PhysRevB[1].63.075102
- 格式:pdf
- 大小:175.45 KB
- 文档页数:12


Excited States of Phosphorescent Platinum(II)Complexes Containing N∧C∧N-Coordinating Tridentate Ligands:Spectroscopic Investigations and Time-Dependent Density Functional Theory CalculationsWataru Sotoyama,*,†Tasuku Satoh,†Hiroyuki Sato,Azuma Matsuura,and Norio SawatariFujitsu Laboratories Ltd.,10-1Morinosato-Wakamiya,Atsugi243-0197,JapanRecei V ed:June22,2005;In Final Form:September4,2005The absorption and emission spectra of the Pt(II)complexes containing N∧C∧N-coordinating tridentate ligands,platinum(II)1,3-di(2-pyridyl)benzene chloride[Pt(dpb)Cl]and platinum(II)3,5-di(2-pyridyl)toluene chloride[Pt(dpt)Cl],together with their corresponding free ligands,1,3-di(2-pyridyl)benzene(dpbH)and3,5-di(2-pyridyl)toluene(dptH),have been analyzed by density functional theory(DFT)for the ground state and time-dependent DFT(TDDFT)for the excited states.T1(A1)and S1(B2)of the complexes(in C2V symmetry)wereassigned on the basis of the calculated excitation energies as well as comparison of the experimentalspectroscopic properties and the calculated states’characteristics.The calculated excitation energies for T1and S1of the complexes as well as those for T1of the free ligands were in good agreement with their observedvalues within600cm-1.The d-π*characters of the excited states were evaluated from the change in electrondensities between the ground and excited states by Mulliken population analysis;values of25%for T1and32%for S1were obtained for both complexes.The calculated values of d-π*character were found to beconsistent with the reported emission lifetimes as well as the observed emission energy shifts from thecorresponding free ligands.Most spectroscopic properties of the complexes and the free ligands,which includesolvatochromic shift,Stokes shifts,methyl substitution shifts,and emission spectra profiles,were well explainedfrom the calculation results.1.IntroductionStudies on phosphorescent transition-metal complexes are ofparticular interest not only from a scientific viewpoint but alsoin development of emissive dopants for phosphorescent organiclight-emitting diodes(OLEDs).1,2The rapid growth of interestin phosphorescent OLEDs has prompted syntheses and photo-physical investigations of a wide variety of phosphorescentcomplexes.3-7Recently,a new class of platinum(II)complexesconsisting of platinum,chloride,and N∧C∧N-coordinatingtridentate ligands[platinum(II)1,3-di(2-pyridyl)benzene chlo-ride,Pt(dpb)Cl,and platinum(II)3,5-di(2-pyridyl)toluene chlo-ride,Pt(dpt)Cl,Figure1]have been synthesized,8,9and highphosphorescence quantum yields ranging from0.58to0.68insolution at room temperature have been reported in these complexes.9Pt(dpt)Cl and its derivative with a phenoxide group that replaces the chloride have been reported to exhibit remark-able performance as emissive dopants of OLEDs.10The lowest excited triplet states(T1)of Pt(dpb)Cl and Pt(dpt)Cl have been assigned to primarily ligand-centered(LC) 3π-π*character from their highly structured emission profile with very small Stokes shifts and relatively long emission lifetimes[Pt(dpb)Cl7.2µs and Pt(dpt)Cl7.8µs in solution at room temperature].9However,these emission lifetimes are not much longer than those of the complexes,which have the triplet-emitting states with metal-to-ligand charge transfer(MLCT) character.For example,in the case of Ir(ppy)3,where ppy represents2-phenylpyridine ligand,the emitting states have been assigned to be mainly3MLCT character by theoretical studies11 and spectroscopic measurements.12The emission lifetimes of this complex were found to be2µs and5µs at room temperature and77K,respectively,in solution.13Taking into account the possible admixture ofπ-π*/d-π*(MLCT)character,which is common for T1of many phosphorescent transition-metal complexes,14-20it is expected that the d-π*character would participate in T1of Pt(dpb)Cl and Pt(dpt)Cl.The d-π* admixture to an excited state,especially to T1,is important even though the state character is primaryπ-π*,since a small amount of d-π*character can dominate the kinetics of the deactivation processes of the excited state by its strong spin-orbit coupling ability to the states with different spin multi-plicities.16,17,21-23In this paper we describe detailed analysis of the excited states of Pt(dpb)Cl and Pt(dpt)Cl by correlating the spectroscopic data with the calculation results based on density functional theory (DFT).The main objectives of this study are assignment of the*To whom correspondence should be addressed:e-mail wataru_sotoyama@fujifilm.co.jp.†Present address:Advanced Core Technology Laboratories,Fuji Photo Film Co.,Ltd.,210Nakanuma,Minamiashigara250-0193,Japan.Figure1.Schematic structure of the two complexes[Pt(dpb)Cl and Pt(dpt)Cl]and the two free ligands[dpbH and dptH]along with the molecular axis.The numbers in the structures designate each carbon atom.9760J.Phys.Chem.A2005,109,9760-976610.1021/jp053366c CCC:$30.25©2005American Chemical SocietyPublished on Web10/11/2005low-lying excited states of the complexes and quantitative evaluation of the d-π*character at the complexes.The free tridentate ligands[1,3-di(2-pyridyl)benzene(dpbH)and3,5-di-(2-pyridyl)toluene(dptH),Figure1]are also investigated to assist in analysis of the excited states of the complexes.Several aspects of the spectroscopic features,such as effect of methyl substitution on the excitation energies and the difference in emission profile between the complexes and the free ligands, will be discussed in light of the calculation results by DFT and time-dependent DFT(TDDFT).2.Experimental Section2.1.Materials and Spectroscopic Measurements.Pt(dpb)-Cl and Pt(dpt)Cl were synthesized from dpbH and dptH, respectively,according to the literature.8,9Absorption and emission measurements in solution were conducted in1cm path length quartz cuvettes at room temperature or in5mm diameter quartz tubes at77K.Absorption spectra were recorded on a Hitachi U-4100spectrophotometer.Emission spectra of Pt(dpb)-Cl and Pt(dpt)Cl were measured on a Minolta CS-1000 spectroradiometer,equipped with an ultra-high-pressure mercury lamp and a visible blocking filter,as the excitation source.The solutions of free ligands(dpbH and dptH)showed phosphores-cence with lifetimes of a few seconds at77K;phosphorescence spectra of these solutions were measured with a phosphoroscope in addition to the above apparatus.putational Details.The ground-state DFT and the excited-state time-dependent DFT(TDDFT)24-26calculations were carried out with the Gaussian98program package.27 TDDFT has been successfully applied to excited-state calcula-tions for many transition-metal complexes.11,28-34All calcula-tions were carried out with the LANL2DZ basis set with the relativistic effective core potential(ECP)for Pt35and the6-31G* basis set for the other elements.The calculation results(orbitals and densities)were plotted with the visualization program(xmo 4.0)in MOPAC2002.36Calculations on the electronic ground state(S0)of the complexes[Pt(dpb)Cl and Pt(dpt)Cl]and the free ligands(dpbH and dptH)were carried out by use of B3LYP density functional theory.37,38The S0geometries were optimized under symmetry constraints of C2V for Pt(dpb)Cl and C s(zx symmetry plane,Figure1)for Pt(dpt)Cl.The optimized planar geometries of these complexes were confirmed by the vibrational frequencies calculations with no imaginary frequencies.The optimized geometry of Pt(dpb)Cl was found to be in good agreement with the X-ray crystallographic data(Table1).8For dpbH and dptH,the geometries optimized within symmetry constraints similar to the complexes(planar forms)were different from those fully optimized without symmetry con-straints(twisted forms).Although the twisted forms have lower S0total energy than the planar forms for both dpbH and dptH, the differences in the S0total energy between the planar and twisted forms were found to be negligible(158cm-1for dpbH and179cm-1for dpbH).On the basis of the minor energy deviations from the twisted forms,we present calculation results at the planar forms of the free ligands in this study for convenience of comparison with the results for the complexes. The geometries for the lowest excited triplet states(T1)of Pt(dpb)Cl and dpbH were optimized at the restricted open-shell B3LYP level.At the respective optimized geometries,TDDFT calculations with the B3LYP functional were carried out.Ten lowest roots for triplet states as well as singlet states were calculated for the complexes and the ligands.In addition to the vertical excitation energies for all calculated roots,the one-particle densities and the dipole moments for the three lowest triplet and singlet roots were obtained for the complexes.3.Results3.1.Absorption and Emission Spectra at Room Temper-ature.Figure2shows absorption and emission spectra of Pt-(dpb)Cl(a)and Pt(dpt)Cl(b)together with the absorption spectra of dpbH(a)and dptH(b)in toluene at room temperature.The spectra of the complexes in Figure2replicate the results reported by Williams et al.9The lowest energy weak peaks in Pt(dpb)Cl and Pt(dpt)Cl absorption spectra are assigned as S0-T1absorp-tion by their mirror-symmetry relation with the emission peak. The strong absorption envelopes at22000-30000cm-1are assigned to the absorptions to the singlet excited states with significant MLCT character involving Pt and Cl orbitals from the comparison with the absorption spectra of the free ligands. The lowest energy peak among these absorptions is considered as the S1peak of the each complex.(Strictly speaking,the absorption spectra do not exclude the possibility of the existence of one or more singlet excited states with very small absorption cross sections at the lower energy side of the S1peak assigned above.However,even if there are such dark states,the discussion about the orbital assignment of the S1peak based on the absorption intensities in section4.1still remains unchanged.)TABLE1:Calculated and Observed X-ray Crystallographic Data for Representative Bond Lengths(Å)and Angles (degree)for Pt(dpb)Cl abond b calcd obsd c Pt-Cl 2.471 2.417Pt-C(1) 1.926 1.907Pt-N 2.065 2.037dC(1)-Pt-N80.4380.5dPt-C(1)-C(2)118.62119.2dPt-N-C(5)113.95114.6da Bond lengths are given in angstroms;bond angles are given in degrees.b See Figure1for numbering of the carbon atoms.c Data from ref8.d Average of two equivalent datapoints.Figure2.Absorption and emission spectra of the complexes alongwith absorption spectra of the free ligands in toluene at roomtemperature:(a)Pt(dpb)Cl and dpbH;(b)Pt(dpt)Cl and dptH.Solidline(green),emission of the complexes;dashed line(purple),absorptionof the complexes;dotted line(red),absorption of the complexes in anexpanded scale(134times);dashed-dotted line(blue),absorption ofthe free ligands.Phosphorescent Platinum(II)Complexes J.Phys.Chem.A,Vol.109,No.43,20059761We observed negative solvatochromic behavior (peak blue shift with increasing solvent polarity)reported in ref 9for S 1and T 1peaks of Pt(dpb)Cl and Pt(dpt)Cl in several solvents.The absorption peak energies in toluene and acetonitrile are presented in Table 2.(See ref 9for further information about spectral data in various solvents including toluene and aceto-nitrile.)According to the theoretical study on solvent effect by Marcus,39,40negative solvatochromic behavior of an absorption peak has been interpreted to be associated with the decrease in dipole moments by excitation.It is observed that the solvato-chromic shifts are fairly larger for S 1peaks than T 1peaks,which would suggest a lager decrease in dipole moment by the S 0-S 1excitation than that of the S 0-T 1excitations of these complexes.3.2.Phosphorescence Spectra at 77K.Figure 3shows the phosphorescence spectra of dpbH and Pt(dpb)Cl (a)and of dptH and Pt(dpt)Cl (b)in toluene at 77K.The highest energy peak of each spectrum (presented in Tables 3and 4)is attributed to the vibrational 0-0origin of the T 1-S 0transition.The phosphorescence origins of the Pt(dpb)Cl and Pt(dpt)Cl have red-shifted 2200and 2580cm -1from those of the corresponding free ligands,respectively.These shifts are much larger than those for complexes with ligand-centered emitting states in the literature,such as [Pt(bpy)2]2+(950cm -1)or [Rh(bpy)3]3+(600cm -1),41where bpy is 2,2′-bipyridine,suggesting significant admixture of the metal character in the emitting state of the complexes.The relative intensity of the origin peak,compared to the peaks of vibrational satellites with frequencies of about 1300cm -1,would become notably larger for the complexes than for the free ligands;these features imply closer equilibrium geometries between S 0and T 1for these complexes compared to those for the free ligands.Similar trends have been observed for complexes having T 1with significant MLCT character,such as [Ru(bpy)3]2+or Pt(thpy)2,where thpy is 2-(2-thienyl)pyridine,in comparison with their corresponding free ligands.17,18,42The characteristics such as the position and profile of the spectraindicate a certain amount of metal d-orbital admixture to T 1of Pt(dpb)Cl and Pt(dpt)Cl.The methyl substitution in place of the hydrogen atom at C(4)of the phenyl ring causes somewhat different effects to the emission spectra between the complexes and the free ligands.Both the methyl-substituted complex [Pt(dpt)Cl]and free ligand (dptH)displayed red-shifted emission from their unsubstituted counterparts [Pt(dpb)Cl and dpbH].However,the spectral shift was found to be substantially greater between the complexes (540cm -1)than between the free ligands (160cm -1).This large difference in methyl-substitution effect would suggest the orbital nature difference of T 1between the complexes and the free ligands.4.Theoretical and Experimental Data Comparison 4.1.Assignments and Excitation Energies of Calculated Roots.Figure 4depicts the four highest occupied and the two lowest unoccupied molecular orbitals (HOMOs and LUMOs)of Pt(dpb)Cl,as well as the HOMO and LUMO of dpbH obtained from the ground-state DFT calculations.The HOMOs and LUMOs for Pt(dpt)Cl and dptH are similar in shape with the corresponding orbitals presented in Figure 4.Every fourTABLE 2:T 1and S 1Absorption Peak Energies for Pt(dpb)Cl and Pt(dpt)Cl in Toluene and Acetonitrile