不同标记荧光基团的稳定性、半衰期介绍说明
- 格式:pdf
- 大小:247.38 KB
- 文档页数:33

时间分辨技术的原理目前最先进的免疫检测技术:1、时间分辨原理:用三价稀土离子及其鳌合剂作为示踪物,代替荧光物质、同位素或酶,标记蛋白质、多胎、激素、抗体、核酸探针或生物流行性细胞,当反应体系发生后,用时间分辨仪器测定最后产物中荧光强度。
根据荧光强度或相对荧光强度比值,来判断反应体系中分析物的浓度,达到定量分析之目的。
2、时间分辨原子标记物的特点:●发射光和激发光有较大的STOKES位移——高特异性●长寿命荧光,降低其他物质的荧光干扰——高灵敏度●半衰期长达几十万年,试剂受干扰小——高稳定性原子标记与大分子标记物的对比3、波长分辨:●标记离子的荧光激发光波长范围较宽,发射光光谱范围较窄,是类线光谱,有利于降低本底荧光强度,提高分辨率。
●激发与发射光之间有一个较大的STOKES位移,有利于排除非特异荧光的干扰,增强测量的特异性。
4、时间分辨:●标记离子螯合物产生的荧光强度高,寿命长,有利于消除样品及环境中荧光物质对检测结果的影响。
●每一秒名检测样品1000次,取其中不受干扰的400次的均值作为测定值,有利于提高检测的准确性。
时间分辨技术取代酶免、放免是免疫检测技术发展的必然趋势!RIA(放免)●放射性(125I),对环境和身体的危害,已经为重视环保的国家逐步取消,如整个欧洲仅尚存几个放免试验室。
●125I半衰期短,导致试剂的有效期短,需每次定标,造成很大的浪费。
●由于标记物(125I)的不断变化,带来药盒批间、批内较大的变异,标准曲线无法保存备用。
ELESA(酶免)●灵敏度、重复性不及放免,易造成漏检和假阳性。
●酶的纯度和反应过程容易受环境因素影响,导致稳定性、重复性不好。
与其它技术的相对优势:(1)、是现有的免疫检测方法中灵敏度最高的(2)、是现有的免疫检测方法中稳定性最好的(3)、多标记检测是目前所有免疫检测技术中独一无二的TRF技术与电化学发光的比较TRF与化学发光的比较时间分辨荧光免疫定量分析简介时间分辨荧光分析(Timeresolved Fluoroimmunoassay,TRFIA)是一种非同位素免疫分析技术,它用镧系元素标记抗原或抗体,根据镧系元素螯合物的发光特点,用时间分辨技术测量荧光,同时检测波长和时间两个参数进行信号分辨,可有效地排除非异荧光的干扰,极大地提高了分析灵敏度。

摘要:在进行基因分离、克隆和核苷酸序列分析等生物过程中,通常会运用到聚合酶链反应技术,即PCR技术。
荧光PCR是近年来科学家和学者常用的手段,本文就对荧光PCR的原理、常用探针的优缺点以及多重荧光PCR技术的进展进行了综述。
关键词:荧光PCR;探针;多重荧光PCR聚合酶链反应( PCR) 技术自1985年问世以来,以其灵敏性高、特异性强和速度快在分子生物学等科研领域得到了广泛应用[1]。
但是,利用传统的PCR 技术进行检测鉴定时,需要进行扩增反应后的电泳分离及染色处理,且不能准确定量,使其应用受到限制[2]。
荧光PCR 技术拥有特异性强、灵敏度高、重复性好、定量准确、速度快、全封闭反应等优点,已成为了分子生物学研究的重要工具[1]。
1、荧光PCR原理荧光PCR的原理是以荧光共振能量转移原理为基础。
荧光共振能量转移原理是当一个荧光分子(供体分子)的荧光光谱与另一个荧光分子(受体分子)的激发光谱重叠时,供体荧光分子自身的荧光强度衰减,受体荧光分子的荧光强度增强。
Ct值是指每个反应管内的荧光信号到达设定的阈值时所经历的循环数。
Ct值是实时荧光PCR 中一个很关键的因素,C 代表循环(Cycle),t代表阈值(Threshold)。
每个模板的Ct 值与该模板的起始拷贝数的对数存在线性关系,起始拷贝数越多,Ct 值越小。
利用已知起始拷贝数的标准品可做出标准曲线。
因此,根据荧光探针的发光基团所发出的荧光强度与PCR 产物的数量呈对应关系,只要对荧光信号进行检测并获得未知样品的Ct值,即可从标准曲线上计算出该样品的起始拷贝数。
与普通PCR相比,荧光PCR可以利用荧光信号实时监测PCR反应过程中每一个循环扩增产物的变化,可以对初始模板量进行定量分析。
概括地说,荧光PCR的基本原理就是样本核酸扩增呈指数增长,在反应体系和条件完全一致的情况下,样本DNA含量与扩增产物的对数成正比,由于反应体系中的荧光染料或荧光标记物(荧光探针)与扩增产物结合发光,其荧光量与扩增产物量成正比,因此通过荧光量的检测就可以测定样本核酸量[2]。
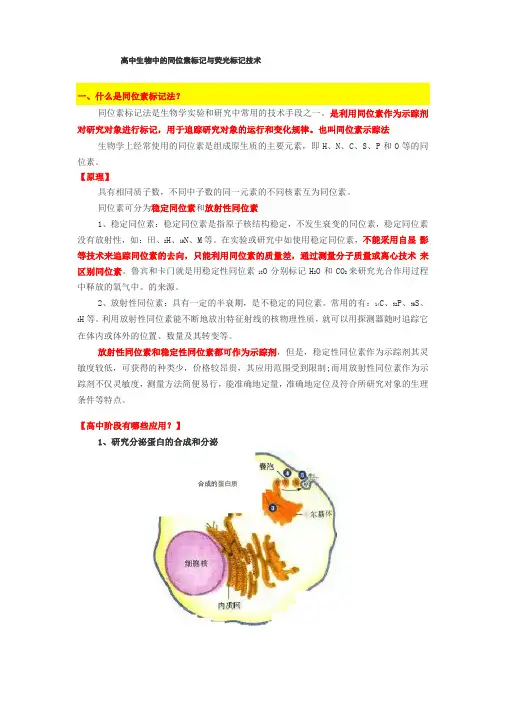
高中生物中的同位素标记与荧光标记技术同位素标记法是生物学实验和研究中常用的技术手段之一。
是利用同位素作为示踪剂对研究对象进行标记,用于追踪研究对象的运行和变化规律。
也叫同位素示踪法生物学上经常使用的同位素是组成原生质的主要元素,即H、N、C、S、P和O等的同位素。
【原理】具有相同质子数,不同中子数的同一元素的不同核素互为同位素。
同位素可分为稳定同位素和放射性同位素1、稳定同位素:稳定同位素是指原子核结构稳定,不发生衰变的同位素,稳定同位素没有放射性,如:田、2H、15N、M等。
在实验或研究中如使用稳定同位素,不能采用自显影等技术来追踪同位素的去向,只能利用同位素的质量差,通过测量分子质量或离心技术来区别同位素。
鲁宾和卡门就是用稳定性同位素18O分别标记H2O和CO2来研究光合作用过程中释放的氧气中。
的来源。
2、放射性同位素:具有一定的半衰期,是不稳定的同位素。
常用的有:14C、32P、35S、3H等。
利用放射性同位素能不断地放出特征射线的核物理性质,就可以用探测器随时追踪它在体内或体外的位置、数量及其转变等。
放射性同位素和稳定性同位素都可作为示踪剂,但是,稳定性同位素作为示踪剂其灵敏度较低,可获得的种类少,价格较昂贵,其应用范围受到限制;而用放射性同位素作为示踪剂不仅灵敏度,测量方法简便易行,能准确地定量,准确地定位及符合所研究对象的生理条件等特点。
【高中阶段有哪些应用?】1、研究分泌蛋白的合成和分泌合成的蛋白质高体研究细胞器在分泌蛋白合成中的作用时,标记某一氨基酸如亮氨酸的3H ,在一次性给予放射性标记的氨基酸的前提下,通过观察细胞中放射性物质在不同时间出现的位置,就可 以明确地看出细胞器在分泌蛋白合成和运输中的作用。
研究手段:观察放射性在不同细胞器1939年,鲁宾和卡门用18O 分别标记与。
和CO 2,然后进行两组对比实验:一组提供H 2O 和CM 2,另一组提供H 2M 和CO 2。
在其他条件相同情况下,分析出第一组释放的氧气全部为 O 2,第二组全部为M 2,有力地证明了植物释放的。

带氨基的荧光分子-概述说明以及解释1.引言1.1 概述在当今科学领域中,荧光分子作为一种重要的工具在生物、化学、材料等领域发挥着重要作用。
氨基的荧光分子,即含有氨基基团的荧光物质,具有独特的性质和应用潜力。
通过合理设计和合成,氨基的荧光分子可以展现出不同的荧光性质和生物活性,有望在生物成像、药物研发等方面发挥重要作用。
本文将着重探讨氨基的荧光分子的特性、合成方法与应用以及分子设计与发展趋势,旨在为读者提供关于这一领域的最新进展和未来发展方向的综合性了解。
通过深入探讨氨基的荧光分子的特性和应用价值,有助于拓展这一领域的研究和应用范围,推动相关领域的发展。
1.2 文章结构文章结构部分描述了整篇文章的组织和内容安排。
本文的结构分为引言、正文和结论三个部分。
引言部分包括概述、文章结构和目的。
在概述中,介绍了带氨基的荧光分子的主题,并简要描述了相关背景和意义。
文章结构部分则是本段,详细说明了整篇文章的目录和各部分之间的关系。
目的部分阐述了本文的目标和意义,指导读者对全文的阅读有一个初步了解。
正文部分包括氨基的荧光分子特性、合成方法与应用和分子设计与发展趋势三个主要内容。
在这部分中,将详细介绍带氨基的荧光分子的特性及其与荧光性质相关的重要性,介绍其合成方法和应用领域,并探讨分子设计和未来发展的趋势。
结论部分包括总结、展望和结论三个部分。
在总结部分,将对全文的主要内容进行回顾和总结,强调本文对于带氨基的荧光分子的研究意义。
展望部分将展望该领域未来的发展方向和潜在的应用前景。
最后,在结论部分将对全文进行一个简明扼要的总结,强调本文的主要贡献和重要意义。
1.3 目的本文的主要目的是探讨带氨基的荧光分子在化学和生物学领域中的重要性和应用。
我们将详细介绍氨基的荧光分子的特性、合成方法和应用领域,并探讨其在分子设计与发展趋势中的潜在价值。
通过深入研究带氨基的荧光分子,我们希望为相关领域的研究和应用提供深入的理解和启示,推动这一领域的持续发展和创新。

荧光定量PCR中探针的荧光修饰基团1. 荧光定量PCR两种方法荧光定量PCR技术是为了测定样本中特异的PCR片段相对及绝对量,是一种测定特异的PCR片段含量的方式。
如测定病人样本中病原体的含量、实验样本中某一特定的mRNA 的含量等。
为了达到高效反应,应将实时PCR应用的引物设计为能生成短的扩增子。
常用的方法有SYBR Green的荧光染料法和TaqMan探针法,两种方法的特点及应用如下表:名称SYBR Green染料法TaqMan探针法引物一对特异性的引物一对特异性的引物和一条探针原理SYBR Green一种DNA结合染料,能非特异地掺入到DNA双链的小沟当中去,在游离状态下,它不发出荧光,但一旦结合到双链DNA中以后,便可以发出荧光。
探针的5’端有一荧光报告基团,3’端有一荧光淬灭基团,当两个基团相互先靠近的时候,由于发生能量传递作用,报告基团不能发出荧光,但随着扩增反应的进行,5’端的报告基团随着探针的水解而脱落下来,不再与淬灭发生能量传递作用,从而能发出荧光,被信号探测器所捕获。
特点从实验成本来讲,SYBR Green是最好的,普通PCR加上一点SYBR Green荧光染料即可,其信号强度也很好,还可以进行融解曲线分析等,但缺点是只能在一个反应管内进行一种PCR反应的检测,出现非特异扩增会影响到定量结果的可靠性与重复性。
由于增加了探针的特异性,因此其扩增曲线反映的就是特异性产物的扩增曲线,不含有非特异性扩增的成分,其敏感性要比SYBR Green高出10倍。
其缺点在于探针成本较高,有时设计的探针并不合适,有造成损失的可能性。
并且要进行较多的实验条件的优化。
扩增长度SYBR Green法不超过500 bp,一般选择250-400bp,过大会影响到PCR扩增的效率,过小则很难通过融解曲线与引物二聚体分开。
TaqMan法的扩增片段都很小,几十或是100多,一般不超过300,通常为80-150bp。


HTRF (homogeneous time resolved fluorescence)HTRF®是均相时间分辨荧光技术的简称,HTRF®技术是对TR-FRET技术的进一步改良,提供了更高的灵敏度和稳定性,为法国Cisbio公司的专利技术。
