DIG试剂盒说明书(CSPD)中文翻译版
- 格式:doc
- 大小:225.00 KB
- 文档页数:32
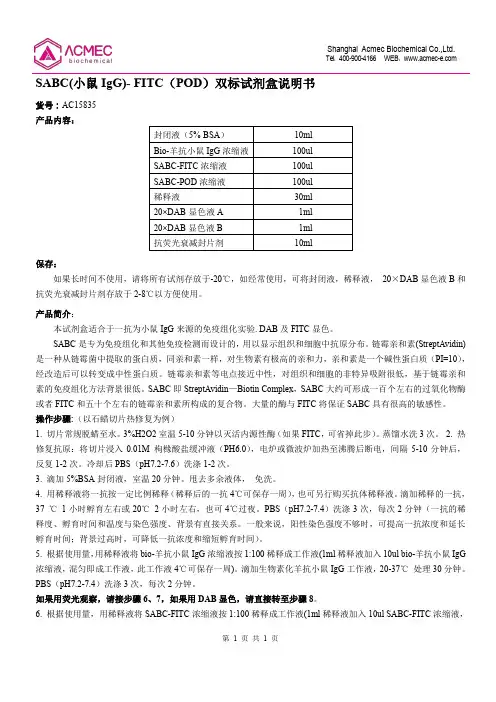
SABC(小鼠IgG)- FITC (POD )双标试剂盒说明书货号:AC15835产品内容:封闭液(5% BSA ) 10ml Bio-羊抗小鼠IgG 浓缩液 100ul SABC-FITC 浓缩液 100ul SABC-POD 浓缩液 100ul 稀释液30ml 20×DAB 显色液A 1ml 20×DAB 显色液B 1ml 抗荧光衰减封片剂10ml 保存:如果长时间不使用,请将所有试剂存放于-20℃,如经常使用,可将封闭液,稀释液, 20×DAB 显色液B 和抗荧光衰减封片剂存放于2-8℃以方便使用。
产品简介: 本试剂盒适合于一抗为小鼠IgG 来源的免疫组化实验. DAB 及FITC 显色。
SABC 是专为免疫组化和其他免疫检测而设计的,用以显示组织和细胞中抗原分布。
链霉亲和素(StreptAvidin)是一种从链霉菌中提取的蛋白质,同亲和素一样,对生物素有极高的亲和力,亲和素是一个碱性蛋白质(PI=10),经改造后可以转变成中性蛋白质。
链霉亲和素等电点接近中性,对组织和细胞的非特异吸附很低,基于链霉亲和素的免疫组化方法背景很低。
SABC 即StreptAvidin —Biotin Complex ,SABC 大约可形成一百个左右的过氧化物酶或者FITC 和五十个左右的链霉亲和素所构成的复合物。
大量的酶与FITC 将保证SABC 具有很高的敏感性。
操作步骤:(以石蜡切片热修复为例)1. 切片常规脱蜡至水。
3%H2O2室温5-10分钟以灭活内源性酶(如果FITC ,可省掉此步)。
蒸馏水洗3次。
2. 热修复抗原:将切片浸入0.01M 枸橼酸盐缓冲液(PH6.0),电炉或微波炉加热至沸腾后断电,间隔5-10分钟后,反复1-2次。
冷却后PBS (pH7.2-7.6)洗涤1-2次。
3. 滴加5%BSA 封闭液,室温20分钟。
甩去多余液体, 免洗。
4. 用稀释液将一抗按一定比例稀释(稀释后的一抗4℃可保存一周),也可另行购买抗体稀释液。

【产品名称】通用名称:他克莫司检测试剂盒(电化学发光法)英文名称:Tacrolimus【包装规格】100测试/盒【预期用途】主要用途主要用于定量测定人全血中他克莫司的含量。
本检测有助于接受他克莫司治疗的心、肝、肾移植病人的治疗。
电化学发光免疫测定试剂“ECLIA”,适用于罗氏Elecsys 和cobas e免疫测定分析仪。
临床应用他克莫司(又称KF506)是一种大环内酯类抗生素,1984年在日本被发现,是链霉菌属的代谢产物1.2,3。
研究表明他克莫司的免疫抑制活性是环孢霉素的10-100倍。
4他克莫司产生免疫抑制效应的主要原理在于它可以抑制T细胞的激活和增殖。
细胞内的他克莫司可结合一种抑免蛋白FK506-结合蛋白(FKBP-12),这种免疫复合物可抑制磷酸酶的活性。
5抑制磷酸酶可限制激活的T细胞(NFAT)的核因子的去磷酸化和核转位,由此调节包括IL-2、IL-4、TNF-α和γ-干扰素在内的多种细胞因子的转录,进一步限制淋巴细胞的激活和增殖。
6,7,8,9,10他克莫司具有很高的亲脂性,吸收不完全并且不稳定。
吸收后,他克莫司高效结合蛋白和红细胞,血浆中99%的药物都与白蛋白或α-1-糖蛋白结合。
11他克莫司的生物利用度和代谢主要受细胞色素P450药物代谢同工酶CYP3A4和CYP3A5以及流出泵P-糖蛋白的影响,在表达和功能方面在同一个体以及不同个体之间都变现出明显的差异。
12,13,14他克莫司在同一病人和不同病人间都表现出很大的变异性,无论剂量高低都可能出现严重不良反应。
他克莫司浓度不足可能导致对抑制器官的排异反应。
浓度过高又可导致严重不良反应。
他克莫司的主要不良反应包括肾毒性、神经毒性、胃肠道损害、糖尿病、高血压病和恶性肿瘤。
15,16为了让每个病人的体内药物浓度都维持在狭窄的治疗窗范围内,治疗药物监测的应用和浓度控制下给药作为标准临床实践的一部分已在临床上应用多年,它也是病人治疗的主要手段。
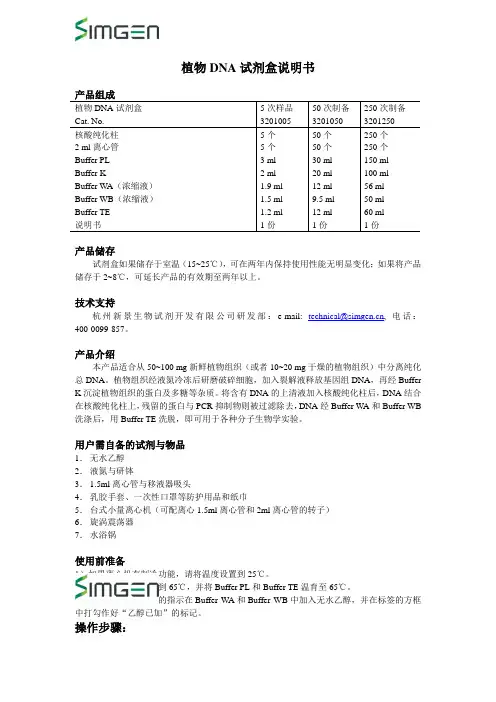
植物DNA试剂盒说明书植物DNA试剂盒Cat. No. 5次样品320100550次制备3201050250次制备3201250核酸纯化柱2 ml离心管Buffer PLBuffer KBuffer WA(浓缩液)Buffer WB(浓缩液)Buffer TE说明书5个5个3 ml2 ml1.9 ml1.5 ml1.2 ml1份50个50个30 ml20 ml12 ml9.5 ml12 ml1份250个250个150 ml100 ml56 ml50 ml60 ml1份产品储存试剂盒如果储存于室温(15~25℃),可在两年内保持使用性能无明显变化;如果将产品储存于2~8℃,可延长产品的有效期至两年以上。
技术支持杭州新景生物试剂开发有限公司研发部:e-mail: technical@, 电话:400-0099-857。
产品介绍本产品适合从50~100 mg新鲜植物组织(或者10~20 mg干燥的植物组织)中分离纯化总DNA。
植物组织经液氮冷冻后研磨破碎细胞,加入裂解液释放基因组DNA,再经Buffer K沉淀植物组织的蛋白及多糖等杂质。
将含有DNA的上清液加入核酸纯化柱后,DNA结合在核酸纯化柱上,残留的蛋白与PCR抑制物则被过滤除去,DNA经Buffer WA和Buffer WB 洗涤后,用Buffer TE洗脱,即可用于各种分子生物学实验。
用户需自备的试剂与物品1.无水乙醇2.液氮与研钵3.1.5ml离心管与移液器吸头4.乳胶手套、一次性口罩等防护用品和纸巾5.台式小量离心机(可配离心1.5ml离心管和2ml离心管的转子)6.旋涡震荡器7.水浴锅使用前准备1)如果离心机有制冷功能,请将温度设置到25℃。
2)将水浴锅温度设置到65℃,并将Buffer PL和Buffer TE温育至65℃。
3)根据试剂瓶标签上的指示在Buffer WA和Buffer WB中加入无水乙醇,并在标签的方框中打勾作好“乙醇已加”的标记。


790000116人份试剂盒Cell Search循环肿瘤细胞检测试剂盒(上皮细胞)IVD使用须知本试剂盒仅用于体外诊断CellSearch循环肿瘤细胞检测试剂盒用于外周血中循环肿瘤细胞(CTC)的计数,这些细胞来源于上皮细胞,即表现为CD45阴性(CD45-),EpCAM阳性(EpCAM+),细胞角蛋白8,18阳性(CK8,18+)和(或者)CK19阳性(CK19+)的细胞。
用CellSearch循环肿瘤细胞检测试剂盒检测的CTC,存在于外周血中,这类细胞与乳腺癌、结直肠癌和前列腺癌*转移患者的无进展生存期和总生存期相关。
因此,CTC可以用来监控乳腺癌、结直肠癌和前列腺癌患者的肿瘤细胞是否发生转移。
CTC应该进行持续检测,并与其他临床诊断方式联合起来监控上述肿瘤细胞的转移。
在肿瘤发生过程中的任何时间段,对CTC数量进行评估,可以预估肿瘤患者治疗后的无进展生存期和总生存期。
*在该研究中,转移性前列腺癌患者定义为血清标志物PSA在标准激素基准上,高于参考水平具有两次连续增加。
这些患者通常描述为具有非雄激素依赖性,激素抗性,或去势抗性的前列腺癌。
摘要说明肿瘤转移是指当肿瘤细胞从原发灶或转移灶剥落后,这些肿瘤细胞进入循环系统并在机体的远端定植生长的情况。
血液循环中的肿瘤细胞源自于上皮细胞,而这类细胞在血液循环系统中是极罕见的。
基于此,本公司推出的细胞自动化捕获仪(CELLTRACKS○R AUTOPREP○R System)通过预设程序并配合使用CellSearch循环肿瘤细胞检测试剂盒(CELLSEARCH○R Kit)可以对样本进行标准化和自动化检测。
用CELLTRACKS II分析仪或者CellSpotter分析仪,一种半自动荧光显微镜,可以对肿瘤细胞进行分析和计数。
这种方法只计具有上皮细胞特性(EpCAM+, CK8,18+,和/或者CK19+)的细胞数量。
检测原理CellSearch循环肿瘤细胞检测试剂盒包括磁流体捕获试剂和荧光免疫试剂。

Q I A G E N-D N A甲基化试剂盒说明中文版-CAL-FENGHAI.-(YICAI)-Company One1QIAGEN EpiTect® Bisulfite•重亚硫酸盐处理DNA时,需要经高盐浓度、高温和低PH条件,会导致DNA断裂和片段化,并在随后的纯化过程中丢失,所以需要大量的input DNA•重亚硫酸盐转化未甲基化的胞嘧啶,转化率超过99%•Bisulfite Mix 足够做8个反应,用时要将Bisulfite Mix溶于800μl RNase-freewater中,溶解的Bisulfite Mix 在-20℃可以保存4周•DNA Protect Buffer 能保护重亚硫酸盐处理的DNA在高温、低ph条件下被片段化•Carrier RNA 能提高小量DNA(小于500pg,体积大于40μl)绑定在回收柱滤膜上的效率,帮助核酸沉淀;总量大于100ng的基因组DNA没有必要加Carrier RNA•EpiTect Bisulfite Kit在-20℃可以保存3年;可以适用于大范围的DNA量,input DNA范围为1ng~2μg;模板DNA片段范围可以在500bp~30kb 之间ProtocolDNA量在1ng~2μg之间,体积20μl以上开始前的准备重点:•Bisulfite Mix 足够做8个反应,如果样本少于8个,可以把溶解的Bisulfite Mix 保存于-20℃,4周内并不会影响其性能•DNA Protect Buffer在加入DNA–Bisulfite Mix后会由绿变蓝(step 2),表明溶液混合很充分且PH值正确•所有的离心分离步骤均在室温(15–25°C)下完成开始前准备的东西:•加入30ml乙醇(96%~100%)到Buffer BW中,室温(15–25°C)保存,使用之前摇匀•加入27ml乙醇(96%~100%)到Buffer BD中,2~8℃保存,使用之前摇匀,使用之后要立即盖上盖子。


ApplicationThe kit is specially designed for the generation of highly sensitive hybridization probes that are suitable for the detection of low- (single-) copy target sequences. Thus, it should be considered as an alternative to random-primed labeling when either the amount of template DNA is limited or only a specific part of the sequence of the template is required for hybridization. The nucleotide concentration in the PCR DIG Probe Synthesis Kit ensures the identification of single-copy genes in the genomic blots following hybridization to DIG-labeled PCR products. Human single-copy genes are detectable in 10 µg of genomic DNA.PCR products can be directly generated and labeled from small amounts of genomic DNA (100 ng–1 µg), and subsequently used as hybridization probes.The PCR DIG Probe Synthesis Kit contains an alkali-labile DIG-11-dUTP formulation. This enables simple removal of the DIG label following chemiluminescent detection, and allows the subsequent rehybridization of blots with multiple DIG-labeled probes.Background InformationThe nonradioactive DIG system uses digoxigenin, a steroid