at Room Temperatureabsorption peak energy/cm -1complex state toluene acetonitrile Pt(dpb)Cl T 12040020700Pt(dpb)Cl S 12400025200Pt(dpt)Cl T 12000020300Pt(dpt)ClS 12340024500TABLE 3:Parameters a from Ground-State DFT and TDDFT Calculations for the Three Lowest Excited Triplet and Singlet Roots of Pt(dpb)Cl and Pt(dpt)Cl at the Ground-State GeometriesPt(dpb)ClPt(dpt)Cl root (assignment b )symmetry cdominant excitation c ,d E /cm -1µz (∆µz )/D f E /cm -1µz (∆µz )/D fground state S 01A 1 6.647.07triplet root 1(T 1)3A 1b 1f b 1*20421 5.51(-1.13)20059 6.69(-0.38)2(T 2)3B 2b 1f a 2*20892 1.03(-5.61)20270 2.66(-4.41)3(T 3)3A 1a 2f a 2*22993 4.41(-2.23)22886 5.09(-1.98)T 1(expt e )2058020040singlet root 1(S 1)1B2b 1f a 2*23489-2.43(-9.07)0.07423120-1.04(-8.10)0.0772(S 2)1A 1b 1f b 1*23707 3.06(-3.58)0.00723342 4.64(-2.43)0.0063(S 3)1A 2b 2f b 1*25928-4.95(-11.58)0.00025899-4.47(-11.54)0.000S 1(expt f )2400023400aE ,vertical excitation energy;µz ,z component of dipole moment;∆µz ,dipole moment differences between ground and excited states;f ,oscillatorstrength.Experimental excitation energies are also listed.b See text.c Symmetry of states and orbitals are designated for Pt(dpb)Cl (C 2V ,see Figure 1).d For designation of the orbitals,see Figure 4.e Phosphorescence 0-0peak energy in toluene at 77K.f Absorption peak energy in toluene at roomtemperature.Figure 3.Emission spectra of the complexes and the free ligands in toluene at 77K:(a)Pt(dpb)Cl and dpbH;(b)Pt(dpt)Cl and dptH.Solid line (green),emission of the complexes;dotted line (blue),emission of the free ligands measured with a phosphoroscope.9762J.Phys.Chem.A,Vol.109,No.43,2005Sotoyama et al.HOMOs of the complexes are characterized by significant admixtures by the Pt d-orbitals.Two of them [b 1(HOMO)and a 2(HOMO-3)]are represented by combinations of d orbitals of the Pt and πorbitals of the tridentate ligand and Cl,while the remainder [b 2(HOMO-1)and a 1(HOMO-2)]have σcharacters that are symmetrical about the molecular plane.On the other hand,the LUMOs correspond to the π*orbitals almost localized in the ligand.Table 3summarizes the TDDFT calculation results of the properties that include dominant orbital excitations,vertical excitation energies (E ),dipole moments (z component,µz ),dipole moment differences from S 0(∆µz ),and oscillatorstrengths (f )of the lowest three triplet and the lowest three singlet roots for Pt(dpb)Cl and Pt(dpt)Cl.Table 3also includes the observed excitation energies for T 1(the origins in the low-temperature phosphorescence spectra)and S 1(the absorption peaks in the room-temperature spectra).The lowest three triplet roots as well as the lowest two singlet roots are represented by dominant excitations from mixed d πorbitals to π*orbitals.The third lowest singlet root is represented by a d σ-π*excitation.Excitations with d σ-π*character appear at the fifth lowest and higher roots for triplet states that are not specified in Table 3.The lowest three triplet as well as singlet roots for Pt(dpt)Cl are identical in characteristics of dominant excitations to the counterparts of Pt(dpb)Cl.In succeeding sections,we discuss only about Pt(dpb)Cl and dpbH,unless the methyl substitution effect is explored.We begin the discussion on the comparison of the calculated and experimental data by assignment of T 1and S 1from the calculated triplet and singlet roots for Pt(dpb)Cl.Experimental evidence is needed for the T 1and S 1assignments because both the calculated energy differences between the two lowest roots of triplet as well as singlet are very small.On the other hand,the third lowest roots for triplet and singlet are excluded from the T 1or S 1assignment because they are calculated to be energetically well above the lowest two roots.Considering that the excitation to S 1has a substantial extinction coefficient in the absorption spectrum (section 3.1),the lowest calculated singlet root (1B 2;f )0.074)is assigned to be S 1since the second lowest root (1A 1;f )0.007)is predicted to have 1order smaller absorption intensity.The calculated S 1has a large ∆µz (-9.07D)that is consistent with the experimentally obtained large negative solvatochromic shift of the S 1peak.39,40On the other hand,T 1of Pt(dpb)Cl has different spectral characteristics from S 1as discussed in section 3.1.Therefore,we assign the lowest calculated triplet root (3A 1)to T 1on the basis of the different dominant orbital excitation from S 1(1B 2)and the small ∆µz (-1.13D)that explain the small negative solvatochromic shift along with the small Stokes shift of the T 1peak.39The second and third lowest calculated triplet (or singlet)roots are assigned to be T 2and T 3(or S 2and S 3),respectively.The calculated T 1excitation energies of Pt(dpb)Cl and Pt(dpt)Cl show excellent agreements with the respective ex-perimental phosphorescence origins at 77K;the differences between the calculated and experimental values are found to be less than 200cm -1.In these comparisons,the calculated energies represent the vertical excitation energies at S 0geom-etries,while the experimental energies correspond to the 0-0transitions.However,these comparisons may be justified by the phosphorescence spectra,which indicate close equilibrium geometries between S 0and T 1of these complexes,as shown in section 3.2.(Geometries of T 1together with the adiabatic excitation energies will be discussed in section 4.3.)The calculated S 1excitation energies of the complexes also show good conformity with the experimental absorption peak energies within 600cm -1.Although TDDFT is known to have irregulari-ties in description of long-range charge-transfer excited states,43-45the above calculation results for excitation energies ensure the applicability of the TDDFT calculations for T 1and S 1of the concerned molecules.Hence we further investigate the charac-teristics of T 1and S 1of the complexes by use of the TDDFT calculation results in the following sections.TDDFT calculations were also carried out for the free ligands.Calculated vertical excitation energies and excitation types for the two lowest excited triplet roots of dpbH and dptH are summarized in Table 4together with the experimental energiesTABLE 4:Vertical Excitation Energies (E )from TDDFT Calculations for Triplet Excited States of DpbH and DptH with Experimental Excitation EnergiesE /cm -1root (assignment a )symmetry bdominant excitation b ,c dpbH dptH 1(T 1)3A 1a 2f a 2*23344232172(T 2)3B 2b 1f a 2*2730427077T 1(expt d )2278022620aSee text.b Symmetry of states and orbitals are designated for dpbH (C 2V ,see Figure 1).c For designation of the orbitals,see Figure 4.dPhosphorescence 0-0peak energy in toluene at 77K.Figure 4.Contour plots of the two lowest unoccupied and the four highest occupied molecular orbitals of Pt(dpb)Cl,along with the LUMO and HOMO of dpbH.Phosphorescent Platinum(II)ComplexesJ.Phys.Chem.A,Vol.109,No.43,20059763of the phosphorescence origin.For dpbH,the lowest calculated triplet root (3A 1)was assigned to be T 1on the basis of the large energy separation between the first and second lowest roots.The small differences between the calculated and experimental T 1excitation energies of dpbH and dptH (560-600cm -1)evidenced the accuracy of the TDDFT calculations.4.2.Evaluation of d -π*Character in the Excited States.Evaluation of the degree of d -π*participation for low-lying excited states of mixed π-π*/d -π*character would be very helpful to understand the photophysical properties of the phosphorescent complexes.The zero-field splittings (ZFS)of sublevels of T 1are shown to be a valuable experimental parameter to measure the metal participation in T 1of com-plexes.17-19Theoretical calculation is considered to be an alternative for the evaluation of the metal participation that can be applied to a variety of complexes.The theoretical studies reported so far treat the evaluation of d -π*participation by several methods:the population analysis of orbitals involved in dominant excitations,11the analysis of distribution of spin densities and atomic charges by SCF calculations in T 1,30and the population analysis of metal atoms 46or the metal d-orbitals for the ground and excited states.47We employed the population analysis of metal orbitals in combination with TDDFT calcula-tions as discussed in the following section.We have noted that T 1and S 1of the concerned complexes are each represented by a dominant excitation from a mixed d πorbital to a π*orbital.More detailed pictures of the excited states depicting the changes in electron density 48are provided in Figure 5,for T 1,S 1,and T 3from S 0of Pt(dpb)Cl along with that for T 1from S 0of dpbH.(The changes for T 2and S 2of thecomplex bear resemblances to those for S 1and T 1,respectively.)T 1and S 1of Pt(dpb)Cl are characterized by d-electron density decrease on Pt and π-electron densities on Cl and some of the carbons in the tridentate ligand and by increases of π-electron densities of the tridentate ligand with respect to S 0,which indicate an admixture of d -π*and π-π*characters in each excited states.Considering that TDDFT is a single excitation theory,49the decrease in d-electron densities calculated by TDDFT ap-proximately gives the percentage of d -π*character in the excited state;that is,one electron decrease in d-electron densities represents 100%d -π*character.The decreases in d-electron densities for T 1and S 1of Pt(dpb)Cl are evaluated by Mulliken population analysis.46,47Table 5summarizes the orbital popula-tions of the Pt consisting of atomic orbitals s,p π,p σ,d π,and d σ(πand σdenote antisymmetric and symmetric orbitals about the molecular plane,respectively)for S 0,T 1,and S 1of Pt(dpb)-Cl and Pt(dpt)Cl.The differences between the two complexes are insignificant,and the s,p σ,and d σdensities remain unchanged by the T 1and S 1excitations.The d πdensities decrease in T 1by 0.25e and in S 1by 0.32e from S 0for both of the complexes,suggesting the percentages of d -π*character in T 1and S 1as 25%and 32%,respectively.On the other hand,the p πdensities on Pt increase in T 1by 0.06e [Pt(dpb)Cl]and 0.05e [Pt(dpt)Cl]from S 0,which indicates some delocalization of the spreading of the π*orbitals over the Pt atom.The emitting states (T 1)of Pt(dpb)Cl and Pt(dpt)Cl with 25%d -π*character seem to fall into the intermediate category of significant LC/MLCT admixture in the sequence of complexes ordered by metal -d character (from pure LC to pure MLCT)of the emitting states.18,19The d -π*character in the emitting state of such a complex dominates the kinetics of the deactiva-tion processes by effective spin -orbit coupling with singlet excited states.The evaluated d -π*characters in T 1of Pt(dpb)-Cl and Pt(dpt)Cl are considered to be consistent with the reported emission lifetimes (7-8µs 9)as well as the emission energy shifts from the corresponding free ligands (2200-2600cm -1).It should be mentioned that the d -π*character in T 1does not lead to large ∆µz ,the difference in dipole moment between the ground and excited state,for the objective complexes despite its charge-transfer nature.The d -π*character combined with the small ∆µz in T 1is clearly shown in Figure 5,which depicts the dominant charge flow from Pt,Cl,and phenyl ring to both pyridine rings in the tridentate ligand that cancel each other in the y -axis,thus exhibiting little change along the dipole moment in the z -axis.On the other hand,S 1of Pt(dpb)Cl was calculated to have a considerably larger ∆µz (-9.07D)than T 1,in contrast to the small differences in the d -π*character between T 1(25%)and S 1(32%).The charge flow in S 1is almost“straightforward”Figure 5.Contour plots of changes in electron densities for T 1,S 1,and T 3from S 0of Pt(dpb)Cl,along with that for T 1from S 0of dpbH.TABLE 5:Orbital Populations on Pt for the Ground and Excited States from Mulliken Population Analysis aorbital population (difference from S 0)state s p πb p σb d πbd σb Pt(dpb)Cl S 0 2.712.094.123.784.74T 1 2.71(0.00) 2.15(0.06) 4.12(0.00) 3.53(-0.25) 4.74(0.00)S 12.71(0.00) 2.09(0.00) 4.12(0.00)3.46(-0.32)4.74(0.00)Pt(dpt)Cl S 0 2.712.094.123.834.69T 1 2.71(0.00) 2.14(0.05) 4.12(0.00) 3.58(-0.25) 4.69(0.00)S 12.71(0.00)2.09(0.00)4.12(0.00)3.51(-0.32)4.70(0.00)aOrbital populations are given in unit electron charge.Inner core electrons (1s through 4f)are not included because they are replaced by the effective core potential (ECP).b σand πrepresent symmetrical and antisymmetrical orbitals about the molecular plane,respectively.9764J.Phys.Chem.A,Vol.109,No.43,2005Sotoyama et al.from the Pt and Cl to the phenyl ring that causes a large dipole moment change parallel to the z-axis.parison of Phosphorescence Spectra of the Complexes and Free Ligands:Methyl Substitution Effects and Spectral Features.The methyl substitution causes a substantial decrease in phosphorescence0-0energy from Pt(dpb)Cl to Pt(dpt)Cl(540cm-1),while the difference between the free ligands(dpbH and dptH)is small(160cm-1),as explained in section3.2.These calculations reproduced the differences in the T1excitation energies(362cm-1between the complexes and127cm-1between the free ligands).The difference in the methyl substitution effect between the com-plexes and the free ligands can be assigned to the different orbital nature of T1in respective species.Although T1of Pt(dpb)Cl and dpbH have identical symmetry(A1),they differ in dominant orbital excitations[b1f b1*for Pt(dpb)Cl;a2f a2*for dpbH].Theπorbitals of a2symmetry in dpbH[as well as Pt(dpb)Cl]have no component at the atoms on the symmetry axis,including C(4),as shown in the MO plots(Figure4).Themethyl substitution at C(4)is expected to have a minimal effect on the orbitals of a2symmetry as well as the a2f a2* excitations.On the other hand,the b1orbital(HOMO)of Pt(dpb)Cl spreads over C(4);hence,it resulted in a substantial difference in T1excitation energies between Pt(dpb)Cl and Pt(dpt)Cl.The difference in the dominant orbital excitations between the complexes and the free ligands stated above gives valuable information about the phosphorescence spectra of these species. The phosphorescence spectra of the free ligands(Figure3)show increased vibrational progressions compared to those of the complexes.These spectral features can be explained from the