HTRF®技术结合了FRET 和时间分辨荧光(TRF''Time Resolved Fluorescence))两种技术。
能量供体:Eu或Tb标记的穴状化合物元素;能量受体:XL665或d2----基于穴状化合物的供体与受体(第二荧光标记物)之间的荧光共振能量转移(FRET)。
在FRET中,受体发射荧光的寿命等同与供体的发射荧光的寿命。
因为Eu 的荧光衰减周期较长,所以含Eu的供体会诱导XL665受体长时间地发射荧光,受体激发后产生的荧光便能持续较长时间,这样通过时间分辨就可以区分那些短寿命的自身散射的荧光,这样从短寿命荧光背景中就很容易区分出FRET信号。
当由于生物分子相互作用导致两个荧光基团接近时,在激发时被穴状化合物捕获的部分能量释放,发射波长为620nm;另一部分能量转移到受体(acceptor)上,发射波长为665nm。
665nm的发射光仅仅由供体(donor)引起的FRET产生。
所以,当生物分子相互作用时,有两个激发光620nm和665nm;当不存在相互作用时,只有620nm一个激发光。
FRET利用两种荧光基团的能量转移,这两种荧光基团分别称为能量供体(Donor)和能量受体(Acceptor)。
Donor 被外来光源激发(例如氙灯或激光),如果它与Acceptor 比较接近,可以将能量共振转移到在Acceptor 上,使其受到激发,发出特定波长的发射光。
将Donor 和Acceptor 分别与相互作用的生物分子结合,生物分子的结合可以将Donor 和Acceptor 拉到足够近的距离,产生能量转移,由于Acceptor 的发射光来自能量转移,所以实验中不需要将未结合与已结合的分子分开,即不需要洗涤步骤。


HTRF 技术介绍快速、稳定、不需洗涤、操作简单、易于自动化和微型化。
上述优势使得Cisbio 的HTRF 技术一直是药物研发领域的领先技术之一,并广泛用于信号转导研究和免疫检测。
该技术已经在知名医药公司、生物技术公司和学术研究机构应用了15年以上。
HTRF (均相时间分辨荧光,Homogeneous Time-Resolved Fluorescence )是用来检测纯液相体系中待测物的一种常用方法。
该技术结合了荧光共振能量转移(FRET ,Fluorescence Resonance Energy Transfer )和时间分辨荧光 (TRF, Time-ResolvedFluorescence))两种技术。
这种结合将FRET的均相实验方式和TRF 的低背景特点融合在一起,使得HTRF 技术拥有如下优势:操作简单、灵敏度高、通量大、实验数据稳定可靠、假阳性率较低。
HTRF 是基于TR-FRET 的化学技术,拥有与其它TR-FRET 技术相似的特征,包括使用镧系元素(铕和铽),具有非常长的半衰期,很大的Stroke's shift (如图1所示,Eu 3+ Stroke’s shift > 300 nm )等。
除此之外,它还有其独特的性质,从而与其它技术区分开来。
这主要表现在HTRF 的镧系元素与络合的穴相结合,而不是像其它所有TR-FRET 技术使用螯合物。
螯合物在溶液中是一种动态平衡,在特定条件下不稳定;而HTRF 中应用的穴与镧系元素是永久地嵌合,非常稳定,可耐受较宽的pH 范围、二价金属离子如Mn 2+等、螯合剂如EDTA 等。
HTRF 的独特之处还包括对数据的专利的比值处理方法,其能校正样品基质不同等带来的干扰。
FRET 技术简介FRET 技术利用了两种荧光基团的能量转移,这两种荧光基团分别称为(能量)供体和(能量)受体,前者的发射光谱与后者的激发光谱重叠。
供体被外来能源激发(例如闪光灯或激光),如果它与受体在足够近的距离之内,可以将能量共振转移到受体上。

蛋白质荧光标记技术简介蛋白质荧光标记技术简介引言:蛋白质是生物体内最重要的大分子类别之一,参与了几乎所有生物过程的调控和执行。
了解蛋白质的定位、表达和相互作用等特性对于生命科学研究具有重要意义。
为了研究蛋白质在细胞中的行为,科学家们发展了多种荧光标记技术,这些技术使得蛋白质能够通过观察荧光信号实现可视化和检测。
本文将介绍蛋白质荧光标记技术的原理、应用以及该领域的最新进展。
一、蛋白质荧光标记技术的原理1. 荧光标记物选择蛋白质荧光标记技术需要选择合适的荧光标记物,通常使用的有荧光染料、荧光蛋白以及量子点等。
这些标记物具有不同的特性,例如染料具有较好的亮度和灵敏度,荧光蛋白具有较长的半衰期和较高的分子量,在选择时需要根据具体实验需求进行评估。
2. 标记物与蛋白质的结合标记物与蛋白质的结合有多种方法,包括共价结合和非共价结合。
共价结合通常采用交联剂或者双硫键等方式将标记物与蛋白质进行连接,而非共价结合则是通过亲和性标记物与蛋白质的特定位点结合。
3. 荧光信号的检测和分析经过标记的蛋白质可以通过荧光显微镜等设备进行观察,获得荧光信号。
这些信号可以通过图像处理和分析软件进行定量和定位分析,从而获得蛋白质在细胞中的空间分布和动态过程等信息。
二、蛋白质荧光标记技术的应用1. 蛋白质定位通过将蛋白质标记为荧光标记,可以直观地观察其在细胞中的定位。
这对于研究蛋白质在细胞器和亚细胞结构中的分布具有重要意义,同时也有助于发现异常定位与相关疾病之间的联系。
2. 蛋白质表达蛋白质荧光标记技术可用于检测蛋白质的表达水平和翻译后修饰等特性。
通过观察荧光信号的强度和分布,可以对蛋白质在细胞中的表达进行直观的观察和比较。
3. 蛋白质相互作用研究荧光标记技术为研究蛋白质之间的相互作用提供了有力的工具。
通过将不同蛋白质分别标记为不同的荧光色素,可以观察它们在细胞中的相互作用过程,深入了解蛋白质相互作用的特点和动力学等。
三、蛋白质荧光标记技术的最新进展1. 单分子荧光标记技术单分子荧光标记技术可以实现对单个蛋白质分子的荧光标记和观察。

常用DNA分子标记类型和特点DNA分子标记是一种广泛应用于生物学研究和诊断领域的技术,用于识别、检测和定量目标DNA序列。
常见的DNA分子标记类型包括荧光染料、酶和放射性同位素等。
每种标记类型都具有其独特的特点和应用场景。
荧光染料是DNA分子标记中最常用的类型之一、它们通过在DNA分子上附着荧光染料,使其在荧光显微镜下可见。
荧光染料具有多种颜色和化学性质,可用于多重标记和多个目标的同时检测。
其主要特点包括:1.高灵敏度:每个荧光染料分子都有较强的荧光信号,因此可以在微量样品中进行检测。
2.高选择性:荧光染料可以针对目标DNA序列进行选择性标记,从而实现目标分子的准确检测。
3.高兼容性:荧光染料可以与不同的DNA分析方法兼容,如凝胶电泳、荧光定量PCR等。
酶也是常用的DNA分子标记类型之一、通过将酶与DNA标记物结合,可以通过酶的催化反应产生可定量的信号。
常用的酶标记包括辣根过氧化物酶(HRP)和碱性磷酸酶(AP)。
其主要特点包括:1.高灵敏度:酶催化反应可以在大量酶底物的参与下放大信号,从而提高检测的灵敏度。
2.稳定性:酶标记的DNA可以在各种条件下稳定存在,并且可以长期保存。
3.可视性:酶催化反应可以产生可见的颜色或发光信号,从而直观地观察到标记物。
放射性同位素是DNA分子标记的传统方式之一、通过将放射性同位素与DNA标记物结合,可以通过放射性测量来定量目标DNA序列。
1.高灵敏度:放射性测量可以实现非常低浓度目标DNA的检测。
2.高特异性:放射性同位素标记DNA具有非常高的特异性,可以准确检测目标序列。
3.长期保存:放射性同位素标记的DNA可以长期保存,方便未来的回溯和再分析。
虽然放射性同位素标记具有较高的灵敏度和特异性,但其使用需要特殊的设备和技术,并且存在较高的辐射风险,因此在现代实验室中较少使用。
总结而言,DNA分子标记在生物学研究和诊断中起着至关重要的作用。
不同类型的DNA标记具有各自的特点和应用场景,研究人员可以根据实验需求选择合适的标记方式,以便实现高灵敏度、高特异性和可视化的目标DNA检测。
荧光衰减曲线荧光衰减曲线(Fluorescence Decay Curve)是一种用于研究特定分子的荧光度变化过程的统计方法。
它使用特定条件下分子荧光衰减的数据,可以被用来表征发射光的光学性质和物理性质。
荧光衰减曲线的原理在于,当特定的分子受到特定的电磁辐射时,其能量会发生变化,导致分子荧光强度的变化。
荧光衰减曲线可以通过测量不同时刻荧光信号的强度变化,来推断出荧光分子在受到电磁辐射后发生的变化情况。
荧光衰减曲线通常是基于直线拟合的,由此可以得到荧光衰减曲线的参数。
其中,最重要的参数是半衰期(Half-Life),即荧光衰减到原始强度的一半所需要的时间。
通过测量半衰期,可以进一步分析分子荧光衰减的物理机制。
另外,荧光衰减曲线的斜率也是一个重要参数,它可以反映分子状态的变化,以及受到电磁辐射后发生的化学反应。
例如,当斜率增大或减小时,可以推断出分子状态和受到的电磁辐射的变化。
荧光衰减曲线的另一个重要参数是初始衰减率(Initial Decay Rate),它可以反映物体受到电磁辐射后分子荧光衰减的速率。
另外,根据物体受到的电磁辐射的不同,可以得出不同的荧光衰减曲线,从而得出不同的参数。
最后,荧光衰减曲线也可以用来评估不同物质的光学性质。
例如,可以利用荧光衰减曲线来比较不同物质的荧光衰减率,从而判断其光学性质的差异。
总之,荧光衰减曲线是一种有效的研究特定分子的荧光度变化的方法。
它可以用来表征发射光的光学性质和物理性质,并且可以根据不同物体受到的电磁辐射的不同,得出不同的荧光衰减曲线,从而得出不同的参数。
此外,也可以用它来评估不同物质的光学性质。
因此,荧光衰减曲线在研究物理和化学领域具有重要的意义。
荧光系列对比操作方法
荧光系列主要分为染料和蛋白质标记两种。
以下是它们的操作方法对比:
一、荧光染料标记:
1. 样品制备:将要标记的生物分子(如蛋白质、核酸等)提取或纯化成一定浓度的溶液。
2. 染料标记:选用合适的荧光染料,按照其说明书中的操作方法将其与生物分子进行反应,使其标记上荧光染料。
3. 纯化:将标记后的生物分子通过某些手段(如凝胶过滤、离心等)分离出未反应的荧光染料和副产物等杂质。
4. 荧光检测:通过荧光光谱仪等仪器检测样品的荧光强度和光谱,判断染料标记的效果以及分子的定量等信息。
5. 保存:将标记好的样品分装保存,避免其被氧化、暴露于光或浓度过度稀释等情况下引起荧光信号的衰减或变异。
二、蛋白质标记:
1. 蛋白质制备:将要标记的蛋白质在无菌条件下制备成一定浓度的溶液。
2. 标记试剂制备:根据标记试剂的类型,选择适当的缓冲液、试剂浓度、反应温度和时间等条件,将标记试剂进行制备。
3. 标记反应:将制备好的标记试剂与蛋白质进行反应,使其标记上标记试剂。
4. 纯化和检测:将标记后的样品进行纯化和检测。