hapten, to label DNA, RNA, or oligonucleotides for hybridization, and subsequent color- or luminescent detection. The digoxigenin is coupled to dUTP via an alkali-labile ester bond. The labeled dUTP can be easily incorporated by enzymatic nucleic-acid synthesis using DNA polymerases.The polymerase chain reaction (PCR) allows the amplification of minute amounts of DNA to levels above 1 µg. The only prerequisite is that some sequence information of the target sequence is needed in order to synthesize the appropriate primers.The combination of nonradioactive labeling with PCR is a powerful tool for the analysis of PCR products, and also for the preparation of labeled probes from small amounts of a respective target sequence. The detection sensitivity of these labeled probes can be adjusted by the ratio of the unlabeled nucleotides to DIG-dUTP. Whereas a low ratio of DIG-dUTP to dTTP of 1:19, as in the PCR DIG Labeling Mix, ensures a high-yield amplification reaction with good sensitivity for direct detection, the hybridization to single-copy genes in complex genomes may not be successful. The PCR DIG Probe Synthesis Kit, therefore, contains a high ratio of DIG-dUTP to unlabeled nucleotides of 1:2, ensuring reliable detection of single-copy genes. However, PCR yield may be decreased by up to 50%. Nevertheless, this ratio will yield enough product for several hybridizations.Contents1.Enzyme Mix, Expand High Fidelity, 30 µl (105 U) Enzyme Mix, 3.5 U/µl in storage buffer, 20 mMTris-HCl, pH 7.5 (25o C), 100 mM KCl, 1 mM dithiothreitol (DDT), 0.1 mM EDTA, 0.5% Tween 20 (v/v), 0.5%Nonidet P40 (v/v), 50% glycerol (v/v)2.PCR DIG Probe Synthesis Mix, 10x conc., 125 µl of a mixture containing dATP, dCTP, dGTP (2 mMeach), 1.3 mM dTTP, 0.7 mM DIG-11-dUTP, alkali-labile, pH 7.03.PCR Buffer with MgCl2, 10x conc., 1 ml Expand High Fidelity Buffer, 10x conc. with MgCl24.dNTP Stock Solution, 10x conc., 125 µl of a mixture containing dATP, dCTP, dGTP, dTTP (2 mM each),pH 7.05.Control Template, 50 µl (1 ng) plasmid DNA (20 pg/µl) in Tris/EDTA buffer, pH 8.0. The 5-kb plasmidcontains the cDNA for the human tissue-type plasminogen activator (tPA)6.Control PCR Primer Mix, 25 µl (containing 50 pmol of each primer) control PCR primer 1 and 2 (2 mMeach)QualityFunction test: Each lot of the PCR DIG Probe Synthesis Kit is function-tested in PCR. Amplification products are assayed in genomic Southern blots.Under PCR conditions described in the pack insert, the control reaction generates an amplification product of 442 bp. Due to multiple incorporations of DIG-dUTP during the PCR process, the molecular weight of the PCR products is significantly increased compared to the unlabeled PCR product.A specific fragment pattern is detected after hybridization of the PCR product to 10 µg human genomic DNA and chemiluminescent detection as described in the pack insert.。

七项呼吸道病毒检测试剂盒(鉴定)使用说明书【产品名称】通用名称:七项呼吸道病毒检测试剂盒英文名称:D3 Ultra DFA Respiratory Virus Screen & ID Kit【包装规格】鉴定试剂:80人份/盒【预期用途】用于临床定性检测7种呼吸道病毒:甲型(A型)流感病毒,乙型(B型)流感病毒,呼吸道合胞病毒,腺病毒,副流感病毒1、2和3型。
【检验原理】荧光素(FITC)标记的单克隆抗体与病毒抗原结合,形成稳定的抗原-抗体复合物,在荧光显微镜下观察,呈现特异性绿色荧光。
【主要组成成份】组份名称规格主要成分1 DFA染色试剂鉴定试剂流感A试剂:2 mL每试剂瓶为一项与病毒抗原相对应的荧光单克隆抗体流感B试剂:2 mL腺病毒试剂:2 mL呼吸道合胞病毒试剂:2 mL副流感1试剂:2 mL副流感2试剂:2 mL副流感3试剂:2 mL2 40X浓缩洗液25 mL PBS3 固定液15 mL 甘油4 阳性质控板5块包被病毒抗原【储存条件及有效期】2ºC-8ºC避光保存18个月。
【适用仪器】荧光显微镜,激发波长490 nm,发射波长520 nm。
【样本要求】鼻咽部取样后保存于生理盐水中,应在24小时内送检。
【检验方法】1、取样:用鼻咽拭子从病人的鼻咽部取样或者使用鼻腔灌洗液,将鼻咽拭子放入储存管中(含有生理盐水)。
2、样本准备步骤:①将样本充分震荡混匀约10-15秒。
②在400-600xg转速下离心5-10分钟。
③弃上清。
④在沉淀中加2-3 mL PBS缓冲液(不能使用试剂盒中的浓缩洗液),充分震荡混匀约10-15秒。
⑤在400-600xg转速下离心5-10分钟。
⑥吸走上清和黏液层,小心不要吸到沉淀。
⑦重复步骤4到6,直至黏液层被完全去除。
⑧在沉淀中加0.5-1 mL的PBS缓冲液。
⑨用移液器反复吹吸来重悬细胞沉淀,形成一个略混浊的悬液。
3、直接样本测试步骤:①在8孔板上的孔内滴加25 µL的细胞悬浊液,每样本需滴加七孔。

使用前注意事项:⏹所有离心操作在室温(15℃-25℃)下进行⏹沉渣用Buffer AL和Buffer ATL重溶⏹在浓缩的Buffer AW1和Buffer AW2中加入乙醇⏹冰冻组织和细胞球在室温下平衡⏹在56℃孵育箱中预处理⏹固定组织、昆虫、细菌或其他材料的预处理请参见使用手册1a. 组织:将组织切碎成小块(脾脏≤10mg,其他组织≤25mg),装入1.5ml离心管中,鼠尾则取1cm(大鼠)或2cm(小鼠)。
加入180μl Buffer ATL。
加入20μl 蛋白激酶K,振荡混匀,56℃孵育至组织完全裂解,孵育过程中间断振荡。
进入步骤2前振荡15秒。
1b. 无核血液:将20μl蛋白激酶K移至1.5ml或2ml离心管,加入50-100μl 抗凝处理过的血液。
用PBS调整体积至220μl,进入步骤2。
1c. 有核血液:将20μl蛋白激酶K移至1.5ml或2ml离心管,加入5-10μl抗凝处理过的血液。
用PBS调整体积至220μl,进入步骤2。
1d. 培养细胞:最多可用5×106个细胞,300×g(190rpm)离心5min。
用200μl重悬,加入20μl蛋白激酶K进入步骤22.加入200μl Buffer AL,振荡混匀,血标本需在56℃下孵育10min。
3.加入200μl 乙醇,振荡混匀。
4.将上述混合物用移液枪移至2ml离心管中的DNeasy Mini 滤柱中,6000×g(8000rpm)离心1min,弃去滤出液及离心管。
5.将滤柱放入新的2ml收集管中,加入500μl Buffer AW1,≥6000×g离心1min,弃去滤出液及收集管。
6.将滤柱放入新的2ml收集管中,加入500μl Buffer AW2,20000×g(14000rpm)离心3min,弃去滤出液及收集管。
7.将滤柱移至新的1.5ml或2ml离心管。
8.将200μl Buffer AE加至滤柱膜中央以洗提DNA,室温(15℃-25℃)下孵育1分钟。
CHIP步骤实验前准备:·处理细胞,将细胞保持在装有20ml培养基的150mm培养皿中。
·冰预冷PBS(第3步)、培养皿(第六步)。
·每个150mm培养皿准备42ml1*PBS(10*PBS 4.2ml+水37.8ml),冰上预冷。
·预热SDS到室温,确保在细胞裂解之前彻底溶解。
·将protease inhibitor cocktail II放置室温待用。
A体内交联和溶解1、加550 μl 37%的甲醛(或1.15ml新鲜的18.5%甲醛)到20 ml 细胞培养基中进行交联反应,轻轻地晃动混匀。
·甲醛的终浓度为1%。
尽量使用新鲜的甲醛(配置方法见附录B)。
2、室温下孵育10 min。
·不要振荡细胞。
3、取2 ml冰冷的1*PBS于分离管中至于冰上(每个培养皿对应一个),每1ml PBS中加5 ul Protease Inhibitor Cocktail II。
4、加2 ml 10×Glycine到培养皿中将未反应的甲醛消除。
5、晃动混匀并在室温下孵育5 min。
6、将培养皿放在冰上。
7、吸出培养基,尽可能的将培养基去除干净,小心不要扰乱细胞。
8、加20 ml冰冷的1*PBS来洗涤细胞。
9、除去PBS,再用PBS洗涤一次。
10、加2 ml含1×Protease Inhibitor Cocktail II的冷PBS到培养皿中(从第3步得)。
11、将细胞从培养皿刮下来放到离心管中。
12、4℃,700 g(900~1000 rpm)离心2~5 min沉淀细胞。
13、准备好1 ml(1*107个细胞推荐1ml) SDS Lysis Buffer(含5ul Protease Inhibitor Cocktail II)。
14、除去上清液(此步中的细胞沉淀可以放在-80℃中保存)。
15、用准备好的1 ml SDS Lysis Buffer(含1* Protease Inhibitor Cocktail II)重悬细胞。