plots of the changes of electron density(Figure5)that show the difference in T1character between Pt(dpb)Cl and dpbH.The changes of the electron density are found on the atoms for Pt-(dpb)Cl,whereas those for dpbH,which show some resemblance to those for T3of Pt(dpb)Cl,are found mainly on the bonds in the phenyl ring and between the phenyl and pyridine rings. These electron-density changes on the bonds,especially in the phenyl ring,are expected to induce significant geometry changes for the free ligands between T1and S0.These geometry changes appear as strong vibrational progressions in the phosphorescence spectra with frequencies of aromatic ring stretching about1300 cm-1.The geometry changes expected by the electron density calculations were confirmed by T1geometry optimizations for Pt(dpb)Cl and dpbH at the restricted open-shell B3LYP level. Geometries were optimized in C2V symmetry and then compared to their S0structures.Particular attention was paid to selection of the initial-guess orbitals for the SCF calculations in T1 geometry optimizations to obtain coherent results with the TDDFT calculations about nature of the singly occupied orbitals. Table6summarizes the changes in bond length between S0and T1.For dpbH,the length of the C(2)-C(3)bond shows a notable increase(0.085Å)by excitation to T1that is consistent with the electron density plot(Figure5),while changes in bond length for Pt(dpb)Cl are rather small compared to those for dpbH,and mainly observed for the bonds between Pt and its neighboring atoms[Cl and C(1)].Additional TDDFT calculations were carried out for both Pt(dpb)Cl and dpbH at the T1optimized geometry to obtain the relaxation energy in T1(the difference between the T1 energies at the S0and T1optimized geometries)and the adiabatic T1excitation energy(the vertical T1excitation energy at the S0 optimized geometry minus the relaxation energy in T1).The obtained T1relaxation energy for Pt(dpb)Cl(631cm-1)is notably smaller than that of dpbH(2070cm-1);this result isconsistent with the above discussion on the phosphorescenceprofiles of Pt(dpb)Cl and dpbH.The adiabatic T1excitationenergies are figured out to be19790cm-1for Pt(dpb)Cl and21274cm-1for dpbH;these values are in good agreement withthe experimental0-0excitation energies in S0-T1transitions.5.ConclusionThe lowest triplet and singlet excited states(T1and S1)ofthe objective complexes Pt(dpb)Cl and Pt(dpt)Cl have beenassigned to3A1(b1f b1*)and1B2(b1f a2*),respectively,from the TDDFT calculations with the assistance of the observedspectroscopic properties.The calculated excitation energies forT1and S1of the complexes show good agreement with theirrespective observed values within600cm-1.These states arepredicted to have mixed characters of d-π*andπ-π*excitations.On the basis of the single excitation nature of theTDDFT approach,the d-π*character in excited states isevaluated from the decrease in the d-electron densities on Ptby excitation;values of25%for T1and32%for S1have beenobtained for both complexes.The emitting states of the freeligands(dpbH and dptH)are predicted to be different in orbitalnature from those of the complexes;this difference in orbitalnature explains the observed difference in methyl-substitutioneffect as well as different emission profiles between thecomplexes and the free ligands.Other spectroscopic featuresof the complexes,such as the negative solvatochromic shift andsmall Stokes shift,are also successfully explained from thecalculation results based on the above assignment.The above results have proved that the TDDFT approach isrational for analysis of the excited-state properties of theobjective complexes.The presented procedure to evaluate thed-π*character may be a useful tool to design highly phos-phorescent complexes.We consider that it is very important toconduct further studies to evaluate d-π*character in the excitedstates of a variety of phosphorescent complexes and then toinvestigate the relationship between the d-π*character andphosphorescence properties of these complexes in order to getan in-depth understanding of the photophysics of the complexes. Acknowledgment.We thank Dr.N.F.Cooray for helpful suggestions and discussions.References and Notes(1)Baldo,M.A.;O’Brien,D.F.;You,Y.;Shoustikov,A.;Sibley,S.; Thompson,M.E.;Forrest,S.R.Nature(London)1998,395,151. TABLE6:Calculated Bond Lengths and Differences between S0and T1of Pt(dpb)Cl and DpbH aPt(dpb)Cl b dpbH b bond c S0T1difference S0T1difference Pt-Cl 2.471 2.444-0.027Pt-C(1) 1.926 1.899-0.027Pt-N 2.065 2.063-0.002C(1)-C(2) 1.403 1.4340.031 1.402 1.397-0.005 C(2)-C(3) 1.402 1.390-0.012 1.405 1.4900.085 C(3)-C(4) 1.401 1.4120.011 1.393 1.386-0.007 C(2)-C(5) 1.469 1.451-0.018 1.492 1.442-0.050 C(5)-C(6) 1.397 1.3970.000 1.406 1.4240.018 C(6)-C(7) 1.392 1.390-0.002 1.392 1.388-0.004 C(7)-C(8) 1.395 1.4080.013 1.393 1.3950.002 C(8)-C(9) 1.391 1.385-0.006 1.396 1.4080.012 C(9)-N 1.343 1.3510.008 1.333 1.321-0.012 C(5)-N 1.379 1.3980.019 1.347 1.3690.022a Bond lengths are given in angstroms.b Geometries are optimized within the symmetry constraint of C2V.c See Figure1for numbering of the carbon atoms.Phosphorescent Platinum(II)Complexes J.Phys.Chem.A,Vol.109,No.43,20059765。
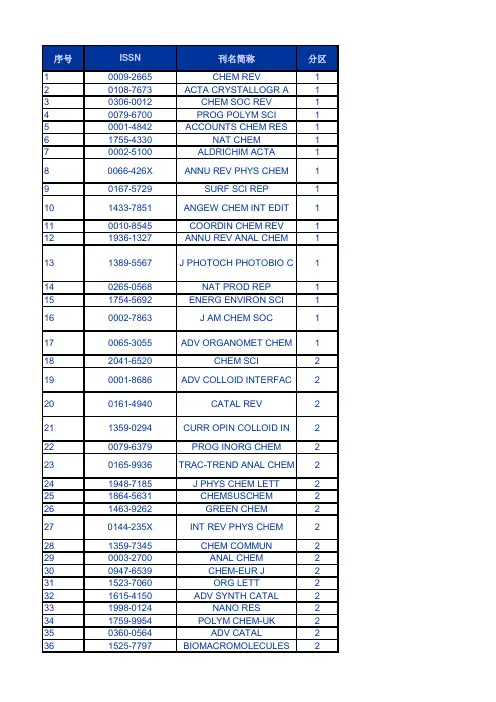
序号ISSN刊名简称分区10009-2665CHEM REV1 20108-7673ACTA CRYSTALLOGR A1 30306-0012CHEM SOC REV1 40079-6700PROG POLYM SCI1 50001-4842ACCOUNTS CHEM RES1 61755-4330NAT CHEM1 70002-5100ALDRICHIM ACTA180066-426X ANNU REV PHYS CHEM1 90167-5729SURF SCI REP1 101433-7851ANGEW CHEM INT EDIT1110010-8545COORDIN CHEM REV1 121936-1327ANNU REV ANAL CHEM1131389-5567J PHOTOCH PHOTOBIO C1140265-0568NAT PROD REP1 151754-5692ENERG ENVIRON SCI1160002-7863J AM CHEM SOC1 170065-3055ADV ORGANOMET CHEM1 182041-6520CHEM SCI2 190001-8686ADV COLLOID INTERFAC2 200161-4940CATAL REV2 211359-0294CURR OPIN COLLOID IN2 220079-6379PROG INORG CHEM2 230165-9936TRAC-TREND ANAL CHEM2241948-7185J PHYS CHEM LETT2 251864-5631CHEMSUSCHEM2 261463-9262GREEN CHEM2270144-235X INT REV PHYS CHEM2281359-7345CHEM COMMUN2 290003-2700ANAL CHEM2 300947-6539CHEM-EUR J2 311523-7060ORG LETT2 321615-4150ADV SYNTH CATAL2 331998-0124NANO RES2 341759-9954POLYM CHEM-UK2 350360-0564ADV CATAL2 361525-7797BIOMACROMOLECULES2370065-3195ADV POLYM SCI2381549-9618J CHEM THEORYCOMPUT2390020-1669INORG CHEM2 401932-7447J PHYS CHEM C2 410070-9778ELECTROANAL CHEM2 421528-7483CRYST GROWTH DES2 431364-5498FARADAY DISCUSS2 441861-4728CHEM-ASIAN J2 450340-1022TOP CURR CHEM2 461527-8999CHEM REC2 471867-3880CHEMCATCHEM2 480021-9673J CHROMATOGR A2 490022-3263J ORG CHEM2 500003-2670ANAL CHIM ACTA2510192-8651J COMPUT CHEM2 521549-9596J CHEM INF MODEL2530743-7463LANGMUIR2 540081-5993STRUCT BOND2 550276-7333ORGANOMETALLICS2 561466-8033CRYSTENGCOMM2 570069-3138CHEM PHYS CARBON2580021-8898J APPL CRYSTALLOGR2 590887-624X J POLYM SCI POL CHEM3601477-9226DALTON T3 610003-2654ANALYST3621044-0305J AM SOC MASS SPECTR3 631463-9076PHYS CHEM CHEM PHYS3 641618-2642ANAL BIOANAL CHEM3 651948-7193ACS CHEM NEUROSCI3 660267-9477J ANAL ATOM SPECTROM3 671570-1794CURR ORG SYNTH3 681477-0520ORG BIOMOL CHEM3690926-860X APPL CATAL A-GEN3 700032-3861POLYMER3 710039-9140TALANTA3 721520-6106J PHYS CHEM B3 730144-8617CARBOHYD POLYM3 741758-2946J CHEMINFORMATICS3 751439-4235CHEMPHYSCHEM3761948-5875ACS MED CHEM LETT3770949-8257J BIOL INORG CHEM3782156-8952ACS COMB SCI3 791076-5174J MASS SPECTROM3 800920-5861CATAL TODAY3810223-5234EUR J MED CHEM3 821040-8347CRIT REV ANAL CHEM3 831350-4177ULTRASON SONOCHEM3 841434-193X EUR J ORG CHEM3850040-4020TETRAHEDRON3 860889-311X CRYSTALLOGR REV3870021-9797J COLLOID INTERF SCI3881387-1811MICROPOR MESOPORMAT3891381-1169J MOL CATAL A-CHEM3901381-1991MOL DIVERS3 911434-1948EUR J INORG CHEM3921385-2728CURR ORG CHEM3 931566-7367CATAL COMMUN3940340-6253MATCH-COMMUN MATHCO3950079-6786PROG SOLID STATE CH3961089-5639J PHYS CHEM A3 970304-4203MAR CHEM3980047-2689J PHYS CHEM REF DATA3 990951-4198RAPID COMMUN MASS SP31000898-8838ADV INORG CHEM3 1010026-3672MICROCHIM ACTA3 1021144-0546NEW J CHEM3 1031040-0397ELECTROANAL3 1040026-265X MICROCHEM J31051572-6657J ELECTROANAL CHEM31060040-4039TETRAHEDRON LETT3 1071615-9306J SEP SCI3 1080936-5214SYNLETT31091661-6596INT J MOL SCI31100957-4166TETRAHEDRON-ASYMMETR 31111542-2119SEP PURIF REV31121432-881X THEOR CHEM ACC3 1130014-3057EUR POLYM J31140141-3910POLYM DEGRAD STABIL3 1151022-1352MACROMOL CHEM PHYS31161022-5528TOP CATAL3 1170260-3594COMMENT INORG CHEM3 1180039-7881SYNTHESIS-STUTTGART31191010-6030J PHOTOCH PHOTOBIO A31200033-4545PURE APPL CHEM3 1210036-021X RUSS CHEM REV+31220165-2370J ANAL APPL PYROL3 1230022-328X J ORGANOMET CHEM3 1240009-2614CHEM PHYS LETT3 1251083-6160ORG PROCESS RES DEV3 1260303-402X COLLOID POLYM SCI3 1271386-2073COMB CHEM HIGH T SCR3 1280022-4596J SOLID STATE CHEM3 1290927-7757COLLOID SURFACE A41300277-5387POLYHEDRON4 1310008-6215CARBOHYD RES4 1321752-153X CHEM CENT J4 1330301-0104CHEM PHYS4 1341571-1013CATAL SURV ASIA4 1351011-372X CATAL LETT4 1361570-193X MINI-REV ORG CHEM4 1371420-3049MOLECULES4 1380959-8103POLYM INT4 1391053-0509J FLUORESC4 1400020-1693INORG CHIM ACTA4 1411610-2940J MOL MODEL4 1420004-9425AUST J CHEM41431387-7003INORG CHEM COMMUN41441061-0278SUPRAMOL CHEM41450108-7681ACTA CRYSTALLOGR B41460039-6028SURF SCI41470268-2605APPL ORGANOMET CHEM4 1480924-2031VIB SPECTROSC4 1490022-1139J FLUORINE CHEM4 1500040-6031THERMOCHIM ACTA4 1511860-5397BEILSTEIN J ORG CHEM4 1521293-2558SOLID STATE SCI4 1531040-0400STRUCT CHEM41540065-2725ADV HETEROCYCL CHEM4 1550894-3230J PHYS ORG CHEM4 1560736-6299SOLVENT EXTR ION EXC4 1570364-5916CALPHAD41580022-0248J CRYST GROWTH4 1591631-0748CR CHIM4 1600267-8292LIQ CRYST4 1611573-4110CURR ANAL CHEM41621388-6150J THERM ANAL CALORIM4 1630065-3160ADV PHYS ORG CHEM41641788-618X EXPRESS POLYM LETT4 1650022-2860J MOL STRUCT41660009-2673 B CHEM SOC JPN4 1670886-9383J CHEMOMETR4 1681574-1443J INORG ORGANOMET P41691735-207X J IRAN CHEM SOC4 1700065-3276ADV QUANTUM CHEM4 1710167-7322J MOL LIQ4 1720366-7022CHEM LETT41730095-8972J COORD CHEM41740749-1581MAGN RESON CHEM4 1751022-9760J POLYM RES41760923-0750J INCL PHENOM MACRO41770910-6340ANAL SCI41780103-5053J BRAZIL CHEM SOC41790026-9247MONATSH CHEM4 1800018-019X HELV CHIM ACTA4 1810033-8230RADIOCHIM ACTA4 1820065-2415ADV CHROMATOGR4 1831878-5352ARAB J CHEM41840095-9782J SOLUTION CHEM4 1850032-1400PLATIN MET REV41860008-4042CAN J CHEM4 1870020-7608INT J QUANTUM CHEM4 1880259-9791J MATH CHEM41892210-271X COMPUT THEOR CHEM4 1900009-4293CHIMIA41911088-4246J PORPHYR PHTHALOCYA41920538-8066INT J CHEM KINET41930032-3896POLYM J4 1940170-0839POLYM BULL41950021-8995J APPL POLYM SCI4 1960044-2313Z ANORG ALLG CHEM41971003-9953J NAT GAS CHEM4 1980892-7022MOL SIMULAT4 1990969-806X RADIAT PHYS CHEM4 2001551-7004ARKIVOC4 2010142-2421SURF INTERFACE ANAL4 2020340-4285TRANSIT METAL CHEM4 2030009-5893CHROMATOGRAPHIA4 2040385-5414HETEROCYCLES4 2050003-2719ANAL LETT4 2060974-3626J CHEM SCI4 2070947-7047IONICS42080022-152X J HETEROCYCLIC CHEM42090954-0083HIGH PERFORM POLYM4 2101318-0207ACTA CHIM SLOV4 2111042-7163HETEROATOM CHEM42121895-1066CENT EUR J CHEM4 2131026-1265IRAN POLYM J4 2140030-4948ORG PREP PROCED INT4 2151023-666X INT J POLYM ANAL CH4 2160010-0765COLLECT CZECH CHEM C42171674-7291SCI CHINA CHEM4 2180039-7911SYNTHETIC COMMUN4 2191751-8253GREEN CHEM LETT REV42200236-5731J RADIOANAL NUCL CH4 2210732-8303J CARBOHYD CHEM4 2221565-3633BIOINORG CHEM APPL42230021-2148ISR J CHEM4 2241040-7278J CLUST SCI42250021-9665J CHROMATOGR SCI4 2260232-1300CRYST RES TECHNOL42271040-6638POLYCYCL AROMATCOMP 42281478-6419NAT PROD RES4 2290253-2964 B KOREAN CHEM SOC42300914-9244J PHOTOPOLYM SCI TEC4 2310933-4173JPC-J PLANAR CHROMAT4 2321082-6076J LIQ CHROMATOGR R T42331385-772X DES MONOMERS POLYM4 2340366-6352CHEM PAP4 2351741-5993J SULFUR CHEM42360932-0776Z NATURFORSCH B4 2371002-0721J RARE EARTH4 2380253-3820CHINESE J ANAL CHEM4 2391060-1325J MACROMOL SCI A42400959-9436MENDELEEV COMMUN4 2411300-0527TURK J CHEM42420376-4710INDIAN J CHEM A4 2431203-8407J ADV OXID TECHNOL42440352-5139J SERB CHEM SOC42451001-604X CHINESE J CHEM4 2461001-8417CHINESE CHEM LETT4 2471570-1786LETT ORG CHEM4 2480100-4042QUIM NOVA42490219-6336J THEOR COMPUT CHEM42500009-3130CHEM NAT COMPD+4 2510011-1643CROAT CHEM ACTA42521109-4028CHEM EDUC RES PRACT42531000-6818ACTA PHYS-CHIM SIN42540922-6168RES CHEM INTERMEDIAT42551233-2356ACTA CHROMATOGR4 2560965-545X POLYM SCI SER A+42570009-4536J CHIN CHEM SOC-TAIP4 2580108-2701ACTA CRYSTALLOGR C42590023-1584KINET CATAL+4 2601468-6783PROG REACT KINET MEC4 2611878-5190REACT KINET MECH CAT4 2621061-9348J ANAL CHEM+42630009-3122CHEM HETEROCYCLCOM+42640256-7679CHINESE J POLYM SCI42651061-933X COLLOID J+4 2661001-4861CHINESE J INORG CHEM42670021-9584J CHEM EDUC4 2680567-7351ACTA CHIM SINICA42690251-0790CHEM J CHINESE U4 2700009-2770CHEM LISTY4 2710193-2691J DISPER SCI TECHNOL4 2721074-1542J CHEM CRYSTALLOGR4 2731042-6507PHOSPHORUS SULFUR42740973-4945E-J CHEM4 2750034-7752REV CHIM-BUCHAREST4 2761024-1221MAIN GROUP CHEM4 2771433-5158HYLE4 2780018-1439HIGH ENERG CHEM+42791070-4280RUSS J ORG CHEM+42801747-5198J CHEM RES4 2811005-281X PROG CHEM42820253-5106J CHEM SOC PAKISTAN42830379-4350S AFR J CHEM-S-AFR T4 2841070-3284RUSS J COORD CHEM+42851857-5552MACED J CHEM CHEM EN42860031-9104PHYS CHEM LIQ4 2870793-0135REV ANAL CHEM4 2881618-7229E-POLYMERS42890037-9980J SYN ORG CHEM JPN42901000-3304ACTA POLYM SIN4 2911063-7745CRYSTALLOGR REP+4 2920009-2223CHEM ANAL-WARSAW42930376-4699INDIAN J CHEM B4 2940040-5760THEOR EXP CHEM+4 2950717-9324J CHIL CHEM SOC4 2960254-5861CHINESE J STRUC CHEM4 2970022-4766J STRUCT CHEM+4 2981542-1406MOL CRYST LIQ CRYST42990039-3630STUD CONSERV4 3000235-7216CHEMIJA4 3010009-2851CHEM UNSERER ZEIT4 3020392-839X CHIM OGGI4 3031870-249X J MEX CHEM SOC4 3041066-5285RUSS CHEM B+43050036-0244RUSS J PHYS CHEM A+4 3061005-9040CHEM RES CHINESE U43070137-5083POL J CHEM4 3080104-1428POLIMEROS4 3091560-0904POLYM SCI SER B+4 3101471-6577LC GC EUR43111070-3632RUSS J GEN CHEM+4 3120036-0236RUSS J INORG CHEM+43130379-153X POLYM-KOREA4 3140525-1931BUNSEKI KAGAKU43151463-9246J AUTOM METHODMANAG43161600-5368ACTA CRYSTALLOGR E43171665-2738REV MEX ING QUIM4 3180334-7575MAIN GROUP MET CHEM43190019-4522J INDIAN CHEM SOC4 3200972-0626RES J CHEM ENVIRON43210012-5016DOKL PHYS CHEM4 3220035-3930REV ROUM CHIM43231011-3924 B CHEM SOC ETHIOPIA43240793-0283HETEROCYCL