纯化过程中,通常采用柱层析等方法去除副产物和未反应的试剂;检测过程中,通常采用SDS-PAGE、western blot等方法对标记后的蛋白质进行检测。
5. 保存:将标记好的样品分装保存,避免其被氧化、暴露于光或浓度过度稀释等情况下引起荧光信号的衰减或变异。
Please refer to STYLE GUIDE.doc for detailed guidelinesColor code legene:Red = Proprietary; Pink = Discontinuation; Green = Anecdotal; Blue = Anything else customers will not see Custom Primers – All ModificationsTABLE OF CONTENTSPRODUCT DESCRIPTIONSHIPPING CONDITIONSSTORAGE CONDITIONSSTABILITYQC SPECIFICATIONSPROTOCOL & APPLICATION NOTESModification/scale/purificationManufacturing DetailsFluoresceinRhodamineHEX/TET/FAMPhosphateBiotinAmineGatewayAlexaOther dyesAlkaline PhosphataseHorse Radish PeroxidasePhosphorothioate“Fully-Phosphorothioated" Custom PrimersAldehydeAcridineThiolDelivery ScheduleOligoPerfect Designed Primers3' Modifications of OligosReconstitution ProtocolA260/A280 ratio of the oligoOligo VisualizationTroubleshooting“Custom Custom” modificationsList of current technology limitationsCOMPETITOR INFORMATIONALTERNATE PRODUCTS & COMPATIBILITYPRODUCT DOCUMENTATIONREFERENCESPRODUCT NAME & CATALOG NUMBERCOMPONENTSDISCONTINUATION NOTICEASSOCIATED PRODUCTSPRODUCT QUALITY ISSUESReturning PrimersLICENSINGINTERNAL CONTACTSPRODUCT DESCRIPTION(back to Table of Content)Custom DNA Primers are synthetic oligonucleotides with specified sequence made to order for use in a variety of applications from PCR and sequencing to probes for gene detection. Custom DNA Primers are available as standard deoxynucleotides, modified bases, 5´ modified nucleotides (including fluorescent dyes, enzyme conjugates), and S-oligos for antisense studies. Five scales and four purity levels are available.A unique and proprietary technology and an optimized production process is used to deliver quality Custom Primers, quickly and efficiently. The process includes easy ordering, linkage to integrated workstations for guaranteed accuracy, high-throughput synthesis, monitoring by a series of stringent quality control steps, useful product packaging, and overnight shipping for rapid, reliable delivery. Custom oligonucleotides are synthesized on automated, proprietary DNA synthesizers using standard cyanoethyl phosphoramidite chemistry. Consistent oligonucleotide quality is ensured by in-process trityl monitoring and quality control analysis. A comprehensive Certificate of Analysis accompanies each order, indicating quantity, molecular weight, extinction coefficient, sequence, and melting temperature.DNA oligos can be ordered in tubes or plates:•Tubes:o Standard tube with cap (Scientific Specialties Inc. (SSI) 2680-00 (tube), 2001-00 (cap)). Volume is 2000µl •Plates:o96-wells:PCR shallow 96-well plate (Abgene AB-0800): 200µl max volume, 180µl working volume; plate code 96N – 180. Shape of wells: VStandard shallow 96-well plate (Corning Costar 3359): 360µl max volume, 250µl working volume;plate code 96B – 250. Shape of wells: U/round. OLD SHALLOW PLATEStandard medium 96-well plate (Abgene AB-0765): 800µl max volume, 650µl working volume; plate code 96L – 650. Shape of well: conical.Cluster tube screwtop 96-well plate (Matrix ScrewTop TrackMates 3742, ): 1000µl max volume, 750µl working volume; plate code 96D – 750. Shape of well: Unknown.SPECIAL IStandard deep 96-well plate (Thompson Instruments, Oceanside distributor, part number 951651B): 1200µl max volume, 800µl working volume (equivalent to Abgene AB-0564); plate code 96C – 800.Shape of well: U/round. OLD DEEP WELL PLATECluster tube capmat 96-well plate (Matrix TrakMates 3791, ): 1400µl max volume, 850µl working volume; plate code 96J – 850. Shape of well: V. SPECIAL II o384 wells:Standard 384-well plate – Thompson Instruments (Oceanside distributor) part number 931504B, 120µl max volume, 75µl working volume (equivalent to Abgene AB-0781); plate code 384E – 75. Shape ofwell: pyramidal.Microarray 384-well plate (Genetix X7020): 65µl max volume, 45µl working volume; plate code 384F – 45. Shape of well: flat.o Seals and caps:Heat Seal – Thompson Instruments (Oceanside distributor) part number 899406 (equivalent to Abgene AB-0745)Adhesive Seal – Thompson Instruments (Oceanside distributor) part number 899405-1Caps – Matrix ScrewTop caps 4472, for use with cluster tube screwtop 96-well plate, 96 caps required for each plateCapmat – Matrix SepraSeal Cap Mats 4463, for use with cluster tube capmat 96-well plate, one mat required for each plateSHIPPING CONDITIONS(back to Table of Content)Please refer to Delivery Schedule.PrimaryRoom Temperature lyophilized for tubes, different options for plates (lyophilized, ambient in solution, frozen in dry ice. SecondarySpecial Handling by request: DNA oligos ordered in plates