DIG Northern Starter Kity Version 8.0Content version: August 2010For transcription-labeling of RNA with digoxigenein and SP6/T7/T3 RNA polymerases and chemiluminescent detection with CDP-Star, ready-to-use.Cat. No. 12 039 672 910Kit for 10 labeling reactions and detection of 10 blots of 10 ϫ 10 cm 2Store the kit at Ϫ15 to Ϫ25°CFor life science research only. Not for use in diagnostics procedures. FOR IN VITROUSE ONLY.1.Preface1.1Table of Contents1. Preface (2)1.1 Table of Contents (2)1.2 Kitcontents (3)2. Introduction (5)2.1 Productoverview (5)3. Procedures and required materials (8)3.1 Before you begin (8)3.2F lowchart (8)3.3DNA template preparation (9)3.4 DIG-RNAlabeling (11)3.5Determination of labeling efficiency (12)3.6Gel system suitable for DIG Northern blots (15)3.7RNA transfer and fixation (16)3.8Hybridization (17)3.9Immunological detection (19)3.10Stripping and reprobing of RNA blots (21)4.Results (22)4.1Typical results (22)5.Appendix (24)5.1Trouble shooting (24)5.2Changes to previous version (25)5.3References (26)5.4Ordering Information (26)1.2Kit contentsBottle/CapLabel Content including function1a Labeling Mix•40 l• 5 × conc. labeling mixture containing optimal concentrations of unlabelednucleotides and DIG -11-UTP•Clear viscous solution•For efficient in vitro transcription of linearized template DNA1b TranscriptionBuffer •40 l• 5 × conc. transcription buffer•Clear solution•For efficient in vitro transcription of linearized template DNA2SP6 RNAPolymerase •20 l•[20 U/l]•Synthezise RNA from a DNA template3T7 RNAPolymerase •20 l•[20 U/l]•Synthezise RNA from a DNA template4T3 RNAPolymerase •20 l•[20 U/l]•Synthezise RNA from a DNA template5Anti-digoxigenin-AP, Fab fragments •60l•[750 U/ml]•Polyclonal sheep anti-digoxigenin, Fab-fragments, conjugated to alkaline phosphatase•Clear solution6DNase I,RNase free •20 l•[10 U/l]•Degrades DNA template after transcription reaction7CDP-Starready-to-use •20 ml•Chemiluminescent substrate for alkaline phosphatase8Actin RNA probe, DIG-labeled •Antisense probe, length 588 bases•[10 ng/l]•Standard for the quantification of DIG-labeled RNA and as a hybridization control9DIG Easy HybGranules • 2 bottles for 100 ml solution each•For the hybridization of the DIG-labeled RNA probe10Blocking solution• 2 × 100 ml•10 × conc.•Yellow, viscous solution•For chemiluminescent detection procedureAdditional equipment and reagents required In addition to the reagents listed above, you have to prepare several solutions, exclusively with DMPC or DEPC treated water. In the table you will find an overview about the equipment which is needed for the different procedures.Detailed information is given in front of each procedure.Procedure Equipment Reagents3.3 DNA TemplatePreparation using3.3.1 standard methodor3.3.1 RT-PCR and PCR Thermocycler •High Pure Plasmid Isolation Kit*•Phenol/Chloroform•Expand High Fidelity PCR System (incl. buffer)*•Primer (sense, antisense)•Nucleotides (dATP, dCTP,dGTP, dTTP, each 10 mM)3.4 Procedure for DIG-RNA labeling •Water-bath or heating block•Ice/water•H2O, sterile, double distilled•EDTA, 0.2 M, pH 8.03.5 Determination of labeling efficiency •Nylon membranes positively charged*•UV-transilluminator or•UV-cross linker or•Oven (120°C or 80°C)•RNA dilution buffer•DIG Wash and Block Buffer Set*or•Washing buffer•Maleic acid buffer•Detection buffer3.6 Suitable gel system for Northern blots3.6.1 Formaldehyde gels•Gel equipment•Heatable water-bath (65° C) or•Heating block (65° C)•Ice •10 × MOPS, pH 7,0 (with NaOH)•Loading buffer•Gel solution•Running buffer3.7 RNA blotting and fixation •Nylon membranes positively charged*•Whatmann 3MM paper•UV- transilluminator or•UV-cross linker or•Oven (120°C or 80°C)Depending on used method:•20 × SSC or•2×SSC3.8 Hybridization•Nylon membranes, positively charged*•Ice/water•Water-bath•Shaking water-bath•or hybridization oven•Hybridization bags*or•Temperature resistant, sealable plasticor glass boxes, petri dishes or rollerbottlesNote: Do not use open trays or boxes withDIG Easy Hyb.For stringency washes:• 2 × SSC, 0.1% SDS •0.1 × SSC, 0.1% SDS3.9 Immunological detection •Hybridization bags* or•Development folders•DIG Wash and Block Buffer Set*or•Washing buffer•Maleic acid buffer•Detection buffer3.10 Stripping and reprobing of RNA blots •Water bath•Hybridization bags*•Double dist.water•Stripping buffer•2×SSC2.Introduction2.1Product overviewTest principle The DIG Northern Starter Kit generates DIG labeled, single stranded RNA probes of defined length by in vitro transcription of template DNA in the presence ofdigoxigenin-UTP, using SP6, T7 or T3 RNA polymerases.Stage DescriptionRNA labeling DIG-labeled RNA probes are generated according to the in vitrotranscription labeling technique. The labeling mix containsoptimal concentrations of nucleotides and DIG-11-UTP and aspecially developed transcription buffer are combined with thelinearized DNA template and the appropriate RNA polymerase.Hybridization DIG-labeled RNA probes are used for hybridization to membraneblotted nucleic acids according to standard methods.Immunological detection The hybridized probes are immunodetected with anti-digoxigenin-AP, Fab fragments and are then visualized with the chemiluminescence substrate CDP-Star, ready-to-use. Enzymatic dephosphorylation of CDP-Star, ready-to-use by alkaline phosphatase leads to a light emission at a maximum wavelength of 465 nm (see fig. 1) which is recorded with the imaging device or on X-ray films. Exposure times are in the range of 5 to 30 min, only.Fig. 1: Reaction of CDP-StarApplication Northern blots on nylon membranesSample material•Linearized plasmid, including the appropriate RNA polymerase promoter sequence (SP6, T3, T7).Note: The length of the region to be transcribed should be in the range of 200 to 1000 bp. To avoid RNase contamination the DNA must be phenolized.