COMMUN4 3251527-5949LC GC N AM43260971-1627INDIAN J HETEROCY CH4 3271070-4272RUSS J APPL CHEM+43280012-5008DOKL CHEM4 3290970-7077ASIAN J CHEM43301433-7266Z KRIST-NEW CRYST ST43310193-4929REV INORG CHEM4 3321990-7931RUSS J PHYS CHEM B+43331063-455X J WATER CHEMTECHNO+43341347-9466SCI TECHNOL ENERG MA43350209-4541OXID COMMUN4 3360324-1130BULG CHEM COMMUN4 3371511-1768J RUBBER RES4 3380001-9704AFINIDAD4 3391473-7604CHEM WORLD-UK4 3400386-2186KOBUNSHI RONBUNSHU4 3411224-7154STUD U BABES-BOL CHE4 3420009-3068CHEM IND-LONDON4 3430151-9093ACTUAL CHIMIQUE4 3442155-5435ACS CATAL4 3452044-4753CATAL SCI TECHNOL4 3462046-2069RSC ADV4刊名全称3年平均IF2011年IFCHEMICAL REVIEWS36.39740.197ACTA CRYSTALLOGRAPHICASECTION A 35.445 2.076CHEMICAL SOCIETY REVIEWS25.14428.76 PROGRESS IN POLYMER SCIENCE23.57424.1ACCOUNTS OF CHEMICALRESEARCH 20.56521.64Nature Chemistry19.22620.524 ALDRICHIMICA ACTA15.52216.091 ANNUAL REVIEW OF PHYSICALCHEMISTRY14.61314.13 SURFACE SCIENCE REPORTS14.58411.696 ANGEWANDTE CHEMIE-INTERNATIONAL EDITION12.67113.455COORDINATION CHEMISTRYREVIEWS 11.11812.11Annual Review of Analytical Chemistry9.7269.048 JOURNAL OF PHOTOCHEMISTRYAND PHOTOBIOLOGY C-PHOTOCHEMISTRY REVIEWS9.70710.36NATURAL PRODUCT REPORTS9.2919.79 Energy & Environmental Science9.1999.61 JOURNAL OF THE AMERICANCHEMICAL SOCIETY9.179.907 ADVANCES IN ORGANOMETALLICCHEMISTRY7.9497Chemical Science7.5257.525 ADVANCES IN COLLOID ANDINTERFACE SCIENCE7.4858.12 CATALYSIS REVIEWS-SCIENCE ANDENGINEERING7.4227.5 CURRENT OPINION IN COLLOID &INTERFACE SCIENCE6.5468.01PROGRESS IN INORGANICCHEMISTRY 6.4819.333TRAC-TRENDS IN ANALYTICALCHEMISTRY6.474 6.273 Journal of Physical Chemistry Letters 6.213 6.213 ChemSusChem 5.973 6.827 GREEN CHEMISTRY 5.876 6.32 INTERNATIONAL REVIEWS INPHYSICAL CHEMISTRY5.832 5.967CHEMICAL COMMUNICATIONS 5.82 6.169 ANALYTICAL CHEMISTRY 5.648 5.856 CHEMISTRY-A EUROPEAN JOURNAL 5.594 5.925 ORGANIC LETTERS 5.511 5.862ADVANCED SYNTHESIS &CATALYSIS 5.495 6.048Nano Research 5.473 6.97Polymer Chemistry 5.321 5.321 ADVANCES IN CATALYSIS 5.135 3.667 BIOMACROMOLECULES 5.103 5.479ADVANCES IN POLYMER SCIENCE 5.071 3.89 Journal of Chemical Theory andComputation5.052 5.215INORGANIC CHEMISTRY 4.528 4.601 Journal of Physical Chemistry C 4.518 4.805 Electroanalytical Chemistry 4.5 4.5 CRYSTAL GROWTH & DESIGN 4.424 4.72 FARADAY DISCUSSIONS 4.4135Chemistry-An Asian Journal 4.354 4.5 TOPICS IN CURRENT CHEMISTRY 4.309 6.568 CHEMICAL RECORD 4.281 4.377 ChemCatChem 4.276 5.207 JOURNAL OF CHROMATOGRAPHY A 4.275 4.531 JOURNAL OF ORGANIC CHEMISTRY 4.224 4.45 ANALYTICA CHIMICA ACTA 4.208 4.555 JOURNAL OF COMPUTATIONALCHEMISTRY4.134 4.583 Journal of Chemical Information andModeling4.126 4.675LANGMUIR 4.118 4.186 STRUCTURE AND BONDING 4.095 3.475 ORGANOMETALLICS 4.018 3.963CRYSTENGCOMM 4.01 3.842CHEMISTRY AND PHYSICS OFCARBON 40JOURNAL OF APPLIEDCRYSTALLOGRAPHY3.988 5.152 JOURNAL OF POLYMER SCIENCEPART A-POLYMER CHEMISTRY3.928 3.919DALTON TRANSACTIONS 3.855 3.838 ANALYST 3.805 4.23JOURNAL OF THE AMERICAN SOCIETY FOR MASSSPECTROMETRY 3.741 4.002PHYSICAL CHEMISTRY CHEMICALPHYSICS3.714 3.573 ANALYTICAL AND BIOANALYTICALCHEMISTRY3.7 3.778ACS Chemical Neuroscience 3.676 3.676 JOURNAL OF ANALYTICAL ATOMICSPECTROMETRY3.676 3.22 CURRENT ORGANIC SYNTHESIS 3.667 3.434 ORGANIC & BIOMOLECULARCHEMISTRY3.636 3.696 APPLIED CATALYSIS A-GENERAL 3.617 3.903 POLYMER 3.613 3.438TALANTA 3.602 3.794JOURNAL OF PHYSICAL CHEMISTRYB 3.59 3.696CARBOHYDRATE POLYMERS 3.419 3.628 Journal of Cheminformatics 3.419 3.419 CHEMPHYSCHEM 3.402 3.412ACS Medicinal Chemistry Letters 3.355 3.355 JOURNAL OF BIOLOGICALINORGANIC CHEMISTRY3.33 3.289ACS Combinatorial Science 3.328 3.408JOURNAL OF MASS SPECTROMETRY 3.323 3.268CATALYSIS TODAY 3.309 3.407 EUROPEAN JOURNAL OFMEDICINAL CHEMISTRY3.269 3.346 CRITICAL REVIEWS IN ANALYTICALCHEMISTRY3.258 3.902 ULTRASONICS SONOCHEMISTRY 3.254 3.567 EUROPEAN JOURNAL OF ORGANICCHEMISTRY3.21 3.329TETRAHEDRON 3.085 3.025 Crystallography Reviews 3.079 2.067 JOURNAL OF COLLOID ANDINTERFACE SCIENCE3.052 3.07 MICROPOROUS AND MESOPOROUSMATERIALS3.052 3.285JOURNAL OF MOLECULARCATALYSIS A-CHEMICAL2.985 2.947MOLECULAR DIVERSITY 2.982 3.153EUROPEAN JOURNAL OFINORGANIC CHEMISTRY2.9673.049 CURRENT ORGANIC CHEMISTRY 2.954 3.064 CATALYSIS COMMUNICATIONS 2.938 2.986 MATCH-COMMUNICATIONS INMATHEMATICAL AND IN COMPUTERCHEMISTRY2.89 2.161PROGRESS IN SOLID STATECHEMISTRY2.88 4.188JOURNAL OF PHYSICAL CHEMISTRYA 2.859 2.946MARINE CHEMISTRY 2.85 3.074 JOURNAL OF PHYSICAL ANDCHEMICAL REFERENCE DATA2.8283.172 RAPID COMMUNICATIONS IN MASSSPECTROMETRY2.777 2.79ADVANCES IN INORGANICCHEMISTRY 2.754 3.048MICROCHIMICA ACTA 2.753 3.033 NEW JOURNAL OF CHEMISTRY 2.747 2.605 ELECTROANALYSIS 2.741 2.872 MICROCHEMICAL JOURNAL 2.702 3.048 JOURNAL OF ELECTROANALYTICALCHEMISTRY2.659 2.905TETRAHEDRON LETTERS 2.654 2.683 JOURNAL OF SEPARATION SCIENCE 2.638 2.733SYNLETT 2.625 2.71 INTERNATIONAL JOURNAL OFMOLECULAR SCIENCES2.598 2.598TETRAHEDRON-ASYMMETRY 2.587 2.652 SEPARATION AND PURIFICATIONREVIEWS2.553 2.615THEORETICAL CHEMISTRYACCOUNTS 2.55 2.162EUROPEAN POLYMER JOURNAL 2.522 2.739 POLYMER DEGRADATION ANDSTABILITY2.506 2.769 MACROMOLECULAR CHEMISTRYAND PHYSICS2.456 2.361TOPICS IN CATALYSIS 2.454 2.624COMMENTS ON INORGANICCHEMISTRY 2.439 1.917SYNTHESIS-STUTTGART 2.433 2.466 JOURNAL OF PHOTOCHEMISTRYAND PHOTOBIOLOGY A-CHEMISTRY2.406 2.421 PURE AND APPLIED CHEMISTRY 2.404 2.789 RUSSIAN CHEMICAL REVIEWS 2.354 2.644 JOURNAL OF ANALYTICAL ANDAPPLIED PYROLYSIS2.344 2.487 JOURNAL OF ORGANOMETALLICCHEMISTRY2.312 2.384CHEMICAL PHYSICS LETTERS 2.303 2.337 ORGANIC PROCESS RESEARCH &DEVELOPMENT2.279 2.391 COLLOID AND POLYMER SCIENCE 2.277 2.331 COMBINATORIAL CHEMISTRY &HIGH THROUGHPUT SCREENING2.274 1.785JOURNAL OF SOLID STATECHEMISTRY 2.253 2.159COLLOIDS AND SURFACES A-PHYSICOCHEMICAL ANDENGINEERING ASPECTS2.118 2.236POLYHEDRON 2.099 2.057 CARBOHYDRATE RESEARCH 2.085 2.332 Chemistry Central Journal 2.08 3.281CHEMICAL PHYSICS 2.063 1.896 CATALYSIS SURVEYS FROM ASIA 2.057 1.897 CATALYSIS LETTERS 2.057 2.242MINI-REVIEWS IN ORGANICCHEMISTRY 2.046 2.406MOLECULES 2.037 2.386 POLYMER INTERNATIONAL 2.032 1.902 JOURNAL OF FLUORESCENCE 2.03 2.107 INORGANICA CHIMICA ACTA 2.022 1.846JOURNAL OF MOLECULARMODELING 2.001 1.797AUSTRALIAN JOURNAL OFCHEMISTRY 1.994 2.342INORGANIC CHEMISTRYCOMMUNICATIONS1.992 1.972 SUPRAMOLECULAR CHEMISTRY 1.992.145 ACTA CRYSTALLOGRAPHICASECTION B-STRUCTURAL SCIENCE1.9722.286SURFACE SCIENCE 1.934 1.994APPLIED ORGANOMETALLICCHEMISTRY 1.897 2.061VIBRATIONAL SPECTROSCOPY 1.888 1.65 JOURNAL OF FLUORINE CHEMISTRY 1.827 2.033 THERMOCHIMICA ACTA 1.818 1.805 Beilstein Journal of Organic Chemistry 1.794 2.517 SOLID STATE SCIENCES 1.786 1.856 STRUCTURAL CHEMISTRY 1.737 1.846 ADVANCES IN HETEROCYCLICCHEMISTRY1.731 1.818 JOURNAL OF PHYSICAL ORGANICCHEMISTRY1.681 1.963 SOLVENT EXTRACTION AND IONEXCHANGE1.6692.024 CALPHAD-COMPUTER COUPLINGOF PHASE DIAGRAMS ANDTHERMOCHEMISTRY1.669 1.669 JOURNAL OF CRYSTAL GROWTH 1.669 1.726 COMPTES RENDUS CHIMIE 1.661 1.803 LIQUID CRYSTALS 1.654 1.858 Current Analytical Chemistry 1.6511 JOURNAL OF THERMAL ANALYSISAND CALORIMETRY1.648 1.604 ADVANCES IN PHYSICAL ORGANICCHEMISTRY1.641 1.923Express Polymer Letters 1.599 1.769JOURNAL OF MOLECULARSTRUCTURE 1.595 1.634BULLETIN OF THE CHEMICALSOCIETY OF JAPAN1.578 1.436JOURNAL OF CHEMOMETRICS 1.54 1.952 JOURNAL OF INORGANIC ANDORGANOMETALLIC POLYMERS1.539 1.452 Journal of the Iranian Chemical Society 1.517 1.689ADVANCES IN QUANTUMCHEMISTRY 1.514 2.275JOURNAL OF MOLECULAR LIQUIDS 1.502 1.58 CHEMISTRY LETTERS 1.483 1.587 JOURNAL OF COORDINATIONCHEMISTRY1.435 1.547MAGNETIC RESONANCE INCHEMISTRY 1.432 1.437JOURNAL OF POLYMER RESEARCH 1.428 1.733JOURNAL OF INCLUSION PHENOMENA AND MACROCYCLICCHEMISTRY 1.424 1.886ANALYTICAL SCIENCES 1.415 1.255 JOURNAL OF THE BRAZILIANCHEMICAL SOCIETY1.412 1.434MONATSHEFTE FUR CHEMIE 1.4 1.532 HELVETICA CHIMICA ACTA 1.399 1.478 RADIOCHIMICA ACTA 1.387 1.575 ADVANCES IN CHROMATOGRAPHY 1.376 1.842 Arabian Journal of Chemistry 1.367 1.367JOURNAL OF SOLUTION CHEMISTRY 1.364 1.415 PLATINUM METALS REVIEW 1.361 1.361CANADIAN JOURNAL OF CHEMISTRY-REVUE CANADIENNEDE CHIMIE 1.346 1.242INTERNATIONAL JOURNAL OFQUANTUM CHEMISTRY1.325 1.357 JOURNAL OF MATHEMATICALCHEMISTRY1.314 1.303Computational and TheoreticalChemistry 1.314 1.437CHIMIA 1.312 1.212 JOURNAL OF PORPHYRINS ANDPHTHALOCYANINES1.302 1.405INTERNATIONAL JOURNAL OFCHEMICAL KINETICS1.26 1.007POLYMER JOURNAL 1.259 1.258POLYMER BULLETIN 1.254 1.532 JOURNAL OF APPLIED POLYMERSCIENCE1.244 1.289 ZEITSCHRIFT FUR ANORGANISCHEUND ALLGEMEINE CHEMIE1.241 1.249 Journal of Natural Gas Chemistry 1.214 1.348 MOLECULAR SIMULATION 1.19 1.328RADIATION PHYSICS ANDCHEMISTRY 1.169 1.227ARKIVOC 1.146 1.252SURFACE AND INTERFACEANALYSIS 1.142 1.18TRANSITION METAL CHEMISTRY 1.137 1.022 CHROMATOGRAPHIA 1.123 1.195HETEROCYCLES 1.0860.999 ANALYTICAL LETTERS 1.084 1.016 JOURNAL OF CHEMICAL SCIENCES 1.083 1.177IONICS 1.08 1.288 JOURNAL OF HETEROCYCLICCHEMISTRY1.064 1.22 HIGH PERFORMANCE POLYMERS 1.0630.884 ACTA CHIMICA SLOVENICA 1.06 1.328 HETEROATOM CHEMISTRY 1.055 1.243 CENTRAL EUROPEAN JOURNAL OFCHEMISTRY1.043 1.073IRANIAN POLYMER JOURNAL 1.0190.936 ORGANIC PREPARATIONS ANDPROCEDURES INTERNATIONAL1.016 1.015INTERNATIONAL JOURNAL OF POLYMER ANALYSIS AND CHARACTERIZATION 1 1.412COLLECTION OF CZECHOSLOVAKCHEMICAL COMMUNICATIONS0.997 1.283Science China-Chemistry0.992 1.104 SYNTHETIC COMMUNICATIONS0.987 1.062 Green Chemistry Letters and Reviews0.9760.976JOURNAL OF RADIOANALYTICALAND NUCLEAR CHEMISTRY0.976 1.52 JOURNAL OF CARBOHYDRATECHEMISTRY0.9610.631 BIOINORGANIC CHEMISTRY ANDAPPLICATIONS0.9610.716 ISRAEL JOURNAL OF CHEMISTRY0.959 1.535 JOURNAL OF CLUSTER SCIENCE0.9530.916 JOURNAL OF CHROMATOGRAPHICSCIENCE0.9420.884CRYSTAL RESEARCH ANDTECHNOLOGY0.930.946POLYCYCLIC AROMATIC COMPOUNDS 0.926 1.023NATURAL PRODUCT RESEARCH0.908 1.009 BULLETIN OF THE KOREANCHEMICAL SOCIETY0.9040.906 JOURNAL OF PHOTOPOLYMERSCIENCE AND TECHNOLOGY0.9040.904JPC-JOURNAL OF PLANAR CHROMATOGRAPHY-MODERN TLC0.8920.767JOURNAL OF LIQUIDCHROMATOGRAPHY & RELATEDTECHNOLOGIES0.8860.706DESIGNED MONOMERS ANDPOLYMERS 0.885 1.444CHEMICAL PAPERS-CHEMICKEZVESTI 0.88 1.096Journal of Sulfur Chemistry0.88 1.009 ZEITSCHRIFT FURNATURFORSCHUNG SECTION B-AJOURNAL OF CHEMICAL SCIENCES0.8780.864JOURNAL OF RARE EARTHS0.8530.901 CHINESE JOURNAL OF ANALYTICALCHEMISTRY0.8430.941 JOURNAL OF MACROMOLECULARSCIENCE-PURE AND APPLIEDCHEMISTRY0.8320.887 MENDELEEV COMMUNICATIONS0.8280.901 TURKISH JOURNAL OF CHEMISTRY0.8190.943 INDIAN JOURNAL OF CHEMISTRYSECTION A-INORGANIC BIO-INORGANIC PHYSICAL THEO0.8090.891JOURNAL OF ADVANCEDOXIDATION TECHNOLOGIES0.8090.806JOURNAL OF THE SERBIANCHEMICAL SOCIETY0.8080.879 CHINESE JOURNAL OF CHEMISTRY0.8060.755 CHINESE CHEMICAL LETTERS0.7990.978 LETTERS IN ORGANIC CHEMISTRY0.7940.822 QUIMICA NOVA0.7890.763 JOURNAL OF THEORETICAL &COMPUTATIONAL CHEMISTRY0.7730.561CHEMISTRY OF NATURAL COMPOUNDS 0.765 1.029CROATICA CHEMICA ACTA0.760.763 Chemistry Education Research andPractice0.7530.855 ACTA PHYSICO-CHIMICA SINICA0.7440.78 RESEARCH ON CHEMICALINTERMEDIATES0.7420.697ACTA CHROMATOGRAPHICA0.7380.76 POLYMER SCIENCE SERIES A0.7280.838 JOURNAL OF THE CHINESECHEMICAL SOCIETY0.6830.678ACTA CRYSTALLOGRAPHICASECTION C-CRYSTAL STRUCTURECOMMUNICATIONS0.6820.518KINETICS AND CATALYSIS0.6790.638 PROGRESS IN REACTION KINETICSAND MECHANISM0.6770.761 Reaction Kinetics Mechanisms andCatalysis0.6670.876JOURNAL OF ANALYTICALCHEMISTRY 0.6670.747Chemistry of Heterocyclic Compounds 0.6460.725 CHINESE JOURNAL OF POLYMERSCIENCE0.6410.919COLLOID JOURNAL0.6390.707 CHINESE JOURNAL OF INORGANICCHEMISTRY0.6350.628JOURNAL OF CHEMICALEDUCATION 0.6320.739ACTA CHIMICA SINICA0.6320.533 CHEMICAL JOURNAL OF CHINESEUNIVERSITIES-CHINESE0.6320.619CHEMICKE LISTY0.6220.529 JOURNAL OF DISPERSION SCIENCEAND TECHNOLOGY0.6220.56JOURNAL OF CHEMICALCRYSTALLOGRAPHY0.6180.566PHOSPHORUS SULFUR AND SILICON AND THE RELATEDELEMENTS 0.6170.716E-Journal of Chemistry0.6160.516REVISTA DE CHIMIE0.6150.599 MAIN GROUP CHEMISTRY 0.610.636HYLE 0.6070.5 HIGH ENERGY CHEMISTRY0.6040.815 RUSSIAN JOURNAL OF ORGANICCHEMISTRY0.6030.648 JOURNAL OF CHEMICAL RESEARCH0.5920.633 PROGRESS IN CHEMISTRY0.5880.556 JOURNAL OF THE CHEMICALSOCIETY OF PAKISTAN0.587 1.377SOUTH AFRICAN JOURNAL OFCHEMISTRY-SUID-AFRIKAANSETYDSKRIF VIR CHEMIE0.5870.764RUSSIAN JOURNAL OFCOORDINATION CHEMISTRY0.5810.547 Macedonian Journal of Chemistry andChemical engineering0.579 1.079PHYSICS AND CHEMISTRY OFLIQUIDS 0.5790.603REVIEWS IN ANALYTICALCHEMISTRY 0.5790.357E-POLYMERS0.5780.515 JOURNAL OF SYNTHETIC ORGANICCHEMISTRY JAPAN0.5690.546ACTA POLYMERICA SINICA0.5620.769 CRYSTALLOGRAPHY REPORTS0.5570.469 CHEMIA ANALITYCZNA0.550.52 INDIAN JOURNAL OF CHEMISTRYSECTION B-ORGANIC CHEMISTRYINCLUDING MEDICINAL0.5490.648 Theoretical and Experimental Chemistry0.5430.509 JOURNAL OF THE CHILEANCHEMICAL SOCIETY0.5420.448CHINESE JOURNAL OFSTRUCTURAL CHEMISTRY0.5360.44JOURNAL OF STRUCTURALCHEMISTRY 0.5290.586MOLECULAR CRYSTALS AND LIQUIDCRYSTALS0.5250.58STUDIES IN CONSERVATION0.5090.4Chemija 0.5060.586 CHEMIE IN UNSERER ZEIT0.4940.517 CHIMICA OGGI-CHEMISTRY TODAY0.4940.593Journal of the Mexican ChemicalSociety 0.4850.413RUSSIAN CHEMICAL BULLETIN0.4750.379 RUSSIAN JOURNAL OF PHYSICALChemistry A0.4670.459 CHEMICAL RESEARCH IN CHINESEUNIVERSITIES0.4650.379 POLISH JOURNAL OF CHEMISTRY0.4520.393 Polimeros-Ciencia e Tecnologia 0.430.522 POLYMER SCIENCE SERIES B0.4230.558 LC GC EUROPE0.4190.494 RUSSIAN JOURNAL OF GENERALCHEMISTRY0.4180.467 RUSSIAN JOURNAL OF INORGANICCHEMISTRY0.4170.415POLYMER-KOREA0.4110.433BUNSEKI KAGAKU0.4060.43JOURNAL OF AUTOMATED METHODS & MANAGEMENT INCHEMISTRY 0.4050.467ACTA CRYSTALLOGRAPHICASECTION E-STRUCTURE REPORTSONLINE0.390.347Revista Mexicana de IngenieriaQuimica 0.3820.578MAIN GROUP METAL CHEMISTRY0.3810.207 JOURNAL OF THE INDIAN CHEMICALSOCIETY0.3470.359 Research Journal of Chemistry andEnvironment0.3310.379 DOKLADY PHYSICAL CHEMISTRY0.3310.458 REVUE ROUMAINE DE CHIMIE0.3310.418 BULLETIN OF THE CHEMICALSOCIETY OF ETHIOPIA0.3220.299 HETEROCYCLIC COMMUNICATIONS0.2930.265 LC GC NORTH AMERICA0.2870.371INDIAN JOURNAL OFHETEROCYCLIC CHEMISTRY0.2810.205 RUSSIAN JOURNAL OF APPLIEDCHEMISTRY0.2740.283DOKLADY CHEMISTRY0.2590.315 ASIAN JOURNAL OF CHEMISTRY0.2420.266ZEITSCHRIFT FUR KRISTALLOGRAPHIE-NEW CRYSTALSTRUCTURES 0.2380.278REVIEWS IN INORGANICCHEMISTRY 0.2310.222Russian Journal of Physical ChemistryB 0.2190.263Journal of Water Chemistry andTechnology 0.2120.205Science and Technology of EnergeticMaterials0.2080.296 OXIDATION COMMUNICATIONS0.2040.123 Bulgarian Chemical Communications0.2030.283 Journal of Rubber Research 0.1990.297 AFINIDAD0.1870.138 CHEMISTRY WORLD0.1850.159 KOBUNSHI RONBUNSHU0.1670.129Studia Universitatis Babes-BolyaiChemia 0.1490.129CHEMISTRY & INDUSTRY0.1360.12 ACTUALITE CHIMIQUE0.1340.115 ACS Catalysis00 Catalysis Science & Technology00 RSC Advances00。