can be requested to a variety of normalization, pooling and aliquoting options free of charge. For TUBE oligos, the standard format is dried down and ship at room temperature. However, oligos ordered in tubes can be requested to be sent with a few pre-selected special handling options. These options are available in the check out section of the online cart. Two of these options can be checked for an extra $1 charge per primer. The options are re-suspension of the oligos in water to a certain concentration (10, 20, 50, 100 or 200 µM), electronic COA, second set of labels, 100% QC (for DNA oligos we only do Mass Spec or CE for 25% of the oligos, this options will make the oligos go through CE/Mass Spec for sure), ship complete order and not partial orders and/or send on dry ice. For other options, a special handling request can be requested through the Invitrogen Account Manager. Re-suspended primers also ship at Room Temperature (they are stable this way), unless instructed otherwise in the “special handling”.Note: The primers from Frederick are shipped held in a cardboard holder called a “Fluted Partition”. This is made by a company called Fluted Partition (203-368-2548). The tube primers from Illumina are sent in a tube holder that holds 6 tubes.HRP Oligo shipping: The storage conditions are 4oC in HRP buffer. We ship on ice packs. The box should be marked with a sticker to refrigerate upon arrival.STORAGE CONDITIONS(back to Table of Content)Recommended storage for lyophilized primers and reconstituted primers is -20°C.AP Conjugate is shipped in AP storage buffer and should be stored at 4°C.HRP Conjugate is shipped in HRP liquid storage buffer on ice packs and should be stored at 4°C.HEX, TET and FAM are very sensitive to light. Store in the dark or use amber tubes. Storage in a black box is recommended. HEX and TET labels are both stable under 30 cycles of standard PCR conditions; however under harsher conditions (high pH, high temp) TET is more stable than HEX.Dye half life (approximately) when exposed to lightHEX 4.5 hoursTET 2.25 hoursFAM 1.125 hoursFor making adapters with 5’modified oligos:We have limited data to show that in high salt solution at 55°C HRP is stable for 1 hour.We have no stability data of HRP at 65°C. It has been stated that HRP is less stable than APSTABILITY(back to Table of Content)Lyophilized primers are stable for one year, and will be stable for several months at 4°C. Reconstituted primers at -20°C are stable for at least six months, and they will be stable for a few weeks at 4°C.AP Conjugate - shipped in AP storage buffer, store at 4°C, stable for at least 12 months.HRP Conjugate - shipped in HRP storage buffer, store at 4°C, stable for at least 12 months.QC SPECIFICATIONS(back to Table of Content)Most of the modified oligos arel be made at Frederick (see General Custom Primer note for details).For 25, 50 and 200 nM desalted and cartridge-purified DNA oligos, there is 100% OD 260 analysis. Random samples of 25% of the oligos produced are tested by either Capillary Electrophoresis (all oligos at Illumina and any oligo longer than 45-mers at Frederick) or Mass Spectrometry (Illumina does not do Mass Spec anymore but Frederick still uses it for oligos smaller than 45-mers). DNA oligos that are desalted and ordered at 25 and 50 nM scales also have 100% real-time digital trityl monitoring during analysis. Customers can request 100% QC by Mass Spec or CE during ordering paying $1 per oligo.Desalted DNA oligos ordered at 1 and 10 uM, DNA oligos at any scale that are purified by HPLC and PAGE, the majority of the DNA oligos with 3 and 5’ modifications and the RNA oligos have also 100% OD260 analysis and Mass Spectrometry or Capillary Electrophoresis.For a PAGE oligo, regardless of length, the main peak on the trace needs to be at least 85% of the product to pass and the N- peaks for mutations must be less than 10% (Jaime 11.30.06, from Sheryl Moles).Effect of modification on ODOligos are quantified by measuring the absorbance at 260 nm. Some fluorophores do absorb at 260 nm as well. For example, fluorescein and TAMRA have an extinction coefficients of 20.96 and 31.98 OD/µmole at 260 nm. For reference, dA, dC, dG and dT are 15.3, 7.4, 11.8, 9.3 OD/ µmole respectively. So, TAMRA is equivalent to about 3 nucleotides. For the dyes that have a published values at 260 nm, the value is incorporated into the extinction coefficient calculation on the C of A. These modifications are: FAM, Fluorescein, HEX, TAMRA, and TET. Others may absorb at 260 nm, but the data is not published and therefore is not included