•Specially prepared PCR product (see chapter 3.3.2)Assay timeThis table lists the reaction time of the single stepsNumber of tests1 kit is sufficient for•10 labeling reactions of 1g DNA, yielding approx. 20 g labeled RNA, each and detection of•10 blots of 10 × 10 cm 2.StepReaction time RNA labeling 1 h 20 min Hybridization6 h or overnight Immunological detection1 h 40 min Chemiluminescent signal detection5-30 minKit storage/ stabilityThe unopened kit is stable at Ϫ15 to Ϫ25° C until the expiration date printed on the label.Once opened, please refer to the following table for appropriate storageSensitivity and specificity Rare mRNAs can be detected in 0.1 g of total RNA AdvantagesThis table describes benefits and features of the kit:Kit component vial Storage/StabilityLabeling Mix andTranscription Buffer 1a 1b Stable at Ϫ15 to Ϫ25° CNote : Repeated freezing and thawing should be avoided. To avoid contamination we recommend to aliquot the solutions and to store in 2-3 portions. RNA polymerases 2-4Stable at Ϫ15 to Ϫ25° C Anti-digoxigenin-AP, Fab fragments 5Stable at +2 to +8° C Note : Do not freeze!CDP-Star , ready-to-use 7Stable at +2 to +8° CNote : Store protected from light!DIG Easy Hyb Granules9•Stable at +15 to +25° C•Once dissolved, the solution is stable for 1 month when kept sterile.Blocking solution (10x concentrated)10Stable at +2 to +8° CNote: We recommend to store the concentrated blocking solution in aliquots at -15°C to -25°C.Benefit FeatureAccurate and fast The use of the application tested and optimized kit components minimizes the hands-on-time required to label RNA probes and increases efficiency and reproducibility.Sensitive Rare transcripts can be detected in total human or plant RNA.Time-savingDIG-labeled RNA probes can be stored at Ϫ15 to Ϫ25° C in ethanol for at least one year. We recommend to freeze the probe in aliquots, and avoid repeated freeze/thaw cycles.3.Procedures and required materials3.1Before you beginGeneral handlingrecommendationsThis table describes general hints for DIG labeling, detection and handling of RNA.3.2Flow chartRecommendation GuidelineWork under clean andRNase-free conditions•Use DMPC or DEPC treated H2O for preparation ofall solutions•Autoclave solutions•Tween 20 should be added to previously sterilizedsolutionsUse clean incubation trays Rigorously clean and rinse laboratory trays with RNase-ZAP (Sigma R-2020) or bake glass trays 8 hoursat 200° C before useMembrane handlingrequirements•Wear powder-free gloves•Handle membrane only on the edges with cleanforcepsSection 3.3DNA template preparation↓Section 3.4DIG-RNA labeling↓Section 3.5Determination of labeling efficiency↓Section 3.6Suitable gel systems for DIG Northern blots↓Section 3.7RNA blotting and fixation↓Section 3.8Hybridization↓Section 3.9Immunological detection↓Section 3.10Stripping and reprobing of RNA blots3.3DNA Template Preparation 3.3.1Standard methodGeneral information •Plasmid DNA should be purified using the High Pure Plasmid Isolation Kit*. •DNA template must be linearized at a restriction site downstream of the cloned insert. The sequence to be transcribed should be 200 to 1000 bp in length.•To avoid transcription of undesirable sequences, use a restriction enzyme that leaves 5’overhang or blunt ends.•After restriction digest, purify the DNA by phenol/chloroform extraction, followed by ethanol precipitation.•Dissolve template DNA in 10 mM Tris, 0,1 mM EDTA, pH 8,0Note: Too much EDTA inhibits the transcription reaction, do not use more than 0.1 mM EDTA in the TE buffer for dissolving the DNA.3.3.2Preparing DNA Template from total RNA using RT-PCR and PCROverview The following table describes an alternative method for generating templates for in vitro transcription labeling of RNA with DIG without cloning.RNA polymerase promoter sequences Please find in the following table the promoter sequences for the differentRNA polymerases.Stage Description1Prepare total RNA with one of the standard methods.2Run a RT-PCR with oligo(dT) Primer using the Expand Reverse Transcriptase System* .3Run the PCR with specially designed primers, including the sequence of the appropriate RNA polymerase promoter.Use standard PCR conditions; we recommend to use Expand High FidelityPCR System for best results.Promoter SequenceSP6 RNA polymerase Note: The use of SP6 promotor consensus sequences cannotbe recommended for PCR generated DNA fragments becauseexperiments have shown that SP6 Polymerase can only initiateefficient transcription if the promoter sequences lies within aplasmid environment (3).T7 RNA polymerase 5'TAATACGACTCACTATAGGG/X-meror5'TAATACGACTCACTATAGGA/X-merT3 RNA polymerase5'AATTAACCCTCACTAAAGGG/X-merAdditional equipment and reagents required•Thermocycler •Expand High Fidelity PCR System, including buffer •Primer (sense, antisense)•Nucleotides (dATP, dCTP, dGTP, dTTP, each 10 mM)ProcedureFind in the following table the recommended protocol using the Expand High Fidelity PCR System*.Step Action1•Add the following reagents to a sterile, RNase-free