Conjugation vs hyperconjugation in molecular structure of acroleinSvitlana V.Shishkina a ,⇑,Anzhelika I.Slabko b ,Oleg V.Shishkin a ,caDivision of Functional Materials Chemistry,SSI ‘Institute for Single Crystals’,National Academy of Science of Ukraine,60Lenina Ave.,Kharkiv 61001,Ukraine bDepartment of Technology of Plastic Masses,National Technical University ‘Kharkiv Polythechnic Institute’,21Frunze Str.,Kharkiv 61002,Ukraine cDepartment of Inorganic Chemistry,V.N.Karazin Kharkiv National University,4Svobody Sq.,Kharkiv 61077,Ukrainea r t i c l e i n f o Article history:Received 4August 2012In final form 16November 2012Available online 29November 2012a b s t r a c tAnalysis of geometric parameters of butadiene and acrolein reveals the contradiction between the Csp 2–Csp 2bond length in acrolein and classical concept of conjugation degree in the polarized molecules.In this Letter the reasons of this contradiction have been investigated.It is concluded that the Csp 2–Csp 2bond length in acrolein is determined by influence of the bonding for it p –p conjugation and antibonding n ?r ⁄hyperconjugation between the oxygen lone pair and the antibonding orbital of the single bond.It was shown also this bond length depends on the difference in energy of conjugative and hyperconjuga-tive interactions.Ó2012Elsevier B.V.All rights reserved.1.IntroductionButadiene and acrolein belong to the most fundamental mole-cules in the organic chemistry.They are canonical objects for the investigation of phenomena of p –p conjugation between double bonds and polarization of p -system by heteroatom [1].According to many experimental [2–16]as well as theoretical studies [13,17–23]the molecular structure of butadiene is determined by conjugation between p -orbitals of two double bonds and may be described as superposition of two resonance structures (Scheme 1).The presence of zwitterionic structure causes the shortening of the central single Csp 2–Csp 2bond as compare with similar unconjugated bond [24].Acrolein differs from butadiene by presence of the oxygen atom instead terminal methylene group.According to classical concepts of organic chemistry such replacement should causes polarization of p -system due to presence of highly polar C @O bond [25].This leads to significant increase of the contribution of the zwitterionic resonance structure (Scheme 1)reflecting strengthening of conju-gation between p -systems of double bonds.Therefore the central Csp 2–Csp 2bond must be shorter in acrolein as compared with one in butadiene.However numerous investigations of acrolein by experimental [26–28]and theoretical methods [26,27,29–36]demonstrate an opposite situation:the Csp 2–Csp 2bond length varies within the range 1.469Ä1.481Åin acrolein as compared with 1.454Ä1.467in butadiene.Based on these data one can conclude that conjugation between double bonds in acrolein is weaker than in butadiene.At that time the rotation barrier obtained from quan-tum-chemical calculations is higher in acrolein [26,27],confirming stronger conjugation between double bonds.Thus,results of experimental and theoretical investigations demonstrate the con-tradiction between the strengthening of the conjugation in acrolein as compare with butadiene and the values of the Csp 2–Csp 2bond length in these molecules.Recently such illogical situation was observed also in derivatives of cyclohexene containing conjugated endocyclic and exocyclic double bonds [37].It was assumed that elongation of the Csp 2–Csp 2bond in cycloxen-2-enone as compare with one in 3-methylene-cyclohexene is caused by the influence of n ?r ⁄hyperconjugation.In this Letter we demonstrate the results of the investigation of intramolecular interactions in butadiene and acrolein which ex-plain the experimentally observed contradiction between the length of the Csp 2–Csp 2bond and degree of conjugation in acrolein.2.Method of calculationsThe structures of all investigated molecules were optimized using second-order Møller-Plesset perturbation theory [38].The standard aug-cc-pvtz basis set [39]was applied.The character of stationary points on the potential energy surface was verified by calculations of vibrational frequencies within the harmonic approximation using analytical second derivatives at the same level of theory.All stationary points possess zero (minima)or one (saddle points)imaginary frequencies.The verification of the calculation method was performed using optimization of butadi-ene and acrolein by MP2/aug-cc-pvqz,CCSD(T)/cc-pvtz and CCSD(T)/6-311G(d,p)methods [40].The geometry of saddle points for the rotation process was lo-cated using standard optimization technique [41].The barrier of the rotation in all molecules was calculated as the difference be-tween the Gibbs free energies at 298K of the most stable s-trans0009-2614/$-see front matter Ó2012Elsevier B.V.All rights reserved./10.1016/j.cplett.2012.11.032Corresponding author.Fax:+3805723409339.E-mail address:sveta@ (S.V.Shishkina).conformer and saddle-point conformation.All calculations were performed using the G AUSSIAN 03program [42].The intramolecular interactions were investigated within the Natural Bonding Orbitals theory [43]with N BO 5.0program [44].Calculations were performed using B3LYP/aug-cc-pvtz wave func-tion obtained from single point calculations by G AUSSIAN 03program.The conjugation and hyperconjugation interactions are referred to as ‘delocalization’corrections to the zeroth-order natural Lewis structure.For each donor N BO (i )and acceptor N BO (j ),the stabiliza-tion energy E (2)associated with delocalization (‘2e-stabilization’)i ?j is estimated asE ð2Þ¼D E ij ¼q iF ði ;j Þ22j À2i;where q i is the donor orbital occupancy,e j and e i are the diagonal elements (orbital energies),and F (i,j )is the off-diagonal N BO Fock matrix element.3.Results and discussionThe equilibrium geometry of s-trans and s-cis conformers of butadiene and acrolein calculated by MP/aug-cc-pvtz method (Ta-ble 2)agrees very well with obtained earlier results [2–23,25–34]and data of higher and more computationally expensive methods (Table 1).It can be noted that the C–C bond length in acrolein(Tables 1and 2)is longer as compared with butadiene in all sta-tionary points on the potential energy surface.Such relation does not agree with the conception of the resonance theory [45–47].Analysis of intramolecular interactions in both molecules using N BO theory indicates that acrolein differs from butadiene by pres-ence of intramolecular interaction between lone pair of the oxygen atom and antibonding orbital of the C–C bond (Figure 1)as well as the polarization of one double bond containing more electronega-tive ually the interactions between lone pair and antibonding orbital of single bond are stronger than the interac-tions between the C–H bond and antibonding orbital [48]and they can influence geometrical characteristics.Such type of interactions is named by anomeric effect and it is studied very well for the case when the central bond between interacted orbitals is single [48,49].It is investigated in details [49]an influence of classical anomeric effect on conformation of the substituents about central single bond as well as on values of bond lengths.In the case of hyperconjugation interactions along double bond in acrolein orien-tation of substituents around it is determined by its double charac-ter.Therefore,n ?r ⁄hyperconjugative interaction can influence on the bond lengths of interacted ones only.It can assume the dou-ble character of the central bond must also promote some strengthening of this influence due to shorter distance between the lone pair of the oxygen atom and antibonding orbital of the C–C bond.Results of N BO analysis of intramolecular interactions in butadi-ene and acrolein demonstrate that the energy of n ?r ⁄interaction between lone pair of the oxygen atom and antibonding orbital of the C–C bond in acrolein is twice as high of the energy of r ?r ⁄interaction between the C–H bond and antibonding orbital of the C–C bond in butadiene (Table 2).At that time the energy of n ?r ⁄interaction is close enough to the energy of p –pTable 2The equilibrium geometries (bond lengths,Åand C @C–C @X (X =CH 2,O)torsion angle,deg.),transition state of the rotation process,bond length alternation (BLA)parameter,related energy (D E rel ,kcal/mol),related stability (D G 298,kcal/mol)and energy of strongest intramolecular interactions (E (2),kcal/mol)for butadiene and acrolein optimized by MP2/aug-cc-pvtz method.The wave function calculated by b3lyp/aug-cc-pvtz method was used for N BO analysis.ConformerBond lengths (Å)C @C–C @X torsion angle deg.BLA (Å)D E rel(kcal/mol)D G 298(kcal/mol)E (2)(kcal/mol)C @CC–C C @X p –pn ?r ⁄(C–C)r ?r ⁄(C–C)Butadiene s-trans 1.341 1.453 1.340180.0+0.1120030.74–8.67gauche 1.340 1.465 1.34036.8+0.125 2.83 2.8921.04–8.94TS 1.3361.4801.336101.8+0.1446.446.102.32–10.66Acrolein s-trans 1.339 1.469 1.219180.0+0.1300027.7518.06–s-cis 1.338 1.481 1.2190.0+0.143 2.26 2.2224.6319.05–TS1.334 1.492 1.21692.5+0.1588.017.46–18.78–Acrolein +BH 3s-trans 1.341 1.449 1.235180.0+0.1080033.12 2.4511.43s-cis 1.340 1.460 1.2340.0+0.120 2.45 2.3829.61 2.6011.43TS1.334 1.476 1.23193.7+0.1429.258.61–2.3911.55Table 1The Csp 2–Csp 2bond length in butadiene and acrolein optimized by different quantum-chemical methods.Method of calculationCsp 2–Csp 2bond length (Å)D (Csp 2–Csp 2)(Å)ButadieneAcrolein MP2/aug-cc-pvtz 1.453 1.4690.016MP2/aug-cc-pvqz 1.451 1.4670.016CCSD(T)/cc-pvtz1.461 1.4780.017CCSD(T)/6–311G(d,p)1.4681.4870.019S.V.Shishkina et al./Chemical Physics Letters 556(2013)18–2219conjugation between two double bonds.Therefore it can assume that the influence of p–p conjugation and n?r⁄hyperconjuga-tion on the C–C bond length should be comparable.However two strongest intramolecular interactions in the acrolein differ from each other:p–p conjugation between double bonds causes the shortening of the C–C bond in contrary to n?r⁄hyperconjugation which leads to the elongation of the C–C bond owing to the popu-lation of its antibonding orbital.Taking into account this situation it is possible to conclude that length of the Csp2–Csp2single bond in acrolein is determined by balance of two opposite factors namely p–p conjugation and n?r⁄hyperconjugation which may be considered as bonding and antibonding interactions for this bond(Figure1).In this case the length of the Csp2–Csp2bond in acrolein depends on the con-tribution of each of these factors.The changing of the delocaliza-tion of the structures due to influence of intramolecular interactions can be analyzed easier by mean of the bond length alternation(BLA)parameter(Table2).The analysis of BLA shows the presence of n?r⁄hyperconjugative interaction in acrolein what results in the increasing of alternation of double bonds as compare with butadiene.Clear estimation of influence of both interactions on geometri-cal parameters of molecule may be performed by comparison of properties of single C–C bond and BLA parameter in equilibrium s-trans conformation and in situations where one or both intramo-lecular interactions are absent.It is well known that p–p conjugation between double bonds decreases appreciably up to disappearing(in acrolein)in the tran-sition state for the rotation around single bond process(Figure2). The data of N BO analysis for butadiene and acrolein in the transition state confirm this evidence(Table2).As expected the absence of p–p conjugation results in the elongation of the Csp2–Csp2bond and increasing of BLA as compare with equilibrium geometry.At that time single C–C bond remains longer in the transition state for acrolein as compare with one for butadiene’s transition state.1.4921.4691.4491.476π−π is present n σ* is presentπ−π is absentn σ* is absent without π−πwithout n σ∗Figure2.Influence of p–p⁄conjugation and n?r⁄hyperconjugation on the C–Cbond length in acrolein.Table3The energy(E(2),kcal/mol)of the conjugative(bonding)and hyperconjugative(antibonding)intramolecular interactions influencing the Csp2–Csp2bond length in butadiene, acrolein and its complex with BH3.Molecule Bonding interactions E(2)(kcal/mol)Antibonding interactions E(2)(kcal/mol)Butadienes-trans BD(2)C1-C2–BD(2)C3-C430.74BD(1)C1-H5–BD(1)C2-C38.67 BD(1)C2-H7–BD(1)C3-H87.68BD(1)C4-H9–BD(1)C2-C38.67 gauche BD(2)C1-C2–BD(2)C3-C421.04BD(1)C1-H5–BD(1)C2-C38.94 BD(1)C2-H7–BD(1)C3-C4 5.36BD(1)C4-H9–BD(1)C2-C39.01BD(1)C3-H8–BD(1)C1-C2 5.36TS BD(1)C1-C2–BD(1)C3-C4 3.5BD(1)C1-H5–BD(1)C2-C310.66 BD(1)C1-C2–BD(2)C3-C4 3.46BD(1)C4-H9–BD(1)C2-C310.66BD(1)C3-C4–BD(2)C1-C2 3.46BD(2)C1-C2–BD(2)C3-C4 2.32BD(1)C3-H8–BD(2)C1-C29.57BD(1)C2-H7–BD(2)C3-C49.57Acroleins-trans BD(1)C1-C2–BD(1)C3-O4 2.73BD(1)C1-H5–BD(1)C2-C38.25 BD(2)C1-C2–BD(2)C3-O427.75LP(2)O4–BD(1)C2-C318.06BD(1)C2-H7–BD(1)C3-H8 5.95s-cis BD(2)C1-C2–BD(2)C3-O424.63BD(1)C1-H5–BD(1)C2-C38.76 BD(1)C2-H7–BD(1)C3-O4 4.05LP(2)O4–BD(1)C2-C319.05BD(1)C3-H8–BD(1)C1-C2 5.01TS BD(1)C1-C2–BD(2)C3-O4 2.98BD(1)C1-H5–BD(1)C2-C39.07 BD(1)C3-O4–BD(2)C1-C2 4.48LP(2)O4–BD(1)C2-C318.78BD(1)C3-H8–BD(2)C1-C2 5.85BD(1)C2-H7–BD(2)C3-O4 6.16Acrolein+BH3s-trans BD(1)C1-C2–BD(1)C3-O4 3.07BD(1)C1-H6–BD(1)C2-C38.09 BD(2)C1-C2–BD(2)C3-O433.12BD(1)C2-C3–BD(1)O4-B511.43BD(1)C2-H8–BD(1)C3-H9 5.97LP(1)O4–BD(1)C2-C3 2.45 s-cis BD(1)C1-C2–BD(1)C3-H9 4.98BD(1)C1-H6–BD(1)C2-C38.51 BD(2)C1-C2–BD(2)C3-O429.61BD(1)C2-C3–BD(1)O4-B511.43BD(1)C2-H8–BD(1)C3-O4 4.41LP(1)O4–BD(1)C2-C3 2.60 TS BD(1)C1-C2–BD(1)C3-O40.55BD(1)C1-H6–BD(1)C2-C39.05 BD(1)C1-C2–BD(2)C3-O4 3.01BD(1)C2-C3–BD(1)O4-B511.55BD(2)C1-C2–BD(1)C3-O4 4.90LP(1)O4–BD(1)C2-C3 2.39BD(1)C3-H9–BD(2)C1-C2 6.13BD(1)C2-H8–BD(2)C3-O4 6.8820S.V.Shishkina et al./Chemical Physics Letters556(2013)18–22It is additional argument about the influence