in the calculation.OD Specifications for AP and HRP Conjugates- Not applicable because pass/ fail based on moles of conjugate primers. The protein and oligo make up the OD value and may not reflect the true OD value of the oligo.AP and HRP ConjugatesBases Scale Desalted Cartridge*HPLC PAGE<20 Bases25 nmol NA NA NA NA50 nmol NA NA NA NA200 nmol NA NA NA NA1 µmol NA NA1-5 nmole (0.5-13OD)1-5 nmole (0.5-13 OD)10 µmol NA NA Inquire Inquire 20 Bases25 nmol NA NA NA NAor greater50 nmol NA NA NA NA200 nmol NA NA NA NA1 µmol NA NA1-5 nmole (0.5-13OD)1-5 nmole (0.5-13 OD)10 µmol NA NA Inquire Inquire*Per Jeremiah Mitchell (03/18/2009) we can't offer any mod on a cartridge purified oligo. The modification interferes in some way with the purification mechanism.QC on Taqman probes:Taqman and other Fluorescent probes will be QC’d by Mass Spectrometry (Mass Spec, MS). “They will be run on mass spec. We try to hit the guaranteed amount but we do not make a guarantee.” – Jeremiah Mitchell (04/02/2009)PROTOCOL AND APPLICATION NOTES(back to Table of Content)Modification/scale/purificationManufacturing DetailsFluoresceinRhodamineHEX/TET/FAMPhosphateBiotinAmineGatewayAlkaline PhosphataseHorse Radish PeroxidasePhosphorothioate“Fully-Phosphorothioated" Custom PrimersAldehydeAcridineThiolDelivery ScheduleOligoPerfect Designed Primers“Fully-Phosphorothioated" Custom Primers3' Modifications of OligosReconstitution ProtocolA260/A280 ratio of the oligoOligo VisualizationTroubleshooting“Custom Custom” modificationsModification, Scale of Synthesis/Purification and Availability Grid (back to Table of Content)(back to Protocol and Application Notes)Mod SynthScale PurificationDesaltedPurificationCartridgePurificationHPLCPurificationGel Purified5' Biotin25 N Yes (10-50 bases)NO NO NO5' Phosphate50 N Yes (10-50 bases)NO NO NO5' Primary200 N Yes (10-50 bases)NO Yes (7 – 60 bases)Yes (7-100 bases) Amine 1 U Yes (10-50 bases)NO Yes (7 – 60 bases)Yes (7-100 bases)25 U Yes (10-50 bases)NO Yes (7 – 60 bases)Yes (7-100 bases)25 N NO NO NO NO50 N Yes (5-100 bases)Yes (7 – 60 bases)Yes (7 – 60 bases)Yes (7-100 bases) 5' Rhodamine200 N Yes (5-100 bases)Yes (7 – 60 bases)Yes (7 – 60 bases)Yes (7-100 bases)1 U Yes (5-100 bases)Yes (7 – 60 bases)Yes (7 – 60 bases)Yes (7-100 bases)25 U Yes (5-100 bases)Yes (7 – 60 bases)Yes (7 – 60 bases)Yes (7-100 bases)25 N Yes (10-50 bases)NO NO NO5' HEX/ TET/FAM50 N Yes (5-100 bases)Yes (7 – 60 bases)Yes (7 – 60 bases)Yes (7-100 bases) 5' Fluorescein200 N NO NO Yes (7 – 60 bases)Yes (7-100 bases) 5' GATEWAY 1 U Yes (5-100 bases)Yes (7 – 60 bases)Yes (7 – 60 bases)Yes (7-100 bases)25 U Yes (5-100 bases)Yes (7 – 60 bases)Yes (7 – 60 bases)Yes (7-100 bases)25 N NO NO NO NOAP Conjugate50 N NO NO NO NOHRP Conjugate**200 N NO NO NO NO1 U NO NO Yes (7 – 60 bases)Yes (7-100 bases)25 U NO NO Yes (7 – 60 bases)Yes (7-100 bases)25 N NO NO NO NO50 N Yes (5-100 bases)Yes 7 – 60 bases)YES (7 – 60 bases)YES (7-100 bases) Phosphorothioates200 N Yes (5-100 bases)Yes (7 – 60 bases)YES (7 – 60 bases)YES (7-100 bases)1 U Yes (5-100 bases)Yes (7 – 60 bases)YES (7 – 60 bases)YES (7-100 bases)25 U Yes NO YES (7 – 60 bases)YES (7-100 bases)Reason for lower limit of 10 mer on the 25 nmol scale: "We have had a high failure rate on synthesis of short oligos at the 25 nmoles scale. We have determined that the problem cannot be corrected. Therefore, we have decided to make the minimum length at the 25 nmole scale to be a 10 mer.The amount of modified oligo specified in the CoA includes the modification. For example, for the biotin modification, the amount of oligo in ‘ug’ and ‘uM’ specified in the CoA includes the oligo, the C6 linker and the biotin.**HRP max oligo length of 25 bases, requires 1 umole scale.Manufacturing Details(back to Table of Content)(back to Protocol and Application Notes)These modifications are added as phosphoramidites on the machine during synthesis.FluoresceinRhodamineHEX/TET/FAMPhosphateBiotinAmineGatewayAlkaline PhosphataseHorse Radish PeroxidasePhosphorothioateAldehydeAcridineThiol5’ FLO Modification (Formerly referred to as Fluorescein)(back to Table of Content)(back to Protocol and Application Notes)(back to Manufacturing Details)5(6)-FAM (5(6)-carboxyfluorescein) is added as a phosphoramidite by B-cyanoethyl chemistry and therefore added on the sugar at the 5' end of the primer NOT on the last base as the last synthesis cycle. This is a mixture of two isomers of FAM (therefore on an HPLC trace, there will be two peaks).Absorption Max (nm)Emission Max (nm)Extinction Coeff.