reaction tube (on ice) in the following order:•Mix gently and centrifuge briefly.2Cycle 30 × as follows:3Use your PCR product directly for labeling (Section 3.4).ReagentVolume Final conc.sterile double dist. water, DMPC or DEPC-treated variable -Expand buffer (10 ×)containing MgCl 2(supplied with the enzyme) 5 l 1.5 mM MgCl 2(1 × )10 mM dATP, dCTP, dGTP, dTTP each 1 l 0.2mM Primer 1 (sense)variable 300 nMPrimer 2 (anti-sense)variable 300 nM Expand High Fidelity 0.75 l 2.6 U cDNA 2 l -Final volume50 lStepT emperatureTime Denaturation 94°C 45 s Annealing 60°C 45 s Elongation72°C90 s3.4DIG-RNA LabelingIntroduction RNA is labeled in an in vitro transcription reaction with digoxigenin-11-UTP using a Labeling mixture and an optimized Transcription Buffer.Additional equipment and reagents required •Water bath for 42° C and 37° C or heating block•Ice/waterThis table lists composition, storage and use of the additionally required reagents.Procedure This procedure is designed for 1 μg of DNA template. Larger amounts can be labeled by scaling up of all components and volumes.Labeling efficiency and size of labeled RNA •In the standard reaction with 1 g DNA per assay 67% of the nucleotides are incor-porated into approx. 20 g of newly synthesized DIG-labeled RNA within 1 h.•The size of the labeled RNA is in the range of 200-1000 bases.Solution Composition Storage/StabilityUseWater Autoclaved, DMPC or DEPC treateddouble distilled water+15 to +25° C,stableDilution ofsolutions EDTA0.2 M ethylenediamino-tetracetic acid,pH 8.0+15 to +25° C,stableStopping thereactionStep Action1Add 1 g linearized plasmid DNA or 4 l PCR product (100-200 ng) and sterile RNase-free, DMPC or DEPC treated, double dist. water to a finalvolume of 10 l to a sterilized reaction vial.2•Add the following on ice:•Mix and centrifuge briefly.•Incubate for 1 h at 42° C.3•Add2l DNase I, RNase-free to remove template DNA.•Incubate for 15 min at 37° C.4Stop the reaction by adding 2 l 0.2 M EDTA (pH 8.0).Reagent VolumeLabeling mix, 5 × (vial 1a) 4 lTranscription buffer 5 × (vial 1b) 4 lRNA polymerase (SP6, T7 or T3) 2 l3.5Determination of labeling efficiencyIntroduction Determination of the yield of DIG-labeled RNA is very important for optimal andreproducible hybridization results. Too high probe concentrations in the hybridizationstep causes background, while too low concentrations leads to weak signals.Test principle The preferential method for determination of labeling efficiency of probes is the direct detection method in comparison to the control RNA (vial 8).Additional equipment required •Nylon membranes, positively charged* •UV-transilluminator or•UV-cross linker or•Oven (120°C or 80°C)Preparation of additional solutions required The Washing buffer, Maleic acid buffer, Blocking solution, and Detection buffer are also available in a ready-to-use form in the DIG Wash and Block Buffer SetDNase- and RNase-free* . These solutions are also used in the detection procedure of Chapter 3.9 and can be prepared in larger amounts.Stage Description1• A series of dilutions of DIG-labeled RNA is applied to a small strip of nylon membrane, positively charged*.•Part of the nylon membrane is preloaded with defined dilutions of control RNA, used as standards.2•The nylon membrane is subjected to immunological detection with anti-digoxigenin-AP and CDP-Star ready-to-use.•The intensities of the dilution series of DIG-labeled RNA and control RNA are compared by exposure to imaging device or X-ray film.Solution Composition / Preparation Storage/Stability UseRNA dilu-tion bufferMix DMPC treated double dist. water:20 × SSC: Formaldehyd (37%) in theratio 5 : 3 : 2preparefreshDilution of RNAWashingbuffer0.1 M Maleic acid, 0.15 M NaCl; pH 7.5;0.3% (v/v) Tween 20+15 to+25° C,stableRemoval ofunspecific boundantibody Maleic acidbuffer0.1 M Maleic acid, 0.15 M NaCl; adjustwith NaOH (solid) to pH 7.5+15 to+25° C,stableDilution of Block-ing solutionDetectionbuffer0.1 M Tris-HCl, 0.1 M NaCl, pH 9.5(20° C)+15 to+25° C,stableAdjustment of pHto 9.5Preparation of kit working solutionsPlease refer to the following table for the preparation of the working solutions.Dilution seriesPrepare a dilution series of your labeled probe and your control RNA as described in the table, where tube 1 is either a dilution of your labeling reaction to 10 ng/l(expected yield of a standard labeling reaction is 20 g of labeled RNA) or the control RNA (vial 8) which also has the concentration of 10 ng/l.Solution Composition / PreparationStorage/StabilityUseBlocking solution Prepare a 1 × working solution by dilu-ting the 10 × Blocking solution (bottle10) 1:10 in Maleic acid buffer.Always pre-pare fresh Blocking of un-specific binding sites on the mem-brane Antibody solutionCentrifuge anti-digoxigenin-AP (vial 5) for 5 min at 10 000 rpm in the original vial prior to each use, and pipet the necessary amount carefully from the surface. Dilute anti-digoxigenin-AP 1:10, 000 (75 mU/ml) in Blocking solu-tion.