Les12 Other Database Objects
- 格式:ppt
- 大小:218.50 KB
- 文档页数:28



基于网络药理学及分子对接技术探究知葛通脉颗粒治疗糖尿病下肢动脉粥样硬化性病变的作用机制徐仁佳1,樊月月2,沈晓喻2,吴坚21.上海中医药大学附属上海市中西医结合医院,上海200082;2.上海市嘉定区中医医院,上海201899【摘要】目的借助网络药理学研究方法,初步探索知葛通脉颗粒治疗2型糖尿病下肢动脉粥样硬化性病变的靶点和机制,为进一步研究提供相关理论支持和线索。
方法通过TCMSP 数据库检索、文献补充,筛选知葛通脉颗粒有效成分及效用靶点,通过GeneCards 、OMIM 等数据库挖掘2型糖尿病LEAD 靶点,取药物与疾病的交集靶点;利用STRING11.5制作PPI 网络图,并通过PPI 网络拓扑关系分析知葛通脉颗粒治疗2型糖尿病LEAD 的关键靶点;构建“药物-成分-靶点”关系图,筛选中药颗粒核心成分。
在Metascape 平台进行GO 、KEGG 富集分析,探索药物治疗疾病的潜在机制,构建“成分-靶点-通路”图。
最后,通过AutoDockTools 1.5.6进行分子对接,验证药物活性成分与疾病靶点之间的反应活性。
结果研究获得知葛通脉颗粒有效成分71种,潜在靶点261个,与DLEAD 交集靶点107个,筛选出核心成分槲皮素、木犀草素、山柰酚、芒柄花黄素、异鼠李素、去水淫羊藿黄素、薯蓣皂苷元、β-谷甾醇、金圣草素、豆甾醇,关键靶点AKT1、IL -6、TNF 、TP53、VEGFA 、IL -1B 。
相关作用通路有183条,作用通路主要涉及MAPK 、糖尿病并发症中的AGE-RAGE 、HIF -1信号通路等。
对接结果显示中药成分与疾病靶点之间可较好地结合。
结论知葛通脉颗粒可通过槲皮素、木犀草素、山柰酚、芒柄花黄素、异鼠李素、去水淫羊藿黄素、薯蓣皂苷元、β-谷甾醇、金圣草素、豆甾醇等关键有效成分作用于AKT1、IL -6、TNF 、TP53、VEGFA 、IL -1B 等关键靶点,调节MAPK 、糖尿病并发症中的AGE-RAGE 、HIF -1等信号通路,多靶点、多通路协同发挥对DLEAD 的治疗作用。

PL/SQL Developer 12使用手册目录一、PL/SQL Developer 12简介二、安装和配置1. 下载PL/SQL Developer 122. 安装PL/SQL Developer 123. 连接到数据库4. 配置PL/SQL Developer 12三、基本功能1. 编写和执行SQL查询2. 编写和调试PL/SQL代码3. 数据建模和设计4. 代码版本控制5. 其他实用工具四、高级功能1. 性能优化和调整2. 自动化任务3. 数据安全和权限管理五、常见问题和解决方法1. 连接数据库失败2. PL/SQL Developer 12崩溃3. 执行SQL语句超时4. 其他常见问题六、总结一、PL/SQL Developer 12简介PL/SQL Developer 12是一款功能强大的集成开发环境,主要用于Oracle数据库的开发和管理。
它提供了丰富的功能和工具,可以帮助开发人员提高工作效率,简化开发流程,提升代码质量。
二、安装和配置1. 下载PL/SQL Developer 12我们需要从冠方全球信息站下载PL/SQL Developer 12的安装包。
在下载之前,需要注册一个账号并购物授权。
下载完成后,可以开始安装过程。
2. 安装PL/SQL Developer 12双击安装包开始安装过程,按照提示逐步进行安装配置。
在安装过程中,可以选择安装路径、创建桌面快捷方式等选项。
安装完成后,可以启动PL/SQL Developer 12。
3. 连接到数据库打开PL/SQL Developer 12后,需要建立与数据库的连接。
在连接对话框中输入数据库位置区域、用户名和密码,选择连接类型(如OCI、Thin等),点击连接按钮即可建立连接。
4. 配置PL/SQL Developer 12在使用PL/SQL Developer 12之前,可以根据个人偏好进行一些配置。
设置代码自动格式化的风格、定义快捷键、配置代码模板等。

2022年安徽医科大学信息管理与信息系统专业《数据库概论》科目期末试卷A(有答案)一、填空题1、在数据库系统封锁协议中,一级协议:“事务在修改数据A前必须先对其加X锁,直到事务结束才释放X锁”,该协议可以防止______;二级协议是在一级协议的基础上加上“事务T在读数据R之前必须先对其加S锁,读完后即可释放S锁”,该协议可以防止______;三级协议是在一级协议的基础上加上“事务T在读数据R之前必须先对其加S锁,直到事务结束后才释放S 锁”,该协议可以防止______。
2、“为哪些表,在哪些字段上,建立什么样的索引”这一设计内容应该属于数据库设计中的______阶段。
3、在SQL语言中,为了数据库的安全性,设置了对数据的存取进行控制的语句,对用户授权使用____________语句,收回所授的权限使用____________语句。
4、对于非规范化的模式,经过转变为1NF,______,将1NF经过转变为2NF,______,将2NF 经过转变为3NF______。
5、设有关系模式R(A,B,C)和S(E,A,F),若R.A是R的主码,S.A是S的外码,则S.A的值或者等于R中某个元组的主码值,或者______取空值,这是规则,它是通过______和______约束来实现的。
6、若事务T对数据对象A加了S锁,则其他事务只能对数据A再加______,不能加______,直到事务T释放A上的锁。
7、采用关系模型的逻辑结构设计的任务是将E-R图转换成一组______,并进行______处理。
8、在RDBMS中,通过某种代价模型计算各种查询的执行代价。
在集中式数据库中,查询的执行开销主要包括______和______代价。
在多用户数据库中,还应考虑查询的内存代价开销。
9、数据库恢复是将数据库从______状态恢复到______的功能。
10、设某数据库中有作者表(作者号,城市)和出版商表(出版商号,城市),请补全如下查询语句,使该查询语句能查询作者和出版商所在的全部不重复的城市。


postgres12主从同步流复制原理PostgreSQL 12支持的主从同步流复制原理如下:
1. WAL (Write-Ahead Logging): PostgreSQL使用WAL记录数据库中的所有修改。
在主服务器上对数据库进行更改时,这些更改首先被写入WAL文件中。
从服务器通过读取主服务器上的WAL文件来获取这些更改,并将它们应用到自己的数据库中,以保持与主服务器同步。
2. 流复制: 流复制是指从主服务器到从服务器的数据传输过程。
主服务器将WAL记录传输到从服务器,从服务器则按顺序应用这些记录。
这种方法确保了数据在传输过程中的一致性和完整性。
3. 同步和异步复制: 在同步复制模式下,主服务器等待至少一个从服务器确认已接收并应用了WAL记录才会继续进行操作。
这确保了主从服务器之间的数据同步,但会增加写入延迟。
在异步复制模式下,主服务器不会等待从服务器的确认,而是立即继续进行操作。
这会减少写入延迟,但可能导致主从服务器之间的数据稍有滞后。
4. 复制槽: 复制槽是一种机制,用于在主服务器和从服务器之间传输WAL记录。
它确保了即使从服务器暂时不可用,主服务器仍然可以继续记录WAL记录,而不会出现因为从服务器无法接收而导致的WAL日志溢出或数据丢失问题。
5. 归档日志: PostgreSQL还支持归档日志,它可以将WAL日志定期归档到持久存储中,以便在灾难恢复或从备份中进行恢复时使用。
这样可以确保即使主服务器发生故障,也可以使用归档日志将从服务器恢复到最新的状态。

plsql developer 12使用手册摘要:1.PL/SQL Developer 12 简介2.安装与配置PL/SQL Developer 123.PL/SQL Developer 12 功能概述4.PL/SQL Developer 12 的使用方法与技巧5.PL/SQL Developer 12 的优势与不足6.总结正文:【PL/SQL Developer 12 简介】PL/SQL Developer 12是一款功能强大的PL/SQL(Procedural Language/SQL)编程工具,它为Oracle数据库的开发者提供了一个高效、便捷的编程环境。
使用PL/SQL Developer 12,开发者可以轻松地进行PL/SQL 代码的编写、调试、运行和维护。
【安装与配置PL/SQL Developer 12】在安装PL/SQL Developer 12 之前,需要确保计算机上已安装了Oracle 数据库。
安装过程相对简单,只需按照安装向导的提示进行操作即可。
安装完成后,需要对PL/SQL Developer 12 进行一些基本配置,如设置Oracle 数据库的连接信息等。
【PL/SQL Developer 12 功能概述】PL/SQL Developer 12 具有以下主要功能:1.提供丰富的代码编辑功能,如自动补全、语法高亮等;2.支持PL/SQL代码的调试,可以进行断点设置、单步执行等操作;3.提供代码模板,方便开发者快速生成PL/SQL 代码;4.支持数据模型设计,可以绘制数据库表结构、关系等;5.支持批处理操作,可以批量执行PL/SQL 程序;6.支持Oracle 数据库的备份与恢复功能。
【PL/SQL Developer 12 的使用方法与技巧】在使用PL/SQL Developer 12 时,可以参考以下方法和技巧:1.善用代码编辑功能,如自动补全、语法高亮等,提高编程效率;2.熟悉PL/SQL代码调试流程,及时发现和排除程序中的错误;3.掌握批处理操作,提高工作效率;4.学会使用数据模型设计功能,便于进行数据库结构设计与优化。
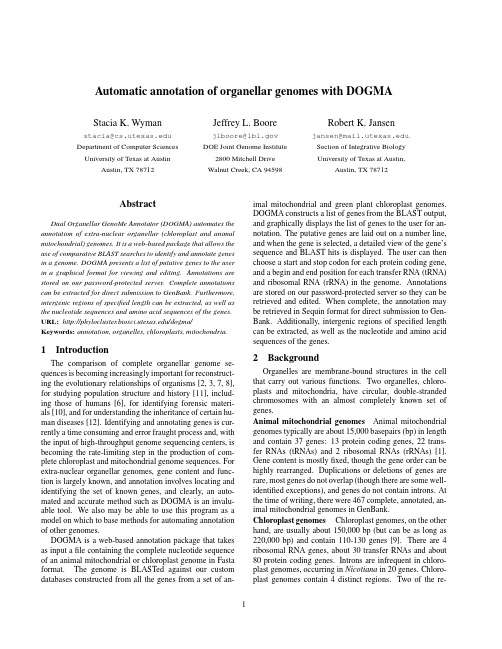
Automatic annotation of organellar genomes with DOGMAStacia K.Wyman stacia@ Department of Computer Sciences University of Texas at AustinAustin,TX78712Jeffrey L.Boorejlboore@DOE Joint Genome Institute2800Mitchell DriveWalnut Creek,CA94598Robert K.Jansenjansen@.Section of Integrative BiologyUniversity of Texas at Austin,Austin,TX78712AbstractDual Organellar GenoMe Annotator(DOGMA)automates the annotation of extra-nuclear organellar(chloroplast and animal mitochondrial)genomes.It is a web-based package that allows the use of comparative BLAST searches to identify and annotate genes in a genome.DOGMA presents a list of putative genes to the user in a graphical format for viewing and editing.Annotations are stored on our password-protected plete annotations can be extracted for direct submission to GenBank.Furthermore, intergenic regions of specified length can be extracted,as well as the nucleotide sequences and amino acid sequences of the genes. URL:/dogma/ Keywords:annotation,organelles,chloroplasts,mitochondria. 