(OD/mole) at maxExtinction Coeff. (OD/mole) at260 nm49452076,00031,500 Fluorescent color is green.Note: 5’ FLO is a mixture of the 5- and 6-isomers of FAM. They are not stereoisomers. The conjugation bond is either on the 5 and 6 position on the phenyl ring. Half the molecules have the bond at the 5 position and half have it at the 6 position**. The two different oligos can be detected when analyzed by HPLC or CE. The extinction coefficients at lambda max and at 260 nm are estimates based on known values of the two isomers. This modification is not available desalted or cartridge purified per Sheryl Moles 3/29/07 (V.Gurtu).The phosphoramidite used for this does not have a trityl group, but the FAM itself is hydrophobic enough to allow HPLC purification. A modified HPLC protocol is used for the 5' phosphate oligos to achieve the separation. Typically, 5' fluorescein oligos will not be as pure as those that can be purified by using the trityl on approach, but close in purity.FAM and Fluorescein are different chemicals. FAM is 5-carboxyfluorescein or 6-carboxyfluorescein (single isomers) or 5(6)-carboxyfluorescein (mixed isomers) – FAM has two carboxyl moieties on the phenyl ring, while Fluorescein has only one carboxyl on the phenyl ring; all reactive groups will be conjugated via the 5- or 6- carboxyl group on FAM but directly to the phenyl ring on Fluorescein. Although these two dyes have very similar spectral properties, the names “FAM” and “Fluorescein” are not interchangeable. FAM, not Fluorescein, is used on ABI sequencers. The two isomer forms of FAM differ in electrophoretic mobility and both FAM isomers differ significantly from the electrophoretic mobility of Fluorescein.Specs for FLO/FAM mods:“We will use 5'FLO amidite (6-FAM)so there is no free dye.If the oligo is desalted it has to have at least 45% full length product (note that these specs are not public knowledge) and contain no N- peaks that are greater than 15%.” – Jeremiah Mitchell (05/19/2009)*[Note: Highly concentrated solutions and the solid forms of fluorescein and FAM appear by eye as yellow or orange colored solutions or solid, this may also be true of lyophilized oligos labeled with 5’FLO].** “Half the molecules have the bond at the 5 position and half have it at the 6 position”. Our mixed isomer FAM products, 5(6)-FAM, can have a lot-to-lot variablility in 5:6 isomer ration from 70:30 to 50:50; is the 5(6)-FAM phosphoramidite SPECIFICALLY provided always a 50:50 ratio of isomers, or is there lot-to-lot variability as well? It is possible, so you can not always tell customers it is 50:50. (O. Cholewa, 8/2008).This modification can be added to phosphorothioated oligos.Rhodamine(back to Table of Content)(back to Protocol and Application Notes)(back to Manufacturing Details)Can suggest as the alternative to 5’ROX modified oligos.All the rhodamine dyes (rhodamine, rhodamine green dye and rhodamine green-X) are HPLC purified as part of the dye price. The web site forces you to order it as desalted for now (8.4.06) but it will be changed in the future.Rhodamine when resuspended in water or TE solution will appear pink-red-purplish.Rhodamine = RHD on email order form.MW(NH4 salt)MW (no salt)EmissionAbs. (nm)Extinct. Coeff. (at lambda max)**Max (nm)743.8726.859657292,000Note that this is for measurements of the labeling dye in methanol. The values in water/TE and attached to an oligo will be different.** We do not have info on what A260 of rhodamine would be.The modification provided is x-rhodamine isothiocyanate (X-RITC) which is a mix of rhodamine-5 and 6-isothiocyanate (5(6))-XRITC). The addition of the rhodamine modification is done on all scales as a post-synthesis modification. To see the structure, click on the link in this paragraph.HEX, TET or 6-FAM(back to Table of Content)(back to Protocol and Application Notes)(back to Manufacturing Details)HEX is 4,7,2`4`5`7` hexachlorofluoresceinTET is 2`7`4,7, tetrachlorofluoresceinFAM is 6-carboxyfluorescein, while fluorescein is the 5 and 6 isomers mixed together.FAM, HEX and TET are added as a phosphoramidite as the last synthesis cycle using B-cyanoethyl chemistry and therefore added on the sugar at the 5'end of the primer NOT on the last base. They are covalently attached to the 5' end of the last sugar via a phosphodiester bond.Point at which modification occurs:In synthesis we can add HEX, TET, FAM or flourescein. All other dyes are added post-synthesis. In synthesis mods do not require purification. (Jeremiah Mitchell, Melissa Almeida; 04/30/2009).Trityl-OFF cartridge purification on confirmation sheet refers to the fact that there is no DMT on the phosphoramidites