+2 to +8° C for 2 hBinding to theDIG-label Tube RNA (l)From tube #RNA Dilution Buf-fer (l)DilutionFinal concentration1-diluted labeling reaction or vial 8--10 ng/l 221181:10 1 ng/l 3221981:10010 pg/l 4153351:3.3 3 pg/l 553451:10 1 pg/l 654451:100.3 pg/l 755451:100.1 pg/l 856451:100.03 pg/l 957451:100.01 pg/l10-50-0Procedure The following table describes the direct detection.Note: The volumes suggested below are for small membrane stripes of approx.3 × 5 cm, processed in a small plastic container, such as a petri dish.Use sufficient buffer volumes to cover the membrane completely during all steps.Analysing the result Compare the intensity of the spots out of your labeling reaction to the control and calculate the amount of DIG-labeled RNA.Step Action1Apply 1 l spots of tubes 3-10 from your labeled probes and the labeled control to the nylon membrane.2Fix the nucleic acid to the membrane by cross linking with UV-light or baking for 30 min at 120° C3•Transfer the membrane into a plastic container with 20 ml Washing buffer.•Incubate under shaking for 2 min at 15-25° C.4Incubate for 30 min in 10 ml Blocking solution.5Incubate for 30 min in 10 ml Antibody solution.6Wash with 20 ml Washing buffer, 2 x 15 min.7Equilibrate 2-5 min in 10 ml Detection buffer.8•Place membrane (with RNA side facing up) on a development folder (or hybridization bag) and apply CDP-Star approx. 4 drops (from the drop-per bottle 7) to the membrane.•Immediately cover the membrane with the second sheet of the folder to spread the substrate evenly and without airbubbles over the membrane.Incubate for 5 min at + 15 to +25° C.9Seal the edges of the development folder around the damp membrane.Note: Drying of the membrane during exposure will result in dark background.10Expose to imaging device or to X-ray film for 5-25 min at +15 to +25° C.3.6Gel System suitable for DIG Northern BlotsGeneral•Standard protocols for gel electrophoresis can be used as described in Sambrook et al. [2].•Gels lacking ethidium bromide are preferential used, because ethidium bromide can cause uneven background problems.•We recommend formaldehyde denaturation and MOPS/formaldehyde gels, because to our experience they are easy to handle and reliable.Target amounts3.6.1Formaldehyde GelSolutions required The following buffers are required for running a formaldehyde gel:Preparation of samples Find the following table the preparation of the samplesIf you are using....Then....total RNA load maximally 1 µg per lanemRNA load 100 ng per laneSolution Composition10 × MOPS, pH 7.0 (with NaOH)200 mM MOPS50 mM NaAc20 mM EDTALoading buffer(always prepare fresh)250 l of 100% Formamide83 l of 37% Formaldehyde50 l 10 × MOPS50 l 100% Glycerol10 l 2.5% Bromphenolblue57 l DEPC/DMPC treated waterGel with Formaldehyd (2%) 1.8 g Agarose141.9 ml 1 × MOPS8.1 ml 37% FormaldehydeRunning buffer 1 × MOPSStep Action1Add 20 µl (or 2-3 volume) of Loading buffer to the RNA probe.2Denature the RNA probe/Loading buffer mix at 65°C for 10 min.3Chill the RNA probe/Loading buffer mix on ice for 1 min.4Run the gel wit 3-4 V/cm in RNase free gel boxes for at least 2 h (preferably over-night) until the RNAs are well separated..5To access the quality of the target RNA after electrophoresis, stain the gel briefly in 0.25 - 0.5 µg/ml ethidium bromide and examine the gel under UV light.3.7RNA transfer and fixationTransfer methods and membranes •When using formaldehyde gels, please rinse gels prior to blotting for 2 × 15 min in20 × SSC•All common types of RNA transfer methods are suitable for subsequent DIG hybridization [7]•Best results are obtained when gels are blotted by capillary transfer with 20 × SSC overnight or at least for 6 hours•Use only nylon membranes, positively chargedNote: We do not recommend to use alkali transfer (e.g., in 0.4 M NaOH) for RNA and DIG-labeled molecular weight markers*.Fixation procedure Fix the RNA to the membrane by any of the following procedures. Note: Use sterile and RNase-free solutions and equipmentStorage of the membrane Find in the following table the storage conditions for the membrane.IF you want to...THEN...UV-crosslinking•place the membrane on Whatman 3MM-paper soakedwith 2 × SSC.•UV-crosslink the wet membrane without priorwashing.•after the UV-crosslinking, rinse the membrane brieflyin double distilled water and allow to air-dry.bake at 120° C•rinse membrane 2 × briefly in 2 × SSC.•bake the nylon membrane at 120° C for 30 min oraccording to the manufacturer´s instructions.bake at 80° C•rinse membrane 2 × briefly in 2 × SSC•bake at 80° C for 2 hIF...THEN...you want to go e the membrane immediately forprehybridization.you want to work later on.store the membrane dry at +2 to +8 °C.3.8HybridizationFactors which influence strin-gency •The degree of homology of the probe to the target RNA (see ”hybridization temperature”) is an important factor for determining the appropriate conditions for hybridization.•Stringency is influenced by temperature: high temperature increases stringency of hybridization, low temperature decreases stringency.•For RNA : RNA hybridizazions with DIG Easy Hyb, we recommend generally a hybridization temperature of 68° C. The temperature may have to be adjusted depending on the GC content and homology of probe to target.Additional equipment required •Nylon membranes, positively charged *•Ice-water•Water-bath•Shaking water-bath•or Hybridization oven•Hybridization bags* or•Temperature resistant plastic or glass boxes, petri dishes, roller bottles or sealable plastic bags.Note: Do not use open trays with DIG Easy Hyb buffer.Additional solutions required •20×SSC •10%SDSPreparation of kit working solution Please refer to the following table to find the preparation protocol for the kit working solution.Note: Use sterile, RNase-free solutions and equipment!Solution Composition / Preparation Storage/StabilityUseDIGEasy