1IntroductionThe comparison of complete organellar genome se-quences is becoming increasingly important for reconstruct-ing the evolutionary relationships of organisms[2,3,7,8], for studying population structure and history[11],includ-ing those of humans[6],for identifying forensic materi-als[10],and for understanding the inheritance of certain hu-man diseases[12].Identifying and annotating genes is cur-rently a time consuming and error fraught process and,with the input of high-throughput genome sequencing centers,is becoming the rate-limiting step in the production of com-plete chloroplast and mitochondrial genome sequences.For extra-nuclear organellar genomes,gene content and func-tion is largely known,and annotation involves locating and identifying the set of known genes,and clearly,an auto-mated and accurate method such as DOGMA is an invalu-able tool.We also may be able to use this program as a model on which to base methods for automating annotation of other genomes.DOGMA is a web-based annotation package that takes as input afile containing the complete nucleotide sequence of an animal mitochondrial or chloroplast genome in Fasta format.The genome is BLASTed against our custom databases constructed from all the genes from a set of an-imal mitochondrial and green plant chloroplast genomes. DOGMA constructs a list of genes from the BLAST output, and graphically displays the list of genes to the user for an-notation.The putative genes are laid out on a number line, and when the gene is selected,a detailed view of the gene’s sequence and BLAST hits is displayed.The user can then choose a start and stop codon for each protein coding gene, and a begin and end position for each transfer RNA(tRNA) and ribosomal RNA(rRNA)in the genome.Annotations are stored on our password-protected server so they can be retrieved and edited.When complete,the annotation may be retrieved in Sequin format for direct submission to Gen-Bank.Additionally,intergenic regions of specified length can be extracted,as well as the nucleotide and amino acid sequences of the genes.2BackgroundOrganelles are membrane-bound structures in the cell that carry out various functions.Two organelles,chloro-plasts and mitochondria,have circular,double-stranded chromosomes with an almost completely known set of genes.Animal mitochondrial genomes Animal mitochondrial genomes typically are about15,000basepairs(bp)in length and contain37genes:13protein coding genes,22trans-fer RNAs(tRNAs)and2ribosomal RNAs(rRNAs)[1]. Gene content is mostlyfixed,though the gene order can be highly rearranged.Duplications or deletions of genes are rare,most genes do not overlap(though there are some well-identified exceptions),and genes do not contain introns.At the time of writing,there were467complete,annotated,an-imal mitochondrial genomes in GenBank.Chloroplast genomes Chloroplast genomes,on the other hand,are usually about150,000bp(but can be as long as 220,000bp)and contain110-130genes[9].There are4 ribosomal RNA genes,about30transfer RNAs and about 80protein coding genes.Introns are infrequent in chloro-plast genomes,occurring in Nicotiana in20genes.Chloro-plast genomes contain4distinct regions.Two of the re-gions(IRA and IRB)involve a large inverted repeat.The other two regions are the large and small single-copy re-gions.In general,gene content and order are highly con-served[9],although in some groups numerous structural rearrangements have been identified[4].Some genes can contain large introns or even contain tRNA genes within the intron.There are currently26complete plant chloroplast genomes in GenBank and18of these are green plants.3DetailsDatabases We created custom databases for select chloroplast and all animal mitochondrial genomes.For the animal mitochondrial database,we downloaded the com-plete genomes for243organisms(the total number in Gen-Bank at the time)and extracted the annotated genes to com-pile a database for each individual protein coding gene. Each database contains the amino acid sequence for a spe-cific gene from each of the genomes in which it appears. There are databases for each of the13protein coding genes, plus one for each of the two rRNA genes.For chloroplast genomes,we created the database from 16complete genomes of green plants.They include Adi-antum,Arabidopsis,Chlorella,Epifagus,Lotus,Marchan-tia,Mesostigma,Nephroselmis,Nicotiana,Oenothera, Oryza,Pinus,Psilotum,Spinacia,Triticum,and Zea. Databasefiles were created for98chloroplast protein cod-ing genes(with two entries for the each of the trans-spliced pieces of rps12).We did not include open reading frames (ORFs)in the database,however,hypothetical chloroplast reading frames(ycfs)were included.There are4rRNA nu-cleotide sequence databasefiles and35nucleotide sequence databasefiles for tRNA genes.Transfer RNA databasefiles were divided up based on pilation of the chloroplast gene databases required correcting many errors in the Genbank entries.Identifying protein coding genes Protein coding genes are identified in the input genome based upon conservation of sequence similarity with that gene in other genomes in the database.The input nucleotide sequence is translated to amino acids and then BLASTed against the database for each gene.Various BLAST parameters may be set by the user based upon their data.Once DOGMA has identified the putative protein coding genes,the user must select start and stop codons for each gene as part of the annotation pro-cess.For each gene in the genome,the program displays to the user the nucleotide sequence for the gene from the in-put genome with the translation to amino acids,along with the amino acid sequences from the BLAST hits.For genes containing introns,DOGMA will identify exon boundaries based upon the BLAST output.DOGMA compiles a list of possible exons for the user to put together in the annotation. This has proven to work quite well,however,genes with very small exons(less than5amino acids)may be difficult to annotate with DOGMA because they can be missed by BLAST.Identifying tRNAs In chloroplast genomes,the nu-cleotide sequences for tRNAs are highly conserved.We have found that sequence similarity is sufficient for detec-tion of tRNAs in chloroplast genomes.Databases of nu-cleotide sequences are used for searching with BLAST.The tRNA databases are divided up based on anticodons and several databases may exist for one amino acid if there is more than one anticodon.In this way,the anticodon of the tRNA is annotated for the user.Transfer RNAs have almost no sequence similarity in an-imal mitochondrial genomes and therefore,sequence simi-larity is not a sufficient criterion to locate the genes,and BLAST cannot be used.Animal mt tRNAs must be identi-fied based on conservation of basepairing in the secondary structure.This is a difficult task,but we have found that methods based on hidden Markov models work best[13] and DOGMA uses COVE[5]to identify sequences and then uses our own program to fold the putative tRNAs. Identifying rRNAs Ribosomal RNAs can be detected through BLAST searches for sequence similarity for both mitochondrial and chloroplast genomes.For mt genomes, the BLAST parameters(such as gap penalty or percent iden-tity)must be adjusted since there can be significant degrada-tion in the quality of the match in the middle of the rRNAs. 