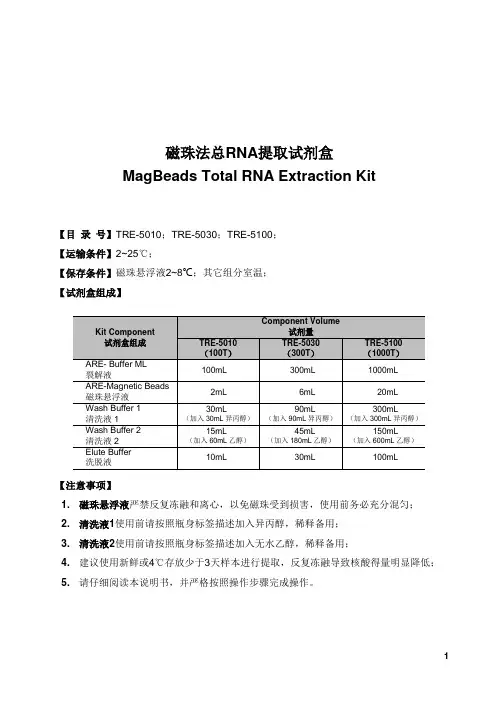
磁珠法总RNA提取试剂盒MagBeads Total RNA Extraction Kit【目录号】TRE-5010;TRE-5030;TRE-5100;【运输条件】2~25℃;【保存条件】磁珠悬浮液2~8℃;其它组分室温;【试剂盒组成】【注意事项】1. 磁珠悬浮液严禁反复冻融和离心,以免磁珠受到损害,使用前务必充分混匀;2. 清洗液1使用前请按照瓶身标签描述加入异丙醇,稀释备用;3. 清洗液2使用前请按照瓶身标签描述加入无水乙醇,稀释备用;4. 建议使用新鲜或4℃存放少于3天样本进行提取,反复冻融导致核酸得量明显降低;5. 请仔细阅读本说明书,并严格按照操作步骤完成操作。
磁珠法·自动化:为生命科学提供自动化磁纳米捕获方案【产品简介】本试剂盒适用于从植物材料、动物组织、各种微生物、培养细胞等样本中提取总RNA。
试剂盒采用独特分离作用的纳米磁珠和缓冲液体系,磁珠表面修饰有特殊化学基团,在一定条件下对核酸具有极强的富集能力,当条件改变时可以可逆的释放所吸附的核酸,从而达到快速分离纯化核酸的目的。
试剂盒经过磁珠与核酸结合、清洗、洗脱等步骤,最大限度的去除了蛋白质及其它杂质,所得RNA产物纯度优良,可直接应用于酶切、PCR、qPCR、文库构建、测序等各种下游分子生物学实验。
【试剂盒说明】仪器自动版英芮诚ETP-300型全自动核酸提取仪、核酸提取仪配套用磁棒套、EP管离心机、涡旋振荡仪、96孔方孔圆底板、EP管、无水乙醇、异丙醇以及氯仿、PBS溶液(1×)。
手动版水浴锅或金属浴、EP管离心机、涡旋振荡仪、EP管、EP管配套用磁力架、无水乙醇、异丙醇、氯仿、PBS溶液(1×)。
【手动版操作步骤】1. 样本前处理及裂解1)贴壁培养细胞倒弃培养液,用PBS溶液(1×)冲洗一次,按照每10cm2生长细胞中加入1mL的比例加入裂解液,轻微晃动使裂解液均匀分布于细胞表面,将内含细胞的裂解液转移至EP管中,涡旋振荡至无明显颗粒物,室温静置5min。
TaKaRa提取RNA说明书
TaKaRa Code N0.9769 TaKaRa MiniBEST Plant RNA Extration Kit 说明书
一、制品说明
本试剂盒是小量提取植物组织T otal RNA 的试剂盒。
试剂盒采用了独特的裂解系统,可以对各种简单的植物材料(叶片、茎、幼苗等)富含多糖多酚的植物组织材料(果实、种子)、真菌等进行RNA的提取,适用范围广。
按照标准流程使用本试剂盒可以有效提取分子量大于200nt的RNA,也可以按照可选步骤提取得到包含Small RNA(<200nt)的T otal RNA。
试剂盒中包含了RNA提取所需的全部是局,无需额外购买其他试剂。
本试剂盒具有高效、快速、方便的特点,组织或自爆裂解后,提取操作仅需要30min 便可完成操作。
提取过程中无需苯酚氯仿抽提等步骤。
利用该试剂盒提取的RNA纯度高,基本不含蛋白质及及基因组DNA污染。
本试剂盒可以从50mg—100mg植物组织中纯化得到多至数十微克的高纯度RNA。
提取得到的RNA可以直接用于Northern 杂交、斑点杂交、mRNA纯化、体外翻译、RNA分解酶的保护分析、RT-PCR、Real Time PCR、构建cDNA文库等各种分子生物学实验。
二、制品内容(50次量)
本试剂盒分两部分|Part 1和Part 2。
RNA 抽提及荧光定量PCR试剂∙RNAiso Plus (Takara , cat. no. 9109) ∙氯仿(国药) ∙异丙醇(国药) ∙75%乙醇(DEPC 水配制) ∙DEPC (生工,cat.no. DB0154) ∙ DEPC 水:向超纯水中加入DEPC 至终浓度0.1%(v/v ),搅拌过夜后,高温高压灭菌,以降解除去DEPC (DEPC 分解为CO2和乙醇)。
Note :DEPC 气味芳香浓烈,强挥发性,有毒,毒性并不是很强,但吸入的毒性是最强的,使用时戴口罩,需在通风橱中操作。
∙M-MLV Reverse Transcriptase (RNase H+)(华大,cat.no. EM-004) ∙Taq DNA Polymeras (tiangen ,cat.no. ET101-02) ∙ SYBR Green I (百唯信)∙ RQ1 RNase-Free DNase (Promega ,cat.no.M6101)操作步骤RNA 抽提1.实验样品的研磨和匀浆A.贴壁培养细胞去除培养液,用1*PBS 清洗一次,加入适量RNAiso Plus (依据培养板的面积来决定所需的RNAiso Plus 量,每10 cm 2加1 ml ,即6-well plate 的一个孔加1ml ),使裂解液均匀分布于细胞表面,将内含细胞的裂解液转移至离心管中,用移液枪反复吹吸直至裂解液中无明显沉淀。