HybGranulesReconstitute granules (bottle 9) by carefullyadding 64 ml sterile double distilled, DEPCor DMPC treated water in two portions tothe plastic bottle, dissolve by stirringat 37° C.Note: DIG Easy Hyb Granules must bedissolved under RNase-free conditions.at+15 to+25° Cfor1 monthAsprehybridizationand hybridizationsolutionProcedure Please refer to the following table.Note: Hybridization temperature for Northern Blots with DIG Easy Hyb is 68 °C (for100% homology of the probe to the target sequence), the DIG Easy Hyb working solu-tion used for prehybridization has to be prewarmed to 68 °C.Storage of hybridization solution In principle, DIG Easy Hyb containing DIG-labeled probe can be stored atϪ15 to Ϫ25° C and be reused when freshly denatured at 65°C for 10 min before use. We do not recommend reusage of DIG Easy Hyb containing RNA probes, because successful reuse depends on inherent probe stability and RNase-free working conditions.Stringency washes Please refer to the following table to find the procedure for stringency washes.Step Action1•Prewarm an appropriate volume of DIG Easy Hyb (10-15 ml/100 cm2 filter) to hybridization temperature (68 °C).•Prehybridize membrane with DIG Easy Hyb for 30 min with gentle agita-tion in an appropriate container.Note: Membranes should move freely, especially if you use severalmembranes in the same prehybridization solution.2Denature DIG-labeled RNA probe by boiling for 5 min and rapidly cooling in ice/water.Note: RNA is degraded by alkali-solutions!3Add denatured DIG-labeled RNA probe (100 ng/ml) to prewarmed DIG Easy Hyb (3.5 ml/100 cm2 membrane) and mix well but avoid foaming(bubbles may lead to background).4•Pour off prehybridization solution and add probe/hybridization mixture to membrane.•Incubate for 6 h or O/N at 68 °C with gentle agitation.Step Action1Wash 2 × 5 minutes in 2 × SSC, 0.1% SDS at +15 to +25 °C under constant agitation.2Wash 2 × 15 minutes in 0.1 × SSC, 0.1% SDS(prewarmed to wash temperature) at 68° C under constant agitation.Note: The stringency of the final wash must be determined empirically. Depending on homologous probes will often require 0.1x wash solution.3.9Immunological detectionGeneral information •Work under sterile, RNase-free conditions•If the membrane is to be reprobed, do not allow the membrane to dry at any time.•Use sufficient volume of all solutions.•Make sure that membranes do not stick to trays during detection.Avoid that membranes stick together.•Shake membranes during the whole procedure.•When using laboratory trays for the detection procedure, they should be rigorously cleaned before use.•Work under clean conditions when handling the chemiluminescent substrate solution and avoid phosphatase contamination.•For chemiluminescent detection, membranes must also be sealed in plastic bags or development folders•Do not use ”Saran Wrap”.Additional reagents required The Washing buffer, Maleic acid buffer, Blocking solution, and Detection buffer are also available in a ready-to-use form in the DIG Wash and Block Buffer Set, DNase and RNase free*.Solution Composition / Preparation Storage/StabilityUseWashing buffer0.1 M Maleic acid, 0.15 M NaCl;pH 7.5 (20° C); 0.3% (v/v) Tween 20+15 to+25° C,stableRemoval ofunspecific boundantibody Maleic acid buf-fer0.1 M Maleic acid, 0.15 M NaCl;adjust with NaOH (solid) topH 7.5 (20° C)+15 to+25° C,stableDilution ofBlocking solutionDetection buffer0.1 M Tris-HCl, 0.1 M NaCl,adjustment of pH to 9.5 (20° C)+15 to+25° C,stableAdjustment of pHto 9.5。
QIAGEN试剂盒提取RNA说明书中⽂翻译1:细菌的溶菌酶破碎2:细菌的溶菌酶破碎和机械破碎3:细菌的机械破碎4:细菌的溶菌酶裂解和蛋⽩酶K的消化5:细菌的溶菌酶裂解、蛋⽩酶K的消化及机械破碎6:固体培养基细菌的裂解7:使⽤RNeasy Mini Kit从细菌溶解物中提纯总RNA。
8:使⽤RNeasy Midi Kit从细菌溶解物中提纯总RNA。
章节说明该⼿册含有两种不同类型⽅案。
我们提供了多种⽅案,其中⽅案1-6是针对制备细菌溶解物,⽅案7-8是针对如何从细菌溶解物中提取全RNA。
你需要在⽅案1-6中选择⼀种操作,⽽后在⽅案7-8中任选其⼀。
制备细菌溶解物的各个⽅案都介绍了破碎细菌时如何固定RNA。
如何选择不同的实验⽅案由细菌细胞壁的稳定性所决定。
细菌细胞壁的稳定性受多个因素影响,包括细菌种类、⽣长阶段和培养基的成分。
细菌必须完全破碎以确保RNA的有效提取。
制备细菌溶解物的各个⽅案都包含了不⽌⼀步。
■酶消化:细菌细胞壁可以被溶菌酶进⾏消化(例如溶菌酶、溶葡球菌酶)。
我们认为溶菌酶的作⽤对于所有的⾰兰⽒阳性、阴性细菌均是有效的。
■蛋⽩酶K消化:在复杂培养基上⽣长的细菌⼤部分蛋⽩质可以被蛋⽩酶K消化以提⾼RNA 的纯化率。
我们认为蛋⽩酶K对于可在复合培养基中⽣长的细菌均是有效的。
蛋⽩酶K常常⽤于提⾼⾰兰⽒阳性细菌RNA。
另外,若从⼤量原材料中提取RNA,蛋⽩酶K可以有效提⾼RNA产率。
■机械破碎:细菌细胞壁的机械破碎可以使⽤组织研磨机和玻璃微珠。
机械破碎法对于绝⼤多数的细菌来说都是适⽤的。
机械破碎法与酶消化法相结合可以极⼤程度的提⾼RNA产率。
尽管本⼿册中的机械破碎⽅案均为使⽤组织研磨机,但采⽤其他的破碎⽅法是完全可⾏的。
考虑到细菌种类的多样性以及培养条件的多样性,细菌溶解物的制备⽅案(⽅案1-6)必须谨慎选择。
在第10页中介绍了制备细菌溶解物的不同⽅案,第11页(表格⼀)则提供了这些⽅案概况以便于参考选择。
土壤基因组DNA提取及PCR报告一、步骤试剂盒22℃--25℃储存使用前:用无水乙醇稀释SPW Wash Buffer 1、0.5g【500ml】玻璃珠加入15ml离心管中(15ml离心管可以更好的涡旋、悬浮),加0.5g【0.4—0.5g】土样,加1ml Buffer SLX Mlus 【裂解菌体】,最高速度涡旋3-5 min;2、加100ul Buffer DS,涡旋混匀;3、70℃水浴10 min,并短暂涡旋;(对于难提取样品可以在90℃水浴)4、3000rpm 室温【25℃】离心3min,转移800ul上清液于一新的2ml离心管中,加入270ul Buffer SP2,涡旋混匀;5、冰浴5 min,4℃,13000xg 离心5 min;6、小心转移上清液于一新的2ml离心管,加0.7体积【约600—900ul】的异丙醇,反复颠倒离心管20-30次以混合,(若样品DNA含量低,可以放置于-20℃1h );7、13000xg 4℃离心10min,获得沉淀DNA;8、小心去除上清液,将离心管在吸水纸上倒置1min;9、加200ul Elution Buffer,涡旋10s,溶解DNA,【预热EB,保存60℃,15min水浴】;10、加100ul HTR Reagent,涡旋10s混匀;(HTR Reagent使用前要摇匀,吸取时将枪头剪大一点儿);11、室温静置2min后13000xg 室温离心2min;12、取上清液于新2ml离心管中(若上清液有黑色则重复步骤10-12);13、加上清液的等体积【300—350ul】XP2 Buffer并涡旋混匀;14、将HiBind DNA柱置于2ml 收集管中,将上一步样品转移到柱中,10000xg 室温离心1min,弃流液,保留收集管;15、将柱移入上一步中的收集管,加入300ul XP2 Buffer,10000xg 离心1min,弃流液、收集管;16、将柱移入新2ml离心管中,加入700ul SPW Wash Buffer(用无水乙醇稀释过的)洗涤,10000xg 离心1min,弃流液保留收集管;17、重复16步;18、弃流液,将柱插入空收集管中,13000xg 室温离心2min,空甩干燥柱子;(是为了除去乙醇,乙醇对后续操作有影响)19、将柱移入干净1.5ml 离心管,将50ul Elution Buffer 直接加入HiBind matrix 中心,65℃水浴10-15min,室温放置3—5min;20、13000xg 离心1min 以洗脱DNA21、加入30-100ul Elution Buffer,重复19-20步22、检测二、结果1、OD值Nanodrop 2000 analysis2、电泳:土样量6ul;1%琼脂糖凝胶分析3、PCR:20ul体系:PCR Mix:10ul 引物:0.3+0.3ul 模板:1ul 加水补齐预变性:94℃10 min变性:94℃20s退火:52℃20s延伸:72℃20s终延伸:72℃5minPCR电泳:上样量:8ul;1%琼脂糖凝胶分析。