of n?r⁄hypercon-jugation on the C–C bond length through the C@O double bond.In contrary to p–p conjugation n?r⁄hyperconjugation is present in all stationary points on the potential energy surface for acrolein(Table2).But this interaction can be shielded by for-mation of dative bond involving lone pair of the oxygen atom and unoccupied orbital of Lewis acid,for example,BH3.The formed O–B bond has r-character and the energy of its interaction with antibonding orbital of the central C–C bond is very close to the en-ergy of similar C–H?r⁄(C–C)interaction in butadiene(Table2). The absence of n?r⁄hyperconjugation results significant short-ening of the Csp2–Csp2bond and decreasing of BLA in all stationary points for acrolein.It is more interesting that the C–C bond in acro-lein becomes shorter and p–p conjugation becomes stronger as compare with ones in butadiene in the case of absence of n?r⁄hyperconjugative interaction(Table2)what agrees well with the resonance theory.This evidence is confirmed also by values of BLA parameter.It is very interesting the situation when both strong intramolec-ular interactions are absent namely acrolein with shielded by BH3 lone pair in the transition state for the rotation process.In absence of p–p conjugative and n?r⁄hyperconjugative interactions the C–C bond length is almost equal to mean value for length of this bond for s-trans and s-cis conformers of acrolein with both interac-tions(Table2).This fact confirms that the C–C bond length in acro-lein in the equilibrium state is determined by balance of p–p conjugation and n?r⁄hyperconjugation.Taking into account the opposite influence of two types of intra-molecular interactions on the C–C bond one may assume that its length depends on the difference in energy of bonding and antibonding interactions for this bond.In such case all bonding for C–C bond and antibonding for it interactions(Table3)must be taken into account.Specially,this is important for transition states where p–p conjugative interaction is minimal and r(c-H)–p interaction appears instead it.This interaction has bond-ing for Csp2–Csp2bond character and it is weaker as compare with p–p interaction.Analysis of relation between C–C bond length and total energy of intramolecular interaction influencing on it demon-strates good correlation between them(Figure3)with correlation coefficient aboutÀ0.93.The barrier of the rotation around ordinary C–C bond is also sensitive to intramolecular interactions.The absence of n?r⁄hyperconjugation in acrolein results the increase of conjugation in molecule what leads to the increase of the rotation barrier (Table2).4.ConclusionsResults of quantum-chemical calculations demonstrate the structure of acrolein does not correspond to conventional views about influence of the polarization of p-system by the oxygen atom.According to classic viewpoint this effect should lead to in-crease of conjugation between double bonds and shortening of central single C–C bond as compared to butadiene.However,anal-ysis of intramolecular interactions shows that the geometry of acrolein is determined by counteraction of p–p conjugation and n?r⁄hyperconjugation.The energies of these interactions are very close but ones influence on the C–C bond lengths in opposite directions.Conjugation promotes the shortening of the central sin-gle bond due to the overlapping of the p-orbitals of two double bonds.In the contrary the n?r⁄hyperconjugation causes the elongation of the C–C bond due to the population of its antibonding orbital.The absence of conjugation in the transition state for the rotation about the C–C bond process results in the elongation of the single bond in conjugated system.In turn the shielding of n?r⁄hyperconjugation by the formation of dative bond between lone pair of oxygen atom and vacant orbital of Lewis acid causes the shortening of the C–C bond in acrolein.The C–C bond length correlates well with the difference between two strong intramolec-ular interactions.The absence of both interactions does not almost change the C–C bond length.Thus,these data clearly indicate that molecular structure of conjugated systems containing heteroatoms is determined by not only p–p conjugation but also by n?r⁄hyperconjugation.References[1]F.A Carey,R.J.Sundberg,Advanced Organic Chemistry.Part A:Structure andMechanisms,Springer,Virginia,2007.[2]Yu.N.Panchenko,Yu.A.Pentin,V.I.Tyulin,V.M.Tatevskii,Opt.Spectrosc.13(1962)488.[3]A.R.H.Cole,G.M.Mohay,G.A.Osborne,Spectrochim.Acta23A(1967)909.[4]K.Kuchitsu,T.Fukuyama,Y.Morino,J.Mol.Struct.1(1967–1968)463.[5]R.L.Lipnick,E.W.Garbisch Jr.,J.Am.Chem.Soc.95(1973)6370.[6]Yu.N.Panchenko,Spectrochim.Acta31A(1975)1201.[7]Yu.N.Panchenko,A.V.Abramenkov,V.I.Mochalov,A.A.Zenkin,G.Keresztury,G.J.Jalsovszky,J.Mol.Spectrosc.99(1983)288.[8]W.Caminati,G.Grassi,A.Bauder,Chem.Phys.Lett.148(1988)13.[9]M.E.Squillacote,T.C.Semple,P.W.Mui,J.Am.Chem.Soc.107(1985)6842.[10]Y.Furukawa,H.Takeuchi,I.Harada,M.Tasumi,Bull.Chem.Soc.Jpn.56(1983)392.[11]B.R.Arnold,V.Balaji,J.W.Downing,J.G.Radziszewski,J.J.Fisher,J.Michl,J.Am.Chem.Soc.113(1991)2910.[12]J.Saltiel,J.-O.Choi,D.F.Sears Jr.,D.W.Eaker,F.B.Mallory,C.W.Mallory,J.Phys.Chem.98(1994)13162.[13]K.W.Wiberg,R.E.Rosenberg,J.Am.Chem.Soc.112(1990)1509.[14]J.Saltiel,D.F.Sears Jr,A.M.Turek,J.Phys.Chem.A105(2001)7569.[15]M.S.Deleuze,S.Knippenberg,J.Chem.Phys.125(2006)104309-1.[16]P.Boopalachandran,N.C.Craig,ane,J.Phys.Chem.A116(2012)271.[17]H.Guo,M.Karplus,J.Chem.Phys.94(1991)3679.[18]R.Hargitai,P.G.Szalay,G.Pongor,G.Fogarasi,J.Mol.Struct.(THEOCHEM)112(1994)293.[19]G.R.De Maré,Yu.N.Panchenko,J.V.Auwera,J.Phys.Chem.A101(1997)3998.[20]J.C.Sancho-García,A.J.Pérez-Jiménez,F.Moscardó,J.Phys.Chem.A105(2001)11541.[21]N.C.Craig,P.Groner,D.C.McKean,J.Phys.Chem.A110(2006)7461.[22]D.Feller,K.A.Peterson,J.Chem.Phys.126(2007)114105.[23]D.Feller,N.C.Craig,A.R.Maltin,J.Phys.Chem.A112(2008)2131.[24]D.Feller,N.C.Craig,J.Phys.Chem.A113(2009)1601.[25]H.-B.Burgi,J.D.Dunitz,Structure Correlation,vol.2,VCH,Weinheim,1994.[26]K.B.Wiberg,R.E.Rosenberg,P.R.Rablen,J.Am.Chem.Soc.113(1991)2890.[27]K.B.Wiberg,P.R.Rablen,M.Marquez,J.Am.Chem.Soc.114(1992)8654.[28]K.Kuchitsu,T.Fukuyama,Y.Morino,J.Mol.Struct.1(1967–1968)463.[29]G.Celebre,M.Concistré,G.DeLuca,M.Longeri,G.Pileio,J.W.Emsley,Chem.Eur.J.11(2005)3599.[30]R.J.Loncharich,T.R.Schwartz,K.N.Houk,J.Am.Chem.Soc.109(1987)14.[31]G.R.DeMare,Yu.N.Panchenko,A.J.Abramenkov,J.Mol.Struct.160(1987)327.S.V.Shishkina et al./Chemical Physics Letters556(2013)18–2221[32]G.R.DeMare,Can.J.Chem.63(1985)1672.[33]Y.Osamura,H.F.Schaefer III,J.Chem.Phys.74(1981)4576.[34]C.E.Bolm,A.Bauder,Chem.Phys.Lett.88(1982)55.[35]B.Mannfors,J.T.Koskinen,L.-O.Pietilä,L.Ahjopalo,J.Mol.Struct.(THEOCHEM)393(1997)39.[36]J.I.García,J.A.Mayoral,L.Salvatella,X.Assfeld,M.F.Ruiz-López,J.Mol.Struct.(THEOCHEM)362(1996)187.[37]S.V.Shishkina,O.V.Shishkin,S.M.Desenko,J.Leszczynski,J.Phys.Chem.A112(2008)7080.[38]C.Møller,M.S.Plesset,Phys.Rev.46(1934)618.[39]R.A.Kendall,T.H.Dunning Jr.,R.J.Harrison,J.Chem.Phys.96(1992)6792.[40]W.H.Hehre,L.Radom,P.V.R.Schleyer,J.A.Pople,Ab initio Molecular OrbitalTheory,Wiley,New York,1986.[41]P.Culot,G.Dive,V.H.Nguyen,J.M.Ghuysen,Theor.Chim.Acta82(1992)189.[42]M.J.Frisch et al.,G AUSSIAN,Inc.,Wallingford CT,2004.[43]F.Weinhold,in:P.V.R.Schleyer,N.L.Allinger,T.Clark,J.Gasteiger,P.A.Kollman,H.F.Schaefer III,P.R.Schreiner(Eds.),Encyclopedia of Computational Chemistry,vol.3,John Wiley&Sons,Chicheste,UK,1998.1792–1792. [44]E.D.Glendening,J.K.Badenhoop,A.E.Reed,J.E.Carpenter,J.A.Bohmann,C.M.Morales,F.Weinhold,N BO5.0Theoretical Chemistry Institute,University of Wisconsin,Madison,WI,2001.[45]E.D.Glendening,F.Weinhold,put.Chem.19(1998)593.[46]E.D.Glendening,F.Weinhold,put.Chem.19(1998)610.[47]E.D.Glendening,J.K.Badenhoop,F.Weinhold,put.Chem.19(1998)628.[48]A.J.Kirby,Stereoelectronic Effects,Oxford University Press,New York,1996.[49]I.V.Alabugin,K.M.Gilmore,P.W.Peterson,WIREs Computational MolecularScience1(2011)109.22S.V.Shishkina et al./Chemical Physics Letters556(2013)18–22。

DOI: 10.1126/science.1257570, 763 (2014);346 Science et al.Bernhard Misof Phylogenomics resolves the timing and pattern of insect evolutionThis copy is for your personal, non-commercial use only.clicking here.colleagues, clients, or customers by , you can order high-quality copies for your If you wish to distribute this article to othershere.following the guidelines can be obtained by Permission to republish or repurpose articles or portions of articles): November 6, 2014 (this information is current as of The following resources related to this article are available online at/content/346/6210/763.full.html version of this article at:including high-resolution figures, can be found in the online Updated information and services, /content/suppl/2014/11/05/346.6210.763.DC1.html can be found at:Supporting Online Material /content/346/6210/763.full.html#ref-list-1, 58 of which can be accessed free:cites 161 articles This article/cgi/collection/evolution Evolutionsubject collections:This article appears in the following registered trademark of AAAS.is a Science 2014 by the American Association for the Advancement of Science; all rights reserved. The title Copyright American Association for the Advancement of Science, 1200 New York Avenue NW, Washington, DC 20005. (print ISSN 0036-8075; online ISSN 1095-9203) is published weekly, except the last week in December, by the Science o n N o v e m b e r 6, 2014w w w .s c i e n c e m a g .o r g D o w n l o a d e d f r o m9.T.A.White et al .,PLOS Pathog.6,e1001249(2010).10.G.Hu,J.Liu,K.A.Taylor,K.H.Roux,J.Virol.85,2741–2750(2011).11.P.D.Kwong et al .,Nature 393,648–659(1998).12.M.Pancera et al .,Proc.Natl.Acad.Sci.U.S.A.107,1166–1171(2010).13.E.E.Tran et al .,PLOS Pathog.8,e1002797(2012).14.Y.Mao et al .,Nat.Struct.Mol.Biol.19,893–899(2012).15.A.Harris et al .,Proc.Natl.Acad.Sci.U.S.A.108,11440–11445(2011).16.C.G.Moscoso et al .,Proc.Natl.Acad.Sci.U.S.A.108,6091–6096(2011).17.S.R.Wu et al .,Proc.Natl.Acad.Sci.U.S.A.107,18844–18849(2010).18.R.Roy,S.Hohng,T.Ha,Nat.Methods 5,507–516(2008).19.Z.Zhou et al .,ACS Chem.Biol.2,337–346(2007).20.C.W.Lin,A.Y.Ting,J.Am.Chem.Soc.128,4542–4543(2006).21.Materials and Methods are available as supplementarymaterials on Science Online.22.Single-letter abbreviations for the amino acid residues are asfollows:A,Ala;C,Cys;D,Asp;E,Glu;F,Phe;G,Gly;H,His;I,Ile;K,Lys;L,Leu;M,Met;N,Asn;P,Pro;Q,Gln;R,Arg;S,Ser;T,Thr;V,Val;W,Trp;and Y,Tyr.23.Q.Zheng et al .,Chem.Soc.Rev.43,1044–1056(2014).24.N.G.Mukherjee,L.A.Lyon,J.M.Le Doux,Nanotechnology 20,065103(2009).25.K.Henzler-Wildman,D.Kern,Nature 450,964–972(2007).26.P.D.Kwong et al .,Nature 420,678–682(2002).27.U.Olshevsky et al .,J.Virol.64,5701–5707(1990).28.F.Qin,Biophys.J.86,1488–1501(2004).29.H.Haim et al .,PLOS Pathog.5,e1000360(2009).30.N.Sullivan et al .,J.Virol.72,4694–4703(1998).31.J.Arthos et al .,J.Biol.Chem.277,11456–11464(2002).32.A.L.DeVico,Curr.HIV Res.5,561–571(2007).33.T.Zhou et al .,Science 329,811–817(2010).34.L.M.Walker et al .,Science 326,285–289(2009).35.L.M.Walker et al .,Nature 477,466–470(2011).36.M.Pancera et al .,Nat.Struct.Mol.Biol.20,804–813(2013).37.J.-P.Julien et al .,Proc.Natl.Acad.Sci.U.S.A.110,4351–4356(2013).38.J.-P.Julien et al .,PLOS Pathog.9,e1003342(2013).39.P.D.Kwong,J.R.Mascola,G.J.Nabel,Nat.Rev.Immunol.13,693–701(2013).40.Z.Li et al .,Antimicrob.Agents Chemother.57,4172–4180(2013).41.J.B.Munro,K.Y.Sanbonmatsu,C.M.Spahn,S.C.Blanchard,Trends Biochem.Sci.34,390–400(2009).42.Subsequent to the submission of this manuscript,the structureof a trimeric prefusion HIV-1Env (43)was determined in complex with broadly neutralizing antibodies PGT122(35)and 35O22(44).To determine the conformational state these antibodies captured in the crystal lattice,we measured smFRET on labeled JR-FL virions in the presence of either PGT122or 35O22or both.These data indicated that PGT122strongly stabilized the ground state.In contrast,35O22had little effect on Env conformation.HIV-1Env complexed with both antibodies exhibited slight ground state stabilization.43.M.Pancera et al .,Nature 514,455–461(2014).44.J.Huang et al .,Nature 10.1038/nature13601(2014).45.J.S.McLellan et al .,Nature 480,336–343(2011).ACKNOWLEDGMENTSWe thank J.Jin,L.Agosto,T.Wang,R.B.Altman,and M.R.Wasserman for assistance;C.Walsh,J.Binely,A.Trkola,and M.Krystal for reagents;and members of the Structural Biology Section,Vaccine Research Center,for critically reading the manuscript.We thank I.Wilson and J.Sodroski for encouraging us to extend our approach to a R5-tropic Env.The data presented in this paper are tabulated in the main paper and in the supplementary materials.This work wassupported by NIH grants R21AI100696to W.M.and S.C.B;P0156550to W.M.,S.C.B.,and A.B.S.;and R01GM098859to S.C.B.;by the Irvington Fellows Program of the Cancer Research Institute to J.B.M;by a fellowship from the China Scholarship Council-Yale World Scholars to X.M.;by grants from the International AIDS Vaccine Initiative ’s (IAVI)Neutralizing Antibody Consortium to D.R.B.,W.C.K.,and P.D.K.;and by funding from the NIH Intramural Research Program (Vaccine Research Center)to P.D.K.IAVI's work is made possible by generous support from many donors including:the Bill &Melinda Gates Foundation and the U.S.Agency for International Development (USAID).This study is made possible by the generous support of the American people through USAID.The contents are the responsibility of the authors and do not necessarily reflect the views of USAID or the ernment.Reagents from the NIH are subject to nonrestrictive material transferagreements.Patent applications pertaining to this work are the U.S.and World Application US2009/006049and WO/2010/053583,Synthesis of JRC-II-191(A.B.S.,J.R.C.),U.S.Patent Application 13/202,351,Methods and Compositions for Altering Photopysical Properties of Fluorophores via Proximal Quenching (S.C.B.,Z.Z.);U.S.Patent Application 14/373,402Dye Compositions,Methods of Preparation,Conjugates Thereof,and Methods of Use (S.C.B.,Z.Z.);and International and US Patent Application PCT/US13/42249Reagents and Methods for Identifying Anti-HIV CompoundsSUPPLEMENTARY MATERIALS/content/346/6210/759/suppl/DC1Materials and Methods Figs.S1to S15Tables S1to S3References (46–56)4April 2014;accepted 