4Web-based Display and Editing Tool The package contains a web-based display and editing tool(Figure1).The tool consists of three panels.The main (middle)panel displays all the genes along a number line. The genes are color coded by strand and gene type and la-beled with the gene name.When a gene in the middle panel is selected,details about that gene appear in the top panel. DOGMA displays both strands of the genome’s nucleotide sequence,with the translation to amino acids lined up above the nucleotides and the BLAST hits lined up above that. The potential start and stop codons for the gene appear as links.To choose a stop codon,the user simply clicks on the codon.The taxon names are displayed to the far right and when clicked,the gene’s amino acid sequence from the database is shown in a separate window.When the gene name is clicked,the original blast output is displayed in a separate window.For annotation of rRNAs and chloroplast tRNAs,DOGMA functions similarly to the protein cod-ing genes,except that nucleotide sequences are displayed rather than amino acid sequences,and the user chooses the start and end of the gene.Animal mitochondrial tRNAs are notoriously difficult to annotate.DOGMA uses Eddy and Durbin’s COVE software to identify a list of putative tR-NAs.When a tRNA is clicked in the middle panel,a list of the possible tRNAs,with its secondary structure is shown in the top panel.The user can choose the tRNA based onthe quality of the secondary structure folding and its COVE score.The bottom panel of DOGMA consists of a set of buttons for performing different tasks.The user can add or delete genes,extract intergenic sequences,extract amino acid or nucleotide sequences for genes,or get a summary of the genes or extract the Sequin format for direct submission of an annotation to GenBank.When a researcherfirst uses the software,they create a user id and password which keeps their(perhaps unpub-lished)data private from other users of the ers may also save and retrieve existing annotations.This is es-pecially important for annotating chloroplast genomes be-cause there are so many genes.5Future WorkIn the future,plant mitochondrial genomes will be added to DOGMA,as well as private custom databases for in-dividuals.Researchers only interested in a subset of the database can identify the genomes they are interested in for comparison.This will also allow people interested in phy-logenetic reconstruction to identify evolutionary similarity among genomes in anticipation of alignment of the whole genomes.It will also allow users to use their own unpub-lished data for annotation.Future work also includes adding a feature to identify the gene order of a genome with respect to another reference genome.This will allow the genome to be automatically added to a phylogenetic reconstruction by gene order. Acknowledgments SKW was supported by National Sci-ence Foundation IGERT grant0114387.RKJ was sup-ported by National Science Foundation grant DEB0120709. Part of this work was performed under the auspices of the U.S.Department of Energy,Office of Biological and Environmental Research,in the University of California, Lawrence Berkeley National Laboratory,under contract No. DE-AC03-76SF00098.References[1]J.Boore.Animal mitochondrial genomes.Nucleic AcidsResearch,27:1767–1780,1999.[2]J.Boore and W.Brown.Big trees from little genomes:Mi-tochondrial gene order as a phylogenetic tool.Curr.Opin.Genet.Dev.,8(6):668–674,1998.[3]Y.Cao,M.Fujiwara,M.Nikaido,N.Okada,andM.Hasegawa.Interordinal relationships and timesecale of eutherian evolution as inferred from mitochondrial genome data.Gene,259:149–158,2000.[4]M.Cosner,R.Jansen,J.Palmer,and S.Downie.The highlyrearranged chloroplast genome of trachelium caeruleum (campanulaceae):Multiple inversions,inverted repeat ex-pansion and contraction,transposition,insertions/deletions, and several repeat families.Current Genetics,31:419–429, 1997.[5]S.Eddy and R.Durbin.RNA sequence analysis using co-variance models.Nucleic Acids Research,22:2079–2088, 1994.[6]M.Ingman,H.Kaessmann,S.P¨a¨a bo,and U.Gyllensten.Mitochondrial genome variation and the origin of modern humans.Nature,408:708–713,2001.[7]M.Martin,T.Rujan,T.Richly,A.Hansen,S.Cornelsen,T.Lins,D.Leister,B.Stoebe,M.Hasegawa,and D.Penny.Evolutionary analysis of arabidopsis,cyanobacterial,and chloroplast genomes reveals plastid phylogeny and thou-sands of cyanobacterial genes in the nucleus.Proceedings of the National Academy of Sciences,99:122246–12251,2002.[8]M.Miya,A.Kawaguchi,and M.Nishida.Mitogenomic ex-ploration of higher teleostean phylogenies:A case study for moderate-scale evolutionary genomics with38newly deter-mined complete mitochondrial DNA sequences.Mol.Biol.Evol.,18:1993–2009,2001.[9]J.Palmer.Plastid chromosomes:structure and evolution.Cell Culture and Somatic Cell Genetics of Plants,7A:5–53, 1991.[10]T.Parsons and M.Coble.Increasing forensic discrimina-tion of mitochondrial DNA testing through the analysis of the entire mitochondrial DNA genome.Croatian Med.J., 42:304–309,2001.[11] D.Rand.The unites of selection on mitochondrial DNA.Ann.Rev.Ecol.Syst.,32:415–448,2001.[12] D.Wallace.Mitochondrial diseases in man and mouse.Sci-ence,283:482–488,1999.[13]S.Wyman and J.Boore.Annotating animal mitochondrialtrnas:an experimental evaluation of four methods.Proc.European Conf.on Computational Biology,pages44–46, 2003.ing the three panels.。

plsql developer 12使用手册
摘要:
1.介绍PL/SQL Developer 12的使用手册
2.安装和配置PL/SQL Developer 12
3.熟悉PL/SQL Developer 12的主要功能和工具
4.掌握PL/SQL Developer 12的常用快捷键和操作技巧
5.深入了解PL/SQL Developer 12的高级特性
6.常见问题及解决方案
7.更新和升级PL/SQL Developer 12
8.总结与建议
正文:
SQL Server是一款由微软公司开发和推广的关系型数据库管理系统,广泛应用于各种企业级应用中。
本文将为您介绍如何使用SQL Server,包括安装配置、管理工具、常用命令、SQL语言基础、数据库管理、数据表和索引创建、数据库备份恢复、安全性管理、存储过程和触发器编写、性能分析以及日常开发中的应用案例等内容。