with the dye.For Eurogentec orders take the MW from the CofA and add the below MW numbers. PrimerTrak includes the MW of the Mod in the MW from the CofA.MW Emission Max(nm)Excitation (Abs)Max (nm)Extinction (at max)Extinction (at 260)FAM554.551749474,85020,960TET692.253852285,55316,255HEX761.155353595,69831,580Molar extinction coefficients above are measured at excitation maximum nm.Individual experiments may vary fluorescence intensity due to minor variations in pH or composition can affect the above numbers. For FAM, take the reading at pH 9 since at this pH the absorbance is highest. At pH 7, this value is reduced by 16% for FAM. And when the fluorophore is attached to DNA, the value is also reduced.We do not know the effect of pH on the absorbance of TET & HEX.At the maximum wavelength, there is a huge change between pH 8 and pH 6.5. For the absorbance at 260, the change between pH8 and pH 6.5 is relatively minor.I tested the water here and it is about 6.5.Also please see /handbook/figs/fig23-2.htmlfor more information on effect of pH.The color of the dye depends on the "Filter-wheel set" of the ABI instruments:Dyes Filter A Filter BHEX Green (560nm)Yellow (560nm)6-FAM Blue (531nm)Blue (531nm)TET-Green (545)Color in solution: HEX is pinkish, FAM the yellow, and TET is orange, but if the oligos are not subjected to light they may appear colorless. 10nm often will have no color due to the small amount of oligo present.The phosphoramidites with these dyes do not have a DMT (trityl) group. They therefore cannot be monitored during the synthesis with trityl analysis. They still must be deprotected, as there are protective groups on the bases in the oligonucleotide. A & C have a benzoyl protective group. G has an isobutyl protective group. T does not have a protective group.Trityl-OFF cartridge purification is required since these do not have a DMT. This purification is performed as a batch HPLC method on a polystyrene reverse phase support.They cannot be ordered as phosphorothioates.Phosphate(back to Table of Content)(back to Protocol and Application Notes)(back to Manufacturing Details)5’- PhosphateThe cartridge purification option for the 5' phosphate oligos is not offered, because it is not technically possible (cartridge purification relies on the DMT group being present which is not true here). DMT has molecular weight of 79.98.The 5' phosphorylation is added by B-cyanoethyl chemistry and therefore added on the sugar at the 5'end of the primer NOT on the last base, as the last cycle with a phosphoramidite. It therefore has a trityl group, which is released and monitored to assess the coupling efficiency. (During the coupling, the phosphate has a protecting group, which is removed prior to cleavage of the oligo from the solid support). Since 5' phosphorylation comprises the final step after all the cycles are complete, only the complete, or full length oligos (which are uncapped) can be phosphorylated on the synthesizer.Gel purification of phosphorylated primers will not purify full length primers from n-1 primers because the phosphate adds an additional negative charge that affects electrophoresis.The 5’ phosphate oligos are purified by HPLC by size. A modified HPLC protocol is used for the 5' phosphate oligos to achieve the separation. Typically, 5' phosphate oligos will not be as pure as those that can be purified by using the trityl on approach. The cartridge purification option cannot be used for the 5' phosphate oligos, as this relies on the DMT group being present.In fact, when the yield of HPLC phosphorylated primers is low it probably was because there were a lot of failure sequences so not as many fractions could be combined. There would be a lot of multiple overlapping peaks in preps with more "n-x" failures. During HPLC purification, 5 Phosphate oligos are not DMT-selected.It’s been noticed that a couple of phosphate, HPLC oligos after speedvac are a bit "dirty", or slightly yellow. This is sometimes a product of the synthesized product. It is usually cleaned up in the ethanol precipitation step as part of QC.3’-PhosphateThe 3’ phosphate is added attached to the solid support so the sequence is actually coupled to it in the first round of base addition. It will block extension by polymerases. Stability: assuming it's DNA, 3'-PO4 is more stable than 3'-BIO. 3'-BIO will come off under basic condition while 3'-PO4 will not.Biotin(back to Table of Content)(back to Protocol and Application Notes)(back to Manufacturing Details)The structure as shown is called Biotin Phosphoramidite. Biotin is on the far