15September 2014Published online 8October 2014;colonize and exploit terrestrial and freshwa-ter ecosystems.They have shaped Earth ’s biota,exhibiting coevolved relationships with many groups,from flowering plants to hu-mans.They were the first to master flight and establish social societies.However,many as-pects of insect evolution are still poorly under-stood (2).The oldest known fossil insects are from the Early Devonian [~412million years ago (Ma)],which has led to the hypothesis that in-sects originated in the Late Silurian with the earliest terrestrial ecosystems (3).MolecularEarly Ordovician origin (4),which implies that early diversification of insects occurred in marine or coastal environments.Because of the absence of insect fossils from the Cambrian to the Silurian,these conclusions remain highly controversial.Fur-thermore,the phylogenetic relationships among major clades of polyneopteran insect orders —including grasshoppers and crickets (Orthoptera),cockroaches (Blattodea),and termites (Isoptera)—have remained elusive,as has the phylogenetic position of the enigmatic Zoraptera.Even the closest extant relatives of Holometabola (e.g.,SCIENCE 7NOVEMBER 2014•VOL 346ISSUE 6210763RESEARCH |REPORTSbeetles,moths and butterflies,flies,sawflies,wasps,ants,and bees)are unknown.Thus,in order to understand the origins of physiological and mor-phological innovations in insects (e.g.,wings andmetamorphosis),it is important to reliably re-construct the tempo and mode of insect diversifi-cation.We therefore conducted a phylogenomic study on 1478single-copy nuclear genes obtained from genomes and transcriptomes representing key taxa from all extant insect orders and other arthropods (144taxa)and estimated divergence dates with a validated set of 37fossils (5).Phylogenomic analyses of transcriptome and genome sequence data (6)can be compromised by sparsely populated data matrices,gene paral-ogy,sequence misalignment,and deviations from the underlying assumptions of applied evolution-ary models,which may result in biased statistical confidence in phylogenetic relationships and tem-poral inferences.We addressed these obstacles by removing confounding factors in our analysis (5)(fig.S2).We sequenced more than 2.5gigabases (Gb)of cDNA from each of 103insect species,which represented all extant insect orders (5).Addition-ally,we included published transcript sequence data that met our standards (table S2)and offi-cial gene sets of 14arthropods with sequenced draft genomes (5),of which 12served as refer-ences during orthology prediction of transcripts (tables S2and S4).Comparative analysis of the reference species'official gene sets identified 1478single-copy nuclear genes present in all these species (tables S3and S4).Functional annotation of these genes revealed that many serve basic cellular functions (tables S14and S15and figs.S4to S6).A graph-based approach using the best reciprocal genome-and transcriptome-wide hit criterion identified,on average,98%of these genes in the 103de novo sequenced transcriptomes,but only 79%and 62%in the previously published transcriptomes of in-and out-group taxa,respec-tively (tables S12and S13).After transcripts had been assigned and aligned to the 1478single-copy nuclear-encoded genes,we checked for highly divergent,putatively misaligned transcripts.Of the 196,027aligned transcripts,2033(1%)were classified as highly divergent.Of these,716were satisfactorily re-aligned with an automated refinement.How-ever,alignments of 1317transcripts could not be improved,and these transcripts were excluded from our analyses (supplementary data file S5,/10.5061/dryad.3c0f1).Nonrandom distribution of missing data among taxa can inflate statistical support for incorrect tree topologies (7).Because we detected a non-random distribution of missing data,we only considered data blocks if they contained infor-mation from at least one representative of each of the 39predefined taxonomic groups of un-disputed monophyly (table S6).In this represent-ative data set,the extent of missing data was still between 5and 97.7%in pairwise sequence com-parisons,with high percentages primarily because of the data scarcity in some previously published out-group taxa (table S19and figs.S7to S10).We inferred maximum-likelihood phylogenetic trees (Fig.1)with both nucleotide (second-codon positions only and applying a site-specific rate model)and amino acid –sequence data (applying a protein domain –based partitioning scheme to improve the biological realism of the applied evolutionary models)from the representative data set (5)(figs.S21,S22,and S23,A and D).Trees from both data sets were fully congruent.The absence of taxa that cannot be robustly placed on the tree (rogue taxa)in the amino acid –sequence data set and the presence of a few rogue taxa that did not bias tree inference in the nucleotide sequence data set (5)indicated a suf-ficiently representative taxonomic sampling.To detect confounding signal derived from nonrandom data coverage,we randomized ami-no acids within taxa,while preserving the dis-tribution of data coverage in the representative data set (5).This approach revealed no evidence of biased node support that could be attributed to nonrandom data coverage (5)(figs.S11and S12and table S20).Phylogenomic data may violate the assumption of time-reversible evolu-tionary processes,irrespective of what partition scheme one applies,which could lead to in-correct tree estimates and biased node support.Because sections in the amino acid –sequence alignments of the representative data set violat-ing these assumptions were present,we tested whether the observed compositional heteroge-neity across taxa biased node support but found no evidence for this (5)(fig.S20).We next dis-carded data strongly violating the assumption of time-reversible evolutionary processes (tables S21and S22,data files S6to S8,and figs.S13to S19).Results from phylogenetic analysis of this filtered data set (5)were fully congruent with those obtained from analyzing the unfiltered representative data set.The nucleotide sequence data of the representative data set containing also first and third codon positions strongly violated the assumption of time-reversible evolutionary processes,but still supported largely congruent topologies (fig.S23,B to D).In summary,our phylogenetic inferences are unlikely to be biased by any of the above-mentioned confounding factors.Our phylogenomic study suggests an Early Ordovician origin of insects (Hexapoda)at ~479Ma [confidence interval (CI),509to 452Ma]and a radiation of ectognathous insects in the Early Silurian ~441Ma (CI 465to 421Ma)(Figs.1and 2).These estimates imply that insects colonized land at roughly the same time as plants (8),in agreement with divergence date estimates on the basis of other molecular data (4).The early diversification pattern of insects has remained unclear (2,7,9).We received support for a monophyly of insects,including Collembola and Protura as closest relatives (10),and Diplura as closest extant relatives of bristletails (Archae-ognatha),silverfish (Zygentoma),and winged in-sects (Pterygota)(Fig.1).Furthermore,our analyses corroborate Remipedia,cave-dwelling crustaceans,as the closest extant relatives of insects (11,12).A close phylogenetic relationship of bristletails to a clade uniting silverfish and winged insects (Dicondylia)is generally accepted.However,the monophyly of silverfish has been questioned,with the relict Tricholepidion gertschi considered more distantly related to winged insects than7647NOVEMBER 2014•VOL 346ISSUE 6210 SCIENCE1Zoologisches Forschungsmuseum Alexander Koenig (ZFMK)/Zentrum für Molekulare Biodiversitätsforschung (ZMB),Bonn,Germany.2China National GeneBank,BGI-Shenzhen,China.3BGI-Shenzhen,China.4Australian National Insect Collection,Commonwealth Scientific and Industrial Research Organization (Australia)(CSIRO),National Research Collections Australia,Canberra,ACT,Australia.5Abteilung Arthropoda,Zoologisches Forschungsmuseum Alexander Koenig (ZFMK),Bonn,Germany.6Department of Entomology,Rutgers University,New Brunswick,NJ 08854,USA.7Department of Biological Sciences,Rutgers University,Newark,NJ 08854,USA.8Scientific Computing,Heidelberg Institute for Theoretical Studies (HITS),Heidelberg,Germany.9Institut für Spezielle Zoologie undEvolutionsbiologie mit Phyletischem Museum Jena,FSU Jena,Germany.10Steinmann-Institut,Bereich Paläontologie,Universität Bonn,Germany.112.Zoologische Abteilung(Insekten),Naturhistorisches Museum Wien,Vienna,Austria.12Department of Integrative Zoology,Universität Wien,Vienna,Austria.13Institut für Spezifische Prophylaxe und Tropenmedizin,Medizinische Parasitologie,Medizinische Universität Wien (MUW),Vienna,Austria.14Manaaki Whenua Landcare Research,Auckland,New Zealand.15Center for Advanced Modeling,Emergency Medicine Department,Johns Hopkins University,Baltimore,MD 21209,USA.16Biozentrum Grindel und Zoologisches Museum,Universität Hamburg,Hamburg,Germany.17Evolutionary Morphology Laboratory,Graduate School of Science and Engineering,EhimeUniversity,Japan.18Sugadaira Montane Research Center/Hexapod Comparative Embryology Laboratory,University of Tsukuba,Japan.19Land and Water Flagship,CSIRO,Canberra,ACT,Australia.20Florida Museum of Natural History,University of Florida,Gainesville,FL 32611,USA.21Entomology,Staatliches Museum für Naturkunde Stuttgart (SMNS),Germany.22Ecology Evolution and Genetics,Research School of Biology,Australian National University,Canberra,ACT,Australia.23National Evolutionary Synthesis Center,Durham,NC 27705,USA.24Department of Biological Sciences,Macquarie University,Sydney,Australia.25Department für Botanik und Biodiversitätsforschung,Universität Wien,Vienna,Austria.26Natural History Museum of Crete,University of Crete,Post Office Box 2208,Gr-71409,Iraklio,and Biology Department,University of Crete,Iraklio,Crete,Greece.27Department of Biological Sciences and Feinstone Center for Genomic Research,University of Memphis,Memphis,TN 38152,USA.28Centro Universitario de Ciencias Biólogicas y Agropecuarias,Centro de Estudios en Zoología,Universidad de Guadalajara,Zapopan,Jalisco,México.29Leibniz Supercomputing Centre of the Bavarian Academy of Sciences and Humanities,Garching,Germany.30Institute of Evolutionary Biology and Ecology,Zoology and Evolutionary Biology,University of Bonn,Bonn,Germany.31Department of Life Sciences,The Natural History Museum London,London,UK.32Abteilung Entomologie,Biozentrum Grindel und Zoologisches Museum,Universität Hamburg,Hamburg,Germany.33Fakultät für Informatik,Karlsruher Institut für Technologie,Karlsruhe,Germany.34California Academy of Sciences,San Francisco,CA 94118,USA.35Department of Entomology,College of Natural Resources and Environment,South ChinaAgricultural University,China.36Yokosuka City Museum,Yokosuka,Kanagawa,Japan.37Department of Entomology,North Carolina State University,Raleigh,NC 27695,USA.38Systematic Entomology,Hokkaido University,Sapporo,Japan.39Department of Biology,University of Copenhagen,Copenhagen,Denmark.40Princess Al Jawhara Center of Excellence in the Research of Hereditary Disorders,King Abdulaziz University,Jeddah,Saudi Arabia.41MacauUniversity of Science and Technology,Avenida Wai Long,Taipa,Macau,China.42Department of Medicine,University of Hong Kong,Hong Kong.43Department of Ecology,Evolution,and Natural Resources,Rutgers University,New Brunswick,NJ 08854,USA.*Major contributors.†Corresponding author.E-mail:xinzhou@ (X.Z.),b.misof.zfmk@uni-bonn.de (B.M.),kjer@ (K.M.K),wangj@ (J.W.)RESEARCH |REPORTSRESEARCH|REPORTSrelationships.maximum-likelihoodoptimized and sampledare labeled indicateorigin of events.SCIENCE 7NOVEMBER2014•VOL346ISSUE6210765estimates.correspondrange estimatesare Additionally,separatelybarsthe inferredcommon course,bewithRESEARCH|REPORTSother silverfish (13).We find that silverfish are monophyletic,consistent with recently published morphological studies (14),and estimate that Tricholepidion diverged from other silverfish in the Late Triassic (~214Ma)(Figs.1and 2).This result implies parallel and independent loss of the ligamentous head endoskeleton,abdominal styli,and coxal vesicles in winged insects and silverfish (5).The diversification of insects is undoubtedly related to the evolution of flight.Fossil winged insects exist from the Late Mississippian (~324Ma)(15),which implies a pre-Carboniferous origin of insect flight.The description of †Rhyniognatha (~412Ma)from a mandible,potentially indicative of a winged insect,suggested an Early Devonian to Late Silurian origin of winged insects (3).Our results corroborate an origin of winged insect lineages during this time period (16)(Figs.1and 2),which implies that the ability to fly emerged after the establishment of complex terrestrial ecosystems.Ephemeroptera and Odonata are,according to our analyses,derived from a common ancestor.However,node support is low for Palaeoptera (Ephemeroptera +Odonata)and for a sister group relationship of Palaeoptera to modern winged in-sects (Neoptera),which indicates that additional evidence,including extensive taxon sampling and the analysis of genomic meta-characters (17),will be necessary to corroborate these relationships.We find strong support for the monophyly of Polyneoptera,a group that comprises earwigs,stone-flies,grasshoppers,crickets,katydids (Orthoptera),Embioptera,Phasmatodea,Mantophasmatodea,Grylloblattodea,cockroaches,mantids,termites,and Zoraptera (18–20).We estimated the origin of the polyneopteran lineages at ~302Ma (CI 377to 231Ma)in the Pennsylvanian (Figs.1and 2),con-sistent with the idea that at least part of the rich Carboniferous neopteran insect fauna was of poly-neopteran origin.Finally,our analyses suggest that the major diversity within living cockroaches,man-tids,termites,and stick insects evolved after the Permian mass extinction.Given that the oldest known fossil hemipterans date to the Middle Pennsylvanian (~310Ma)(21),it had been thought that the stylet marks on liv-erworts from the Late Devonian (~380Ma)(22)could not have been of hemipteran origin.Our study indicates that true bugs (Hemiptera)and their sister lineage,thrips (Thysanoptera),all of which possess piercing-sucking mouthparts,orig-inated ~373Ma (CI 401to 346Ma),which