一、了解SQL Server数据库的基本概念
1.关系型数据库:基于关系模型的数据库,以表格形式存储数据。
2.数据库实例:包含一个或多个数据库的数据存储容器。
3.数据库表:存储数据的基本单元,由行(记录)和列(字段)组成。

DataWorksSOFTWARE FOR THE MANAGEMENTOF PROJECT DATADataWorks is software of the Product Data Management (PDM) type: it manages product data, increases planning productivity, allows management and control of, and access to the data relevant to design processes, planning and production.Thanks to this safe and controlled activity, DataWorks allows the company to efficiently issue high quality products onto the market.This method interacts with the whole life cycle of a product and gives the opportunity to have access to the right information in each phase of its development.High productivityDataWorks gives a connection between the developmentactivities of the product and those that support its construction.The functionality of DataWorks revolves around themanagement of project data (revisions, control of access,management of documentation) allowing the definition of flowfor management of revisions, issue and process variations ofproduct data, all this thanks to the greater definition and thebetter management of correlated files. With DataWorks it is easyto answer questions such as•Where are the files of the manual of a product?•When and by whom has a detail been modified?•Have the assemblies that contain a modified detailbeen updated?•How many flanges are contained in that group?•Where is the support that i have to change used?DataWorks is the productive and efficient answer to all thesequestions: it exploits and rationalizes the wealth of informationof the company, making it organized and rapidly useable,speeding up the development cycles, thanks to the parallelindustrialization processes; moreover it allows a severe control ofdata and ensures its automatic distribution.Information: the wealth of the companyWith DataWorks information is administered by just onerelational data base, this guarantees its integrity and rapidavailability. Anyone can work simultaneously on the data, alwaysfinding it up-to-date.At the same time the access to data correlated files takesplace under the control of DataWorks, which establishes theform of use, the possibility of overwriting and the simultaneousaccess by more than one authorized user. In this way documentsare immediately usable within the team of work,optimizing time and drastically diminishing the risk of loss ofimportant information.Management of group use, through administration tools andthe centralization of archives, introduces new levels ofinformation security, guaranteeing controlled access to the latestrevisions of company project data.Different applicationsDataWorks is an open environment that allows managementof information originating from different applications.It is possible to organize and have easy access to CAD data,word processing files, part program, data sheets and any otherdocumentation data of a project. This product is integrated with:•Autocad ®•SolidWorks ®•CoCreate ME10 ®•Microsoft Office ®•Acrobat Reader ®Structure of DataWorksDataWorks has at its disposal a series of functions for themanagement of technical data: three-dimensional models,drawings, documents and information.Technical data created by the user is saved in a relational database, while files connected to a company part number areinserted in one or more file system directory (memorization area)controlled by the application. The functionality can be summedup in:•Definition and creation of product families•Dynamic association of specific attributes for eachproduct family•Management of technical part list data•Management of coding•Recording of documents and management of revisions•Management of B.O.M. using a multilevel iconeditorQueries on part numbers, documents, B.O.M.structure and “where used” of part numbers•Control of access to data•Release of whole B.O.M. or single part numbers•Possibility of creation of procedures finalized in theautomation of routine work, for example automaticcodingData flowThe first step towards making the information necessary for management of a product available to DataWorks, is the creation of a part number.During the coding phase it is possible to use a code search to identify the first one available.When there is an existing part number in the data base of DataWorks it is possible to:•use the editor of B.O.M. to create one that puts together the various part number codes•visualize the correlation between codes and make eventual changes and memorize these variations inthe data base•connect to the part numbers created, a series of documents (three-dimensional model, technicalmanual, spreadsheet, Part Program) so that it ispossible to associate all the technical informationconcerning it.DataworksWebIn the Web version, DataWorks allows the consultation, in real time, of data produced by the technical department from remote places connected to the server via Internet.This makes possible all activities of interrogation of data bases: research, “where used” and Bill of Material (B.O.M.) structure and local printing of documents connected to part numbers.DataWorksWeb is an indispensable tool for those who have productive units or offices distant from their central officesERPThe ERP procedure of DataWorks, allows the connection between different company sectors by means of automatic data transfer, guaranteeing continuous, safe up-dating of the data. System RequirementClient•Microsoft Windows XP Professional x32 or x64 Edition•Windows 7 Professional x32 or x64 EditionServer•Microsoft Windows Server 2008 R2DatabaseRelational databases containing information are based onDBMS applications. Supported databases:•Microsoft SQL Server 2008 R2•Oraclethey offer the company extreme flexibility of choice in functionof the available resources, presence of other applicationpackages and the nature and dimensions of the data base to bemanaged.23870 Cernusco Lombardone (LC) Italytel.: +39 039 99 09 703fax. +39 039 99 05 125E-mail: *****************www.aebtechno.it。