right ending with the oxygen group next to the amine group in the chain. MW of biotin is 405. Biotin oligos are quantified as normal by OD260 (biotin does not absorb at 260). Comparing 3’ Mod Stability: 3'-NH2 is slightly more stable than 3'-PO4. It is 3'-NH2>3'-PO4>>3'-BIOThe biotin that we use for our modifications is from Glen Research, 10-5950-95 (Jeremiah Mitchell, 04/27/2009).5’- BiotinPROPRIETARY INFORMATION: It is purchased from Glen Research and Catalog number 10-5950-02./ Please see the biotin structure below.Biotin dT The Biotin dT has the Biotin is attached to a dT base, so they would only use it if the 5' base can be T. I really know no reason why one would choose it over the regular biotin.General info on Biotinylated primers:5' biotinylated primers may work for chemiluminescent detection, but TdT tailing would be better.The biotin is added during synthesis, before the purification step. A biotinylated oligo (or a PCR product made with biotinylated oligos) will be retained for a longer time on a reverse phase HPLC column than an unbiotinylated oligo of the same size. This is due to the hydrophobicity of the biotin moiety.The biotin modification should be fairly resistant to heat and pH (as it has been boiled to 100°C and exposed to pH 2-10). More extreme conditions may be tolerated, but have not been tested. Presence of reducing agents may not be a problem, but exact limits are unknown.The chemical structure of 3'-Biotin predicts that 3'-Biotin would come off/degrade at two conditions: 1 over the time, 2 the oligo was exposed to basic conditions such as dissolved in basic buffer. The reason is that 3'-Biotin has an adjacent hydroxy group which makes it a RNA-like structure.5'-Biotin though does not have this adjacent hydroxy group, so it is pretty stable, more stable than 3'-Biotin.The standard biotin that we use has a C6 linker. We have used the TEG biotin linker several times in the past because of special requests from customers but it is not our “standard” biotin modification (Jaime 5/10/06 from Joseph Hayes).Recover 5’ biotin labeled oligo from C-18 column: An oligo with biotin is more hydrophobic than an unlabeled oligo. Therefore, the biotin oligo would stick better to a C-18 column. I haven’t used a C-18 column for years since the polystyrene columns are more durable for production. To put it in perspective, on the polystyrene I elute the unlabeled oligos in 6 - 12 percentACN/TEAA. For the biotin oligos I use 10 - 15 %. The customer may be able to use this to extrapolate the percentages. Since C-18 is more hydrophobic than polystyrene, their percentages should be a little higher.Frederick QC’s the oligos by Mass Spec or CE and they check the peaks to see if the coupling reaction of the biotin to the molecule went as expected. For a DSL oligo, we will discard the oligo if there is a peak indicating more than 15% of the oligo not coupled to biotin. For HPLC or PAGE purified oligos, we will ask for less than 10% of uncoupled oligo during QC (Jaime from Sheryl Moles, 12.14.06).Biotinylated DNA will successfully transform bacteria, e.g. DH5 alpha.3’- Biotin3’ Biotin is added via biotin solid support.Primers carrying a 3’-biotin modification can not be longer than 99 nucleotides. Since the 3’-biotin is provided as a CPG modification (linked onto the solid support) the system interprets the 3’-biotin as the last nucleotide of the sequence. (Sheryl Moles 02/08/07)Comparing 3’ Mod Stability: 3'-NH2 is slightly more stable than 3'-PO4. It is 3'-NH2>3'-PO4>>3'-BIOGlen Research Catalog Number: 20-2955-xx /Primary Amine(back to Table of Content)(back to Protocol and Application Notes)(back to Manufacturing Details)Primary amine –The amino group is linked to the 5' end via a 6 carbon spacer arm. i.e. Off of the 5’ hydroxyl there is a phosphate. Attached to this via a phosphodiester bond is a C6 linker (CH2-CH2-CH2-CH2-CH2-CH2) followed by a primary amine NH2. It is added as a phosphoramidite at the last synthesis cycle by B -cyanoethyl chemistry and therefore added on the sugar at the 5'end of the primer NOT on the last base.For other scales, we can also do 3’ amine modification as a custom “custom” through PMO request (Jeremiah Mitchell). 3’ Amine mod has a C7 linker. We can now do 3’ amine modification as a catalog item for 50 nmol and 200 nmol scale. Customers will be able to order online, or using the email/fax form in North America. The website will be updated with the proper drop-downs for the mod, and the forms will be updated with the code. Please note that the 3’ amine mod is only compatible with certain 5’。