B.悬浮培养细胞离心收集细胞,加入适量RNAiso Plus (每5-10×106个细胞加1ml 的RNAiso Plus ),枪反复吹吸直至裂解液中无明显沉淀。
破裂某些酵母菌和细菌可能需要使用匀浆器破细胞壁。
C.组织样品将组织在液氮中磨碎,每50~100mg 组织加入1ml RNAiso Plus ,用匀浆仪进行匀浆处理。
样品体积不应超过RNAiso Plus 体积的10℅。
Note :1.所用RNAiso Plus 的量要适当,量多浪费,不足时可导致抽提的RNA 中污染有DNA 。
RNA提取:用TaKaRa RNAiso Reagent试剂提取RNA步骤2007-12-10 13:52试验前准备的物品(1).试剂盒之外所需准备试剂:◆氯仿◆异丙醇◆ 75%乙醇(DEPC 处理水配制)◆ RNase-free 水(制备方法:使用 RNase-free 的玻璃瓶,向超纯水中加入 DEPC 至终浓度 0.01%(v/v),过夜搅拌后,高温高压灭菌。
(2).尽量使用一次性塑料器皿,若用玻璃器皿,应在使用前按下列方法进行处理。
a)用 0.1% DEPC(焦碳酸二乙酯)水溶液在37℃下处理 12 小时。
b)然后在120℃下高压灭菌 30 分钟以除去残留的 DEPC。
RNA 实验用的器具建议专门使用,不要用于其它实验。
(3) 用于 RNA 实验的试剂,须使用干热灭菌(180℃,60 分钟)或使用上述方法进行 DEPC 水处理灭菌后的玻璃容器盛装(也可以使用 RNA 实验用的一次性塑料容器),使用的无菌水须用 0.1%的 DEPC 处理后再进行高温高压灭菌。
RNA 实验用的试剂和无菌水都应专用,避免混用后交叉污染。
实验操作1. RNAiso Reagent 的使用量情况如下。
2. 实验样品的研磨和匀浆。
A. 贴壁培养细胞①倒出培养液,用1×PBS 清洗一次。
②每10 cm2生长的培养细胞中加入 2 ml的RNAiso Reagent,水平放置片刻,使裂解液均匀分布于细胞表面并裂解细胞,然后使用移液枪吹打细胞使其脱落(对于贴壁牢固的培养细胞可用细胞刮勺剥离细胞)。
③将内含细胞的裂解液转移至离心管中,用移液枪反复吹吸直至裂解液中无明显沉淀。
④室温静置 5 分钟。
B. 悬浮培养细胞①将悬浮培养细胞连同培养液一起倒入离心管中,8,000 g 4℃离心 2 分钟,弃上清。
②向每5×106个细胞中加入l ml的RNAiso Reagent。
③用移液枪反复吹吸直至裂解液中无明显沉淀。
takara反转录试剂盒说明书关键信息项:1、试剂盒名称:Takara 反转录试剂盒2、适用样本类型:____________________________3、反应体系:____________________________4、反应条件:____________________________5、储存条件:____________________________6、有效期:____________________________1、产品简介11 Takara 反转录试剂盒是一种用于将 RNA 反转录为 cDNA 的高效、可靠的工具。
111 本试剂盒经过精心设计和优化,能够提供高质量的反转录产物,适用于各种下游分子生物学实验。
2、试剂盒组成21 反转录酶211 具有高活性和热稳定性,确保反转录反应的高效进行。
22 反转录缓冲液221 包含各种必要的成分,为反转录反应提供适宜的环境。
23 RNA 酶抑制剂231 有效抑制 RNA 酶的活性,防止 RNA 降解。
24 随机引物或 oligo(dT)引物241 可根据实验需求选择合适的引物进行反转录。
25 dNTP 混合物251 提供四种脱氧核糖核苷酸,用于 cDNA 合成。
3、适用样本类型31 总 RNA311 包括从细胞、组织、血液等样本中提取的总 RNA。
32 mRNA321 经过纯化或富集的 mRNA 样本。
4、反应体系41 建议的反应体系如下:411 RNA 模板:X μL(根据 RNA 浓度和实验需求确定)412 随机引物或 oligo(dT)引物:Y μL413 dNTP 混合物:Z μL414 反转录缓冲液:A μL415 RNA 酶抑制剂:B μL416 反转录酶:C μL417 无 RNase 水:补充至总体积D μL5、反应条件51 反转录反应的温度和时间可根据引物类型和RNA 质量进行调整。
511 使用随机引物时,反应条件通常为:25°C 孵育 10 分钟,然后42°C 孵育 30 60 分钟。