plsqldeveloper12使用方法PL/SQL Developer 12 使用方法PL/SQL Developer 12 是一款功能强大的集成开发环境(IDE),专门用于开发和调试Oracle 数据库中的PL/SQL 程序。
本文将介绍如何正确使用 PL/SQL Developer 12,以帮助开发人员更高效地编写和管理 PL/SQL 代码。
一、安装和配置在开始使用PL/SQL Developer 12 之前,首先需要下载并安装该软件。
安装过程相对简单,只需按照安装向导的指示进行操作即可。
安装完成后,打开软件,会出现一个欢迎界面,点击"Next"按钮进入配置界面。
在配置界面中,需要设置连接到Oracle 数据库的相关信息,包括数据库的主机名、端口号、用户名和密码等。
确保输入的信息准确无误,并点击"Test"按钮测试连接是否成功。
如果连接成功,点击"Next"按钮进行下一步配置。
二、界面介绍PL/SQL Developer 12 的界面分为多个区域,包括菜单栏、工具栏、对象浏览器、编辑器等。
下面将逐一介绍这些区域的功能和使用方法。
1. 菜单栏和工具栏:菜单栏提供了各种功能的选项,可以通过点击菜单项来执行相应的操作。
工具栏上的图标提供了常用功能的快捷方式,方便快速访问。
2. 对象浏览器:对象浏览器位于左侧,用于显示数据库中的对象,如表、视图、存储过程等。
可以通过展开相应的节点来查看和管理这些对象。
3. 编辑器:编辑器是PL/SQL Developer 12 的核心功能之一,用于编写和编辑PL/SQL 代码。
在编辑器中,可以进行代码的输入、修改和保存等操作。
同时,编辑器还提供了代码自动补全、语法高亮等功能,大大提高了编码效率。
三、基本操作1. 新建文件:点击菜单栏的"文件"选项,选择"新建",或者使用快捷键"Ctrl + N"来新建一个空白文件。

plsql developer 12使用手册**一、介绍PL/SQL Developer 12的使用手册**PL/SQL Developer 12是一款强大的Oracle数据库开发工具,这款工具为开发者提供了丰富的功能,使得编写、调试和维护PL/SQL代码变得更加简单。
本使用手册将详细介绍如何安装、配置以及高效使用PL/SQL Developer 12。
**二、安装和配置PL/SQL Developer 12**1.下载PL/SQL Developer 12安装程序并根据安装向导完成安装。
2.安装完成后,启动PL/SQL Developer 12,按照提示完成初始配置。
**三、掌握PL/SQL Developer 12的主要功能和工具**PL/SQL Developer 12提供了丰富的功能和工具,包括项目管理、代码编辑、调试、测试、性能监测等。
在本手册中,我们将重点介绍如何使用这些功能和工具。
**四、详细介绍PL/SQL Developer 12的编辑器和工作区**1.编辑器:PL/SQL Developer 12的编辑器支持代码高亮、自动补全、语法检查等功能,帮助开发者快速编写高质量的代码。
2.工作区:PL/SQL Developer 12的工作区允许开发者轻松管理项目文件、数据库对象和版本控制。
**五、掌握PL/SQL Developer 12的调试和测试功能**1.调试:PL/SQL Developer 12提供了强大的调试功能,帮助开发者定位和修复代码中的问题。
2.测试:PL/SQL Developer 12支持编写和执行单元测试,确保代码质量。
**六、了解如何使用PL/SQL Developer 12进行团队协作**1.版本控制:PL/SQL Developer 12支持多种版本控制工具,如Git、SVN等。
2.团队协作:PL/SQL Developer 12提供共享项目功能,实现团队成员之间的协同工作。



数据库常见命令及解释数据库是用于存储和管理数据的系统,常见的关系型数据库包括MySQL、Oracle、SQL Server等。
在使用数据库时,需要用到一些常见的数据库命令进行数据的操作。
本文将介绍一些常见的数据库命令及其解释,以帮助读者更好地理解和使用数据库。
1. SELECTSELECT命令用于从数据库中检索数据。
它可以检索特定的列、行或表中所有的数据。
语法如下:SELECT列名称FROM表名称2. INSERTINSERT命令用于向数据库中插入新的数据。
它可以将数据插入到一个表中的指定列或所有列。
语法如下:INSERT INTO表名称(列1,列2,列3,...) VALUES (值1,值2,值3,...)3. UPDATEUPDATE命令用于修改数据库中现有数据。
它可以更新一个表中的指定列或所有列。
语法如下:UPDATE表名称SET列名称=新值WHERE条件4. DELETEDELETE命令用于从数据库中删除数据。
它可以删除一个表中的指定行或所有行。
语法如下:DELETE FROM表名称WHERE条件5. CREATE TABLECREATE TABLE命令用于创建一个新的表。
它定义了表的名称和列名,并指定了每列的数据类型和约束条件。
语法如下:CREATE TABLE表名称(列1数据类型,列2数据类型, ...)6. DROP TABLEDROP TABLE命令用于删除一个表。
它将永久删除表的结构和数据。
语法如下:DROP TABLE表名称7. ALTER TABLEALTER TABLE命令用于修改一个表的结构。
它可以添加新的列、修改列的数据类型、删除列等。
语法如下:ALTER TABLE表名称ADD列名称数据类型8. CREATE INDEXCREATE INDEX命令用于创建索引。
索引可以加快数据检索的速度。
语法如下:CREATE INDEX索引名称ON表名称(列名称)9. DROP INDEXDROP INDEX命令用于删除索引。

Required privileges and permissionsTable of contentsDocument summary ADSelfService Plus overview Required permissions Configuring permissions 1 1 2 3 3 8 12 13 14 1518 19 21To delegate full control in ADUC to access all ADSelfService Plus features To delegate the right to reset user passwords in ADUCTo delegate the right to unlock user accounts in ADUCTo delegate the right to modify user attributes in ADUCTo delegate the right to read user PSO in ADUCTo delegate the right to modify members of a group in ADUCTo delegate the right to create a computer account in ADUCTo delegate the right to modify user logon script path in ADUCTo view deleted users reportTo install Windows login agent To perform other actions 21 22 17To synchronize AD user objects with ADSelfService PlusDocument summaryThis guide will walk you through the process of delegating an Active Directory user account with the required permissions for using the self-service features in ADSelfService Plus. ADSelfService Plus does not require "Domain Admin" membership in order to allow users to reset their passwords, unlock their accounts, update their profiles, or access any of its other features. Based on the principle of least privilege, you can delegate only the permissions required for the self-service operations to a user account manually.If you don't provide any authentication details while adding domains, ADSelfServicePlus will get its privileges one of two ways:●If ADSelfService Plus is installed to run as a console application and no credentialsare provided, then by default it uses the permissions of the user who installed theproduct.●If ADSelfService Plus is installed to run as a service and no credentials are provided,then by default it uses the permissions of the account used to run the service. ADSelfService Plus overviewManageEngine ADSelfService Plus, an integrated Active Directory self-service password man-agement and single sign-on solution, helps reduce password reset tickets and spares end users the frustration caused by computer downtime. It o ers,Self-service password reset and account unlockPassword and account expiration notifierPassword policy enforcerEnterprise single sign-on and password synchronizerEndpoint multi-factor authentication for machine loginsDirectory self-update and employee searchThese features, designed to strike a balance between ensuring network security andease-of-access, warrants improved ROI, and a productive IT workforce.Configuring permissionsTo access all ADSelfService Plus featuresFor users to access all features of ADSelfService Plus, you'll need to grant the ADSelfService Plus service account the following permissions:1.Right-click the domain in ADUC and select Delegate Control from the context menu.2.Click Next in the welcome dialog box.3.Click Add to select the user account or service account, then click OK followed by Next.4.Select Delegate the following common tasks and check the Reset user passwords andforce password change at next logon, Read all user information, and Modify themembership of a group boxes, then click Next.5.Click Finish and repeat steps 1-3.6.Select Create a custom task to delegate and click Next.7.Select Only the following objects in the folder. In the given list,select UserObjects.8.Select the General box. Under Permissions,check the boxes for Read and Write beforeclicking Next.9.Click Finish and repeat steps 1-3.10.Select Create a custom task to delegate and click Next.11.Select Only the following objects in the folder. In the given list, select ComputerObjects and Create selected objects in this folder.12.Select the General box. Under Permissions, check Read before clicking Next.13.Click