大鼠超氧化物歧化酶(SOD)酶联免疫分析试剂盒使用说明书本试剂盒仅供研究使用。
检测范围:48T2.5 U/mL - 80 U/mL使用目的:本试剂盒用于测定大鼠血清、血浆及相关液体样本中超氧化物歧化酶(SOD)含量。
实验原理本试剂盒应用双抗体夹心法测定标本中大鼠超氧化物歧化酶(SOD)水平。
用纯化的大鼠超氧化物歧化酶(SOD)抗体包被微孔板,制成固相抗体,往包被单抗的微孔中依次加入人超氧化物歧化酶(SOD),再与HRP标记的超氧化物歧化酶(SOD)抗体结合,形成抗体-抗原-酶标抗体复合物,经过彻底洗涤后加底物TMB显色。
TMB在HRP酶的催化下转化成蓝色,并在酸的作用下转化成最终的黄色。
颜色的深浅和样品中的超氧化物歧化酶(SOD)呈正相关。
用酶标仪在450nm波长下测定吸光度(OD值),通过标准曲线计算样品中大鼠超氧化物歧化酶(SOD)浓度。
1.标本采集后尽早进行提取,提取按相关文献进行,提取后应尽快进行实验。
若不能马上进行试验,可将标本放于-20℃保存,但应避免反复冻融2.不能检测含NaN3的样品,因NaN3抑制辣根过氧化物酶的(HRP)活性。
操作步骤1.标准品的稀释:本试剂盒提供原倍标准品一支,用户可按照下列图表在2.加样:分别设空白孔(空白对照孔不加样品及酶标试剂,其余各步操作相同)、标准孔、待测样品孔。
在酶标包被板上标准品准确加样50μl,待测样品孔中先加样品稀释液40μl,然后再加待测样品10μl(样品最终稀释度为5倍)。
加样将样品加于酶标板孔底部,尽量不触及孔壁,轻轻晃动混匀。
3.温育:用封板膜封板后置37℃温育30分钟。
4.配液:将20倍浓缩洗涤液用蒸馏水20倍稀释后备用5.洗涤:小心揭掉封板膜,弃去液体,甩干,每孔加满洗涤液,静置30秒后弃去,如此重复5次,拍干。
6.加酶:每孔加入酶标试剂50μl,空白孔除外。
7.温育:操作同3。
8.洗涤:操作同5。
9.显色:每孔先加入显色剂A50μl,再加入显色剂B50μl,轻轻震荡混匀,37℃避光显色15分钟.10.终止:每孔加终止液50μl,终止反应(此时蓝色立转黄色)。
8羟基脱氧鸟苷(8-OHdG)酶联免疫分析试剂盒使用说明书检测范围:96T1.2 ng/L -45 ng/L使用目的:本试剂盒用于测定大肠杆菌样本中8羟基脱氧鸟苷(8-OHdG)含量。
实验原理本试剂盒应用双抗体夹心法测定标本中8羟基脱氧鸟苷(8-OHdG)水平。
用纯化的8羟基脱氧鸟苷(8-OHdG)抗体包被微孔板,制成固相抗体,往包被单抗的微孔中依次加入8羟基脱氧鸟苷(8-OHdG),再与HRP标记的8羟基脱氧鸟苷(8-OHdG)抗体结合,形成抗体-抗原-酶标抗体复合物,经过彻底洗涤后加底物TMB显色。
TMB在HRP酶的催化下转化成蓝色,并在酸的作用下转化成最终的黄色。
颜色的深浅和样品中的8羟基脱氧鸟苷(8-OHdG)呈正相关。
用酶标仪在450nm波长下测定吸光度(OD值),通过标准曲线计算样品中8羟基脱氧鸟苷(8-OHdG)浓度。
试剂盒组成1.标本采集后尽早进行提取,提取按相关文献进行,提取后应尽快进行实验。
若不能马上进行试验,可将标本放于-20℃保存,但应避免反复冻融2.不能检测含NaN3的样品,因NaN3抑制辣根过氧化物酶的(HRP)活性。
操作步骤1.标准品的稀释:本试剂盒提供原倍标准品一支,用户可按照下列图表在小试管中进行稀2.加样:分别设空白孔(空白对照孔不加样品及酶标试剂,其余各步操作相同)、标准孔、待测样品孔。
在酶标包被板上标准品准确加样50μl,待测样品孔中先加样品稀释液40μl,然后再加待测样品10μl(样品最终稀释度为5倍)。
加样将样品加于酶标板孔底部,尽量不触及孔壁,轻轻晃动混匀。
3.温育:用封板膜封板后置37℃温育30分钟。
4.配液:将30倍浓缩洗涤液用蒸馏水30倍稀释后备用5.洗涤:小心揭掉封板膜,弃去液体,甩干,每孔加满洗涤液,静置30秒后弃去,如此重复5次,拍干。
6.加酶:每孔加入酶标试剂50μl,空白孔除外。
7.温育:操作同3。
8.洗涤:操作同5。
9.显色:每孔先加入显色剂A50μl,再加入显色剂B50μl,轻轻震荡混匀,37℃避光显色15分钟.10.终止:每孔加终止液50μl,终止反应(此时蓝色立转黄色)。
1 只用于生命科学研究,不能用于诊断程序 仅用于体外实验 DIG High Prime DNA Labeling and Detection Starter Kit Ⅱ 采用地高辛-dUTP进行随机引物DNA标记,碱性标记,利用CSPD化学发光检测,随时即用。 货号:11 585 614 910 此试剂盒储存条件:-15℃到-25℃。 此试剂盒可对10ng-3μgDNA进行12次标记反应, 可检测10×10cm2面积的杂交膜24张。
指导手册 2005.12 版 1 前言 1.1 内容表 1 前言 1.1 内容表 1.2 试剂盒成分 2.介绍 2.1 产品简介 3. 操作步骤和所需材料 3.1 在你开始前 3.2 流程图 2
3.3 地高辛标记DNA 3.4 标记效率的确定 3.5 DNA的转移和固定 3.6 杂交 3.7 免疫检测 3.8 DNA印记的洗脱和再杂交 4. 附录 4.1 故障排除 4.2 参考文献 4.3 订购信息 1.2 试剂盒的成分
瓶号 标记 内容 包括功能
1 DIG-高效引物 50μL 地高辛标记的高效引物 5×浓度的标记混合物:优化的浓度的随机引物,核苷酸,地高辛标记的dUTP(碱标记的),Klenow酶和缓冲液成分
随时可用 澄清的粘液 用于DNA的高效随机引物标记 3
2 DIG标记的对照DNA
20μL 5μg/mL 质粒pBR328 DNA (用Bam HI线性化)
澄清溶液 用于确定标记效率
3 DNA稀释缓冲液 3管1mL 50μg/mL鱼精DNA,10mM Tris-HCl,1mM EDTA, pH8.0 在25℃
澄清溶液
4 抗地高辛的碱性磷酸酶结合物
50μL 750U/mL 从羊中获取的,Fab段,结合碱性磷酸酶
澄清溶液
5 CSPD 随时可用 50mL CSPD
澄清溶液
6 封闭液 4×100mL 4
10×浓度 黄色,粘液
7 地高辛杂交颗粒 100mL地高辛杂交液共4瓶,用于DNA杂交
附加仪器与所需试剂 除了上表中列出的试剂外,你必须准备一些溶液。在下表中,你可以找到不同操作程序需要准备的设备概要。 在每个操作程序的前面提供了详细的信息。
操作程序 仪器设备 试剂 3.3 DIG-DNA标记 水浴 灭菌的双蒸水 0.2M pH8.0 灭菌的EDTA
3.4 标记效率的半定量 带正电荷的尼龙膜* 地高辛洗涤和封阻缓冲组合*或者
洗涤缓冲液 马来酸缓冲液 检测缓冲液 5
3.5 DNA转移和固定 紫外光盒 或者商业化可用于紫外交联的其他设备 20×SSC 或者10×SSC
3.6 杂交 尼龙膜,带正电荷* 杂交袋*, 或者耐热的塑料袋或者滚筒瓶 注意:当用地高辛杂交缓冲液工作时,不要用开口的托盘。
3.7 免疫检测 耐热的塑料袋或者滚筒瓶 杂交袋* 地高辛洗涤和封阻缓冲液组合*,或者
洗涤缓冲液 马来酸缓冲液 检测缓冲液 3.8 DNA 斑点的脱色和重新标记探针 大的托盘 水浴 10×SSC 10%SDS 0.2M NaOH 6
*标记的产品可以从Roche Applied Science 获得。 2.介绍 2.1 产品概况 实验原则 此试剂盒采用DIG(地高辛),一种甾类半抗原,steroid hapten去标记DNA探针用于杂交和后续的免疫检测[1,2,3]。
步骤 描述
DNA 标记 根据随机引物标记技术采用地高辛标记引物获得了地高辛标记的DNA探针。地高辛标记的高效引物是一种专门生产的反应混合物,其中含有地高辛dUTP,碱性标记(图1)和所有的试剂,包括随机引物标记所必须的酶,已预先混合优化的5×浓度反应缓冲液。
杂交 根据标准方法,地高辛标记的探针可以与固定在膜上的核酸杂交。采用碱标记形式的地高辛-11-dUTP能够更加容易和有效的被洗脱下来,使固定在膜上的核酸可以与其他的地高辛标记探针再次杂交。
免疫检测 杂交探针可以用抗地高辛的碱性磷酸酶进行免疫检测,Fab段,然后采用即时可用的化学发光底物CSPD可以看到杂交结果。采用碱性磷酸酶可以对 7
CSPD进行,酶的脱磷酸化,导致在最大波长477nm处有光的发射(图2),这一现象可以用适合的图像仪或者X光片进行记录。胶片的曝光时间范围只在5-30分钟内。
图1 DIG-dUTP ,碱性标记 图2 CSPD反应 应用 地高辛标记DNA探针可以用于: 所有类型的滤膜杂交 总基因组DNA中单拷贝基因的检测,甚至对于高度复杂的生物体也可以进行检测,如人,大麦和小麦。
样品材料 至少100bp的DNA片段 线性的质粒,cos质粒或者DNA 超螺旋的DNA 实验时间 此表格列出了每一步实验所需的反应时间。
步骤 反应时间 DNA标记 1小时-过夜 8
杂交 6小时或者过夜 免疫检测 1.5小时 化学荧光信号检测 5-30分钟
检测数量 一个试剂盒足够用于: 12个标准的标记反应,每个反应的膜板DNA不超过3μg;并可以检测24张10×10cm2的杂交膜。
质量控制 根据操作步骤中的说明对未标记的对照DNA pBR328进行标记,并按照下面的标准检测操作方法在点杂交中用50ng的异源的DNA稀释0.1pg 同源DNA,用即时可用的CSPD,X光片曝光30分钟进行检测。
试剂盒的储存和稳定性 未开封的试剂盒在保质期内可以稳定的存放在-15℃到-25℃(保质期印在标签上)。在干冰中运输。
一旦开封,请根据下表选择适宜的储存条件。 试剂盒成分 储存条件 抗地高辛碱性磷酸酶结合物(4号管) 2-8℃稳定。注意:不要冻存 9
CSPD 随时即用 (5号管) 2-8℃,避光保存。 封阻液(6号管) 未开封时 15℃-25℃ 稳定。 一旦开封,应分装并储存在-15℃到-25℃或者无菌条件下2-8℃间可以稳定保存1个月。 工作溶液都应是新鲜配制的。 地高辛杂交颗粒 储存在15℃-25℃ 一旦开封,无菌条件下溶液可以稳定存在1个月。 地高辛高效引物混合物 保质期12个月,-15℃到-25℃。 避免反复冻融。 灵敏度和特异性
采用southern杂交从0.3μg人胎盘 DNA(经Bgl Ⅱ或EcoRⅠ酶切)中检测到单拷贝基因(组织纤溶酶原激活因子tPA)。
优点 此表描述了这个试剂盒的优点和特征 优点 特征 精确、快速 预先已混好的地高辛高效引物混合物减少了标记DNA 时手动操作的时间,同时提高了产量,重复性好 灵敏 在复杂的总人DNA及植物基因组中能够检测到单拷贝基因。 10
省时 地高辛标记的探针可以至少储存一年。 杂交溶液可以重复利用3-5次,重复次数主要取决于每次杂交中产生信号的标记探针的量。 3. 实验步骤和所需材料
3.1 在你开始前 主要的操作要求 此表中描述了地高辛标记和检测中主要的提示:
要求 指引 在清洁的条件下进行操作 高压灭菌地高辛系统的试剂 过滤灭菌的溶液包括:SDS,吐温20,并应该加入到已灭菌的溶液中。
用干净的孵育托盘 每次使用前都要严格地清洗实验托盘 处理膜的要求 带无粉手套 处理膜的时候,只能够用干净的镊子夹膜的边缘
3.2 流程图 11
3.3部分 地高辛标记DNA ↓ 3.4部分 标记效率的检测 ↓ 3.5部分 DNA固定 ↓ 3.6部分 杂交 ↓ 3.7部分 免疫检测 ↓ 3.8部分 洗脱和DNA斑点与另一探针的杂交
3.3 地高辛标记DNA 介绍 随机引物标记的DNA带有地高辛-11-dUTP,这一标记采用的是地高辛高效引物试剂,这是一种含有随机六碳糖,dNTP混合物的5×浓度标记混合物,其中dNTP混合物包括碱性标记的地高辛-11-dUTP, 12
标记级的Klenow酶和适合的反应缓冲液。 附加设备和所需试剂 此表中列出了成分,储存条件和除了试剂盒成分外所需试剂的用途。
溶液 成分 储存条件/稳定性 用途
水 高压灭菌的双蒸水 15-25℃,稳定 稀释DNA EDTA(乙二胺四乙酸) 0.2M EDTA,pH 8.0 15-25℃,稳定 终止标记反应 模板DNA 下表中列出了模板DNA所需特征:
特征 详细信息
纯度 模板DNA应该采用the High Pure Plasmid Isolation Kit进行制备。
当采用其他商业化的纯化试剂盒进行纯化时,我们建议额外进行一次苯酚/氯仿抽提以去除残余的蛋白质。