Finish and repeat steps 1-3.14.Select Create a custom task to delegate and click Next.15.Select Only the following objects in the folder. In the given list, selectmsDS-PasswordSettings objects and msDS-PasswordSettingsContainer objects. Click Next.16.Select the General box. Under Permissions, check Read before clicking Next.17.Click Finish.Self-service password resetI n order for this feature to work, you need to delegate the permission to reset users' passwords in the ADUC console. To do this, follow the steps below:1.Right-click the OU or domain in ADUC and select DelegateControl from2.Click Next in the welcome dialog box.3.Click Add to select the ADSelfService Plus user account or serviceaccount, then click OK.4.Click Next.5.Select Create a custom task to delegate and click Next.6.Select Only the following objects in the folder. In the given list, selectand click NextUser objects7.Check the General and Property-specific boxes.8.Under Permissions, check the boxes for Reset password,Read pwdLastset,and Write pwdLastset before clicking Next.9.Click Finish.Note: This permission only enables password reset.Self-service account unlockFor this feature to work, you need to delegate the permission to unlock users' accounts in the ADUC console. To do this:1.Right-click the OU or domain in ADUC and select Delegate Control from thecontext menu.2.Click Next in the welcome dialog box.3.Click Add to select the ADSelfService Plus user account or service account,then click OK.4.Click Next.5.Select Create a custom task to delegate and click Next.6.Select Only the following objects in the folder. In the given list, check Userobjects and click Next.7.Check the General and Property-specific boxes.8.Under Permissions, check the Read lockoutTime and Write lockoutTime boxesand click Next.9.Click Finish.Note: This permission only enables account unlock.Directory self-updateTo utilize this feature, you need to delegate the permission to modify user attributes in the ADUC console. Follow the steps below to do so:1.Right-click the OU or domain in ADUC and select Delegate Control from thecontext menu.2.Click Next in the welcome dialog box.3.Click Add to select the user account or service account, then click OK .4.Click Next .5.Select Create a custom task to delegate and click Next .6.Select Only the following objects in the folder . In the given list, select User objectsand click Next .7.Check the General and Property-specificboxes.Under Permissions, check the Read and Write boxes (or select the Read and Writeboxes of specific attributes that need to be available for end user self-update), andclick Next.Click Finish .Note: This permission only enables self-update.8.9.Display fine-grained password policyIf you want to display the exact password policy requirements in the reset/ change password screen for users with fine-grained password policy enabled, then you need to delegate the permission to read the users' Password Settings Objects (PSOs) in ADUC. Follow the steps below to do this:1.Right-click the OU or domain in ADUC and select Delegate Control from thecontext menu.Click Next in the welcome dialog box.2.Click Add to select the user account or service account, then click OK.3.Click Next.4.Select Create a custom task to delegate and click Next.5.Select Only the following objects in the folder. In the given list, selectmsDS-PasswordSettings objects and msDS-PasswordSettingsContainer objects before clicking Next.7.Check the General box.8.Under Permissions, select Read and click Next.9.Click Finish.Note: This permission only fetches the password policy.Self-service mail group subscriptionTo use this feature, you need to delegate the permission to modify members of a group in the ADUC console. Follow the steps below to do so:1.Right-click the OU or domain where the group containing members that requiremodification belongs in ADUC, and select Delegate Control from the context menu.2.Click Next in the welcome dialog box.3.Click Add to select the user account or service account, then click OK. Click Next.4.Select Create a custom task to delegate and click Next.5.Select Only the following objects in the folder. In the given list, select Group objectsand click Next.Synchronizing AD user objects with ADSelfService PlusTo synchronize Active Directory objects with ADSelfService Plus without any issue,you need to provide the Replicate Directory Changes permission to the user or serviceaccount used in ADSelfService Plus. To do this, follow the steps below:1.In the ADUC console, right-click the domain or OU and select Properties.2.Under the Security tab, click Add to select the user or service account.3.In the Permissions section, Allow the Replicate Directory Changes permission.4.Click OK.8.Click Finish.Note: This permission only enables mail group subscription.Single sign-on to ADSelfService Plus via NTLMv2For this feature to work, you need to delegate the permission to create and readcomputer accounts in the ADUC console. To do this, follow the steps below:1.Right-click the Computers OU or domain in ADUC and select Delegate Control from thecontext menu.2.Click Next in the welcome dialog box.3.Click Add to select the ADSelfService Plus user account or service account, then click OK.4.Click Next.5.Select Create a custom task to delegate and click Next.6.Select Only the following objects in the folder. In the given list, select Computer objects andCreate selected objects in this folder and click Next.7.Check the General box.8.Under Permissions, check the Read box and clickNext.9.Click Finish.Note: This permission only enables you to configure NT LAN Manager (NTLMv2) SSO to ADSelfService Plus.Force enrollment using a logon scriptTo use this feature, you need to delegate the permission to modify the user scriptPath in the ADUC console. Follow the steps below to do this:1.Right-click the OU or domain in ADUC and select Delegate Control from thecontext menu.2.Click Next in the welcome dialog box.3.Click Add to select the user account or service account, then click OK.4.Click Next.5.Select Create a custom task to delegate and click Next.6.Select Only the following objects in the folder. In the following list, check Userobjects and click Next.7.Check both the General and Property-specific boxes.8.Under Permissions, check the Read scriptPath and Write scriptPath boxes andclick Next.9.Click Finish.Note: This permission only enables logon script path modification.To view a deleted users reportThe minimum requirement to view this report is membership in the Domain Admins group.To perform GINA installationThe minimum requirement to perform GINA installation from ADSelfService Plus' web console is membership in the Domain Admins group.If Domain Admin credentials are not available for use,you can install GINA manually via Group Policy Objects (GPOs) or using System Center Configuration Manager (SCCM).Folder permissions for other actionsThe service account used to run ADSelfService Plus and the local user account used to start ADSelfService Plus must be granted full control permission to the product installation folder. Otherwise, you won't be able to:●Install service packs●Generate reports●Start up the product●Apply licenses●Update dashboard graphs●Back up and restore data●Display employee photos and offer users self-update optionsTo configure high availabilityThe minimum requirement to configure high availability in ADSelfService Plus is membership in the Domain Admins group. Domain Admin privileges are only mandatory during the initial setup of high availability. Once high availability has been configured, the service account can be changed to one with lesser privileges based on other features configured. Ensure that folder sharing between both the instances is uninterrupted.Our ProductsAD360 | Log360 | ADManager Plus | ADAudit Plus | RecoveryManager Plus | M365 Manager Plus https:///products/self-service-password。

Oracle基础(习题卷8)第1部分:单项选择题,共63题,每题只有一个正确答案,多选或少选均不得分。
1.[单选题]下列()不是一个角色A)CONNECTB)DBAC)RESOURCED)CREATE SESSION答案:D解析:2.[单选题]在以下命令中,一个使用哪一个来激活(开启)一个角色?( )A)SET ROLLB)ALTER USERC)CREATE ROLED)ALTER SYSTEM答案:A解析:3.[单选题]当一个段需要额外的磁盘空间时,应该增加哪一种数据库的逻辑组件?( )A)区段( extents)B)表空间C)数据库块D)操作系统块答案:A解析:4.[单选题]存储数据的逻辑单位,按大小依次为()。
A)表空间、数据块、区和段B)区、表空间、数据块和段C)段、区、表空间和数据块D)表空间、段、区和数据块答案:D解析:5.[单选题]A non-correlated subquery can be defined as . (Choose the best answer.)A)A set of one or more sequential queries in which generally the result of the inner query is used as the search value in the outer query.B)A set of sequential queries, all of which must return values from the same table.C)A set of sequential queries, all of which must always return a single value.D)A SELECT statement that can be embedded in a clause of another SELECT statement only.答案:A解析:C)alter system remove user brent cascade;D)drop user brent cascade;答案:D解析:7.[单选题]系统事件触发器共支持5种系统事件,下列()事件不会激发触发器。

Package‘ABPS’October12,2022Title The Abnormal Blood Profile Score to Detect Blood DopingVersion0.3Date2018-10-18Description An implementation of the Abnormal Blood Profile Score(ABPS,part of the Athlete Biological Passport program of the World Anti-DopingAgency),which combines several blood parameters into a single score inorder to detect blood doping(Sottas et al.(2006)<doi:10.2202/1557-4679.1011>).The package also contains functions tocalculate other scores used in anti-doping programs,such as theOFF-score(Gore et al.(2003)</content/88/3/333>),as well as example data.Imports kernlabLicense GPL(>=2)Encoding UTF-8LazyData trueRoxygenNote6.0.1Suggests testthatNeedsCompilation noAuthor Frédéric Schütz[aut,cre],Alix Zollinger[aut]Maintainer Frédéric Schütz<*****************>Repository CRANDate/Publication2018-10-1812:30:10UTCR topics documented:ABPS (2)bloodcontrol (3)blooddoping (4)OFFscore (5)Index712ABPS ABPS A function for calculating the Abnormal Blood Profile ScoreDescriptionThe ABPS function computes the Abnormal Blood Profile Score from seven haematological markers.Higher values of this composite score are associated with a higher likelihood of blood doping. UsageABPS(haemdata=NULL,HCT=NULL,HGB=NULL,MCH=NULL,MCHC=NULL,MCV=NULL,RBC=NULL,RETP=NULL)Argumentshaemdata a vector or data frame containing(at least)the7haematological variables,either with the same names as the parameters below,or(not recommended)withoutnames but in the same order as the parameters.HCT haematocrit level[%]HGB haemoglobin level[g/dL]MCH mean corpuscular haemoglobin[pg]MCHC mean corpuscular haemoglobin concentration[g/dL]MCV mean corpuscular volume[fL]RBC red blood cell count[10^6/uL]RETP reticulocytes percent[%]DetailsThe ABPS uses the seven haematological variables(HCT,HGB,MCH,MCHC,MCV,RBC,RETP) in order to obtain a combined score.This score is more sensitive to doping than the individual markers,and allows the detection of several types of blood doping using a single score.The combined score is based on two classification techniques,a naive Bayesian classifier and an SVM(Support Vector Machine).The two models were trained using a database of591blood profiles(including402control samples from clean athletes and189samples of athletes who abused of an illegal substance);the two scores were then combined using ensemble averaging to obtain the final ABPS score.The ABPS is part of the Athlete Biological Passport program managed by the World Anti-Doping Agency.While it is not a primary marker of doping,it has been used as corroborative evidence(seee.g.https:///Shared%20Documents/2773.pdf)Valuea vector containing the ABPS score(s).Scores between0and1indicate a possible suspicion ofdoping;a score above1should only be found in1in1000male athletes.bloodcontrol3NoteThe values for the markers can be specified using either a data frame containing(at least)the7 haematological variables,or using seven named parameters,but not both at the same time.The calculation of the ABPS depends on two sets of parameters,for the two machine learning tech-niques(naive Bayesian classifier and Support Vector Machine),which are provided in the package.Each parameter must be in a prespecified range;parameters outside this range are constrained to the min(respectively max)values.Note that several versions of the ABPS were developed(including several different combinations of parameters).The version provided in this package provides the same results as the W ADA version included in their ADAMS database.However,some values calculated with other versions of the software have also been distributed(see the help page for the blooddoping dataset for an example). ReferencesSottas,P.E.,N.Robinson,S.Giraud,et al.,Statistical classification of abnormal blood profiles in athletes.Int J Biostat,2006.2(1):p.1557-4679.https:///Shared%20Documents/2773.pdfExamplesABPS(HCT=43.2,HGB=14.6,MCH=31.1,MCHC=33.8,MCV=92.1,RBC=4.69,RETP=0.48)ABPS(data.frame(HCT=43.2,HGB=14.6,MCH=31.1,MCHC=33.8,MCV=92.1,RBC=4.69,RETP=0.48)) ABPS(c(43.2,14.6,31.1,33.8,92.1,4.69,0.48))data(blooddoping);ABPS(blooddoping)data(bloodcontrol);ABPS(bloodcontrol)bloodcontrol Blood samples from different individuals.DescriptionA dataset containing the result of the analysis of13blood samples from different individuals. UsagebloodcontrolFormatA data frame with13rows and12haematological variables:HCT haematocrit[%]HGB haemoglobin[g/dL]IRF immature reticulocyte fraction[%]MCH mean corpuscular haemoglobin[pg]4blooddopingMCHC mean corpuscular haemoglobin concentration[g/dL]MCV mean corpuscular volume[fL]RBC red blood cell count[10^6/uL]RDW.SD red blood cell distribution width[fL]RETC reticulocyte count[10^6/uL]RETP reticulocyte percentage[%]OFFscore OFF-scoreABPS Abnormal Blood Profile ScoreThese samples are assumed to represent normal population.NoteOne of the rows actually belongs to one of the authors of this package,who promises that he was not doped.SourceSwiss Laboratory for Doping Analyses(LAD),with some calculations performed by the World Anti-Doping Agency(W ADA).blooddoping Blood samples from an athlete convicted of doping.DescriptionA dataset containing the result of the analysis of13blood samples,taken over a period of5years,of a female athlete who was convicted of doping on the basis of the Athlete Biological Passport. UsageblooddopingFormatA data frame with13rows and11variables(including10haematological variables):date the date of testHCT haematocrit[%]HGB haemoglobin[g/dL]MCH mean corpuscular haemoglobin[pg]MCHC mean corpuscular haemoglobin concentration[g/dL]MCV mean corpuscular volume[fL]RBC red blood cell count[10^6/uL]RETC reticulocyte count[10^6/uL]RETP reticulocyte percentage[%]OFFscore OFF-scoreABPS Abnormal Blood Profile ScoreIn November2012,the athlete was convicted of doping by the Court of arbitration for Sport,the evidence showing at least two occurences of doping,in summer2009(shortly before the test of2 July2009)and in June2011,likely using an agent such as recombinant erythropoietin(rhEPO).Doping is indicated in particular by high haemoglobin values associated with very low reticulocyte %and high OFF scores(a combination of these two variables).NoteThe data tables published in the original source contain several typos,as confirmed by the World Anti-Doping Agency(W ADA).In particular,values for RETP in rows8and10were swapped and the MCHC value for row7was incorrect.This package’s source code contains both the original data and the details of the corrections that were applied.The ABPS values provided are very close(within<2%)to those obtained using the ABPS function, but some of them were likely calculated using different(older)versions of the ABPS code,which may explain some of these differences.Sourcehttps:///Shared%20Documents/2773.pdfOFFscore function OFFscoreDescriptionThe OFFscore function computes the value of the OFF-hr score(or OFF score),a combination of the haemoglobin level and the percentage of reticulocytes,used for detecting blood doping. UsageOFFscore(haemdata=NULL,HGB=NULL,RETP=NULL)Argumentshaemdata a vector or data frame containing(at least)the2haematological variables,either with the same names as the parameters below,or(not recommended)withoutnames but in the same order as the parameters.HGB level of haemoglobin HGB[g/dL]RETP percentage of reticulocytes[%]DetailsThe OFF-hr score is defined as10*HGB[g/dL]-60*sqrt(RETP[%]).It is one of the parameters of the Athlete Biological Passport(ABP)program managed by the World Anti-Doping Agency(W ADA),and is routinely used to identify athletes who use a sub-stance prohibited by anti-doping rules(see e.g.https:///Shared% 20Documents/4006.pdf).The rationale for using this score for detecting blood doping is the following:if a manipulation (use of transfusion or of erythropoietic stimulating agents(ESA)such as recombinant human ery-thropoietin(rhEPO))increases the number of circulating red cells(and thus increases the level of haemoglobin),the organism will react by stopping its own production of red blood cells.This nega-tive feedback will be observed in the reduced percentage of reticulocytes(immature red blood cells).Such a combination of elevated HGB and reduced RETP,which will produce a high OFF score,is found neither naturally,nor as the consequence of a medical condition,and is thus indicative of doping.The OFF score will pick up both withdrawal of blood(which induces a reduction in haemoglobin,a rise in reticulocytes,and thus a rise of RETP and a reduction of OFF score),and its re-infusion(HGB concentration increases,number of reticulocytes and RETP decrease,and OFF score in-creases).Valuevalue of the OFF score.NoteThe values for HGB and RETP can be specified using either a data frame containing(at least)the2 haematological variables,or using two named parameters,but not both at the same time.The original OFF-hr score,as described by Gore et al.,expects the haemoglobin level HGB to be specified in g/L.In order to be coherent with other functions in the package,this function assumes that HGB is specified in g/dL,and will multiply the value by10.A warning will be emitted if units used seem wrong.ReferencesGore,C.J.,R.Parisotto,M.J.Ashenden,et al.,Second-generation blood tests to detect erythropoi-etin abuse by athletes.Haematologica,2003.88(3):p.333-44.ExamplesOFFscore(HGB=14.6,RETP=0.48)data(blooddoping)OFFscore(HGB=blooddoping$HGB,RETP=blooddoping$RETP)Index∗datasetsbloodcontrol,3blooddoping,4ABPS,2bloodcontrol,3blooddoping,4OFFscore,57。