gives support to the possibility of a hemipteroid origin of Early Paleozoic stylet marks.True bugs,thrips,bark lice (Psocoptera),and true lice (Phthiraptera)(together called Acercaria)were thought to be the closest extant relatives of Holometabola (Acercaria +Holometabola =Eumetabola)(10).However,convincing morpho-logical features and fossil intermediates support-ing a monophyly of Acercaria are lacking (13).We recovered bark and true lice (Psocodea)as likely closest extant relatives of Holometabola (5),which suggests that both groups started to diverge in the Devonian-Mississippian ~362Ma (CI 390to 334Ma)(Figs.1and 2).However,this result did not receive support in all statistical tests and,therefore,should be further investigated in future studies that embrace additional types of characters (17).We estimated that the radiation of parasitic lice occurred ~53Ma (CI 67to 46Ma),which implies that they diversified well after the emer-gence of their avian and mammalian hosts in the Late Cretaceous –Early Eocene and contradicts the hypothesis that parasitic lice originated on fea-thered theropod dinosaurs ~130Ma (23).Within Holometabola,our study recovered phylogenetic relationships fully congruent with those suggested in recent studies (2,24,25).Although we estimated the origin of stem lineages of many holometabolous insect orders in the Late Carboniferous,we dated the spectacular diver-sifications within Hymenoptera,Diptera,and Lepidoptera to the Early Cretaceous,contem-porary with the radiation of flowering plants (21,26).The almost linear increase in interordinal insect diversity suggests that the process of di-versification of extant insects may not have been severely affected by the Permian and Cretaceous biodiversity crises (Fig.2).With this study,we have provided a robust phylogenetic backbone tree and reliable time estimates of insect evolution.These data and analyses establish a framework for future com-parative analyses on insects,their genomes,and their morphology.REFERENCES AND NOTES1.The term “insects ”is used here in a broad sense andsynonymous to Hexapoda (including the ancestrally wingless Protura,Collembola,and Diplura).2.M.D.Trautwein,B.M.Wiegmann,R.Beutel,K.M.Kjer,D.K.Yeates,Annu.Rev.Entomol.57,449–468(2012).3.M.S.Engel,D.A.Grimaldi,Nature 427,627–630(2004).4.O.Rota-Stabelli et al.,Curr.Biol.23,392–398(2013).5.Materials and methods are available as supplementarymaterial on Science Online.6.Transcriptome refers to the sequencing of all of the mRNAs ofan individual or many individuals present at the time of preservation.7. E.Dell ’Ampio et al .,Mol.Biol.Evol.31,239–249(2014).8. bandeira,Arthro Syst.Phylo.64,53–94(2006).9.K.Meusemann et al .,Mol.Biol.Evol.27,2451–2464(2010).10.W.Hennig,Die Stammesgeschichte der Insekten(Kramer,Frankfurt am Main,1969).11.With the notable exception of Cephalocarida,for which RNA-sequencing data were unavailable to us.12.B.M.von Reumont et al .,Mol.Biol.Evol.29,1031–1045(2012).13.N.P.Kristensen,Annu.Rev.Entomol.26,135–157(1981).14.A.Blanke,M.Koch,B.Wipfler,F.Wilde,B.Misof,Front.Zool.11,16(2014).15.J.Prokop,A.Nel,I.Hoch,Geobios 38,383–387(2005).16.These estimates are robust whether or not †Rhyniognatha(Pragian stage,~412Ma)is used as a calibration point.17.O.Niehuis et al .,Curr.Biol.22,1309–1313(2012).18.H.Letsch,S.Simon,Syst.Entomol.38,783–793(2013).19.K.Ishiwata,G.Sasaki,J.Ogawa,T.Miyata,Z.-H.Su,Mol.Phylogenet.Evol.58,169–180(2011).20.K.Yoshizawa,Syst.Entomol.36,377–394(2011).21.A.Nel et al .,Nature 503,257–261(2013).bandeira,S.L.Tremblay,K.E.Bartowski,L.VanAllerHernick,New Phytol.202,247–258(2014).23.V.S.Smith et al .,Biol.Lett.7,782–785(2011).24.R.G.Beutel et al .,Cladistics 27,341–355(2011).25.B.M.Wiegmann et al .,BMC Biol.7,34(2009).26.J.A.Doyle,Annu.Rev.Earth Planet.Sci.40,301–326(2012).ACKNOWLEDGMENTSThe data reported in this paper are tabulated in the supplementary materials and archived at National Center for BiotechnologyInformation,NIH,under the Umbrella BioProject ID PRJNA183205(“The 1KITE project:evolution of insects ”).Supplementary files are archived at the Dryad Digital Repository /10.5061/dryad.3c0f1.Funding support:China National GeneBank and BGI-Shenzhen,China;German Research Foundation (NI 1387/1-1;MI 649/6,MI 649/10,RE 345/1-2,BE1789/8-1,BE 1789/10-1,STA 860/4,Heisenberg grant WA 1496/8-1);Austria Science Fund FWF;NSF (DEB 0816865);Ministry of Education,Culture,Sports,Science and Technology of Japan Grant-in-Aid forYoung Scientists (B 22770090);Japan Society for the Promotion of Science (P14071);Deutsches Elektronen-Synchrotron (I-20120065);Paul Scherrer Institute (20110069);SchlingerEndowment to CSIRO Ecosystem Sciences;Heidelberg Institute for Theoretical Studies;University of Memphis-FedEx Institute ofTechnology;and Rutgers University.The authors declare no conflicts of interest.SUPPLEMENTARY MATERIALS/content/346/6210/763/suppl/DC1Materials and Methods Supplementary Text Figs.S1to S27Tables S1to S26Data File Captions S1to S14References (27–187)17June 2014;accepted 23September 201410.1126/science.1257570SCIENCE 7NOVEMBER 2014•VOL 346ISSUE 6210767RESEARCH |REPORTS。

Remnant PbI2, an unforeseen necessity in high-efficiency hybrid perovskite-based solar cells?a)Duyen H. Cao, Constantinos C. Stoumpos, Christos D. Malliakas, Michael J. Katz, Omar K. Farha, Joseph T. Hupp, and Mercouri G. KanatzidisCitation: APL Materials 2, 091101 (2014); doi: 10.1063/1.4895038View online: /10.1063/1.4895038View Table of Contents: /content/aip/journal/aplmater/2/9?ver=pdfcovPublished by the AIP PublishingArticles you may be interested inParameters influencing the deposition of methylammonium lead halide iodide in hole conductor free perovskite-based solar cellsAPL Mat. 2, 081502 (2014); 10.1063/1.4885548Air stability of TiO2/PbS colloidal nanoparticle solar cells and its impact on power efficiencyAppl. Phys. Lett. 99, 063512 (2011); 10.1063/1.3617469High efficiency mesoporous titanium oxide PbS quantum dot solar cells at low temperatureAppl. Phys. Lett. 97, 043106 (2010); 10.1063/1.3459146Near-IR activity of hybrid solar cells: Enhancement of efficiency by dissociating excitons generated in PbS nanoparticlesAppl. Phys. Lett. 96, 073505 (2010); 10.1063/1.3292183Effects of molecular interface modification in hybrid organic-inorganic photovoltaic cellsJ. Appl. Phys. 101, 114503 (2007); 10.1063/1.2737977APL MATERIALS2,091101(2014)Remnant PbI2,an unforeseen necessity in high-efficiency hybrid perovskite-based solar cells?aDuyen H.Cao,1Constantinos C.Stoumpos,1Christos D.Malliakas,1Michael J.Katz,1Omar K.Farha,1,2Joseph T.Hupp,1,band Mercouri G.Kanatzidis1,b1Department of Chemistry,and Argonne-Northwestern Solar Energy Research(ANSER)Center,Northwestern University,2145Sheridan Road,Evanston,Illinois60208,USA2Department of Chemistry,Faculty of Science,King Abdulaziz University,Jeddah,Saudi Arabia(Received2May2014;accepted26August2014;published online18September2014)Perovskite-containing solar cells were fabricated in a two-step procedure in whichPbI2is deposited via spin-coating and subsequently converted to the CH3NH3PbI3perovskite by dipping in a solution of CH3NH3I.By varying the dipping time from5s to2h,we observe that the device performance shows an unexpectedly remark-able trend.At dipping times below15min the current density and voltage of thedevice are enhanced from10.1mA/cm2and933mV(5s)to15.1mA/cm2and1036mV(15min).However,upon further conversion,the current density decreases to9.7mA/cm2and846mV after2h.Based on X-ray diffraction data,we determinedthat remnant PbI2is always present in these devices.Work function and dark currentmeasurements showed that the remnant PbI2has a beneficial effect and acts as ablocking layer between the TiO2semiconductor and the perovskite itself reducingthe probability of back electron transfer(charge recombination).Furthermore,wefind that increased dipping time leads to an increase in the size of perovskite crys-tals at the perovskite-hole-transporting material interface.Overall,approximately15min dipping time(∼2%unconverted PbI2)is necessary for achieving optimaldevice efficiency.©2014Author(s).All article content,except where otherwisenoted,is licensed under a Creative Commons Attribution3.0Unported License.[/10.1063/1.4895038]With the global growth in energy demand and with compelling climate-related environmental concerns,alternatives to the use of non-renewable and noxious fossil fuels are needed.1One such alternative energy resource,and arguably the only legitimate long-term solution,is solar energy. Photovoltaic devices which are capable of converting the photonflux to electricity are one such device.2Over the last2years,halide hybrid perovskite-based solar cells with high efficiency have engendered enormous interest in the photovoltaic community.3,4Among the perovskite choices, methylammonium lead iodide(MAPbI3)has become the archetypal light absorber.Recently,how-ever,Sn-based perovskites have been successfully implemented in functional solar cells.5,6MAPbI3 is an attractive light absorber due to its extraordinary absorption coefficient of1.5×104cm−1 at550nm;7it would take roughly1μm of material to absorb99%of theflux at550nm.Further-more,with a band gap of1.55eV(800nm),assuming an external quantum efficiency of90%,a maximum current density of ca.23mA/cm2is attainable with MAPbI3.Recent reports have commented on the variability in device performance as a function of perovskite layer fabrication.8In our laboratory,we too have observed that seemingly identicalfilmsa Invited for the Perovskite Solar Cells special topic.b Authors to whom correspondence should be addressed.Electronic addresses:j-hupp@ and m-kanatzidis@2,091101-12166-532X/2014/2(9)/091101/7©Author(s)2014FIG.1.X-ray diffraction patterns of CH3NH3PbI3films with increasing dipping time(%composition of PbI2was determined by Rietveld analysis(see Sec.S3of the supplementary material for the Rietveld analysis details).have markedly different device performance.For example,when ourfilms of PbI2are exposed to MAI for several seconds(ca.60s),then a light brown coloredfilm is obtained rather than the black color commonly observed for bulk MAPbI3(see Sec.S2of the supplementary material for the optical band gap of bulk MAPbI3).23This brown color suggests only partial conversion to MAPbI3and yields solar cells exhibiting a J sc of13.4mA/cm2and a V oc of960mV;these values are significantly below the21.3mA/cm2and1000mV obtained by others.4Under the hypothesis that fully converted films will achieve optimal light harvesting efficiency,we increased the conversion time from seconds to2h.Unexpectedly,the2-h dipping device did not show an improved photovoltaic response(J sc =9.7mA/cm2,V oc=846mV)even though conversion to MAPbI3appeared to be complete.With the only obvious difference between these two devices being the dipping time,we hypothesized that the degree of conversion of PbI2to the MAPbI3perovskite is an important parameter in obtaining optimal device performance.We thus set out to understand the correlation between the method of fabrication of the MAPbI3layer,the precise chemical compositions,and both the physical and photo-physical properties of thefilm.We report here that remnant PbI2is crucial in forming a barrier layer to electron interception/recombination leading to optimized J sc and V oc in these hybrid perovskite-based solar cells.We constructed perovskite-containing devices using a two-step deposition method according to a reported procedure with some modifications.4(see Sec.S1of the supplementary material for the experimental details).23MAPbI3-containing photo-anodes were made by varying the dipping time of the PbI2-coated photo-anode in MAI solution.In order to minimize the effects from unforeseen variables,care was taken to ensure that allfilms were prepared in an identical manner.The composi-tions offinal MAPbI3-containingfilms were monitored by X-ray diffraction(XRD).Independently of the dipping times,only theβ-phase of the MAPbI3is formed(Figure1).9However,in addition to theβ-phase,allfilms also showed the presence of unconverted PbI2(Figure1,marked with*) which can be most easily observed via the(001)and(003)reflections at2θ=12.56◦and38.54◦respectively.As the dipping time is increased,the intensities of PbI2reflections decrease with a concomitant increase in the MAPbI3intensities.In addition to the decrease in peak intensities of PbI2,the peak width increases as the dipping time increases indicating that the size of the PbI2 crystallites is decreasing,as expected,and the converse is observed for the MAPbI3reflections.This observation suggests that the conversion process begins from the surface of the PbI2crystallites and proceeds toward the center where the crystallite domain size of the MAPbI3phase increases and that of PbI2diminishes.Interestingly,the remnant PbI2phase can be seen in the data of other reports, but has not been identified as a primary source of variability in cell performance.8,10 Considering that the perovskite is the primary light absorber within the device,we wantedto further investigate how the optical absorption of thefilm changes with increasing dipping timeFIG.2.Absorption spectra of CH3NH3PbI3films as a function of unconverted PbI2phase fraction.FIG.3.(a)J-V curves and(b)EQE of CH3NH3PbI3-based devices as a function of unconverted PbI2phase fraction.(Figure2).11,12The pure PbI2film shows a band gap of2.40eV,consistent with the yellow color of PbI2.As the PbI2film is gradually converted to the perovskite,the band gap is progressively shifted toward1.60eV.The deviation of MAPbI3’s band gap(1.60eV)from that of the bulk MAPbI3 material(1.55eV)could be explained by quantum confinement effects related with the sizes of TiO2and MAPbI3crystallites and their interfacial interaction.13,14Interestingly,we also noticed the presence of a second absorption in the light absorber layer,in which the gap gradually red shifts from1.90eV to1.50eV as the PbI2concentration is decreased from9.5%to0.3%(Figure2—blue arrow).Having established the chemical compositions and optical properties of the light absorberfilms, we proceeded to examine the photo-physical responses of the corresponding functional devices in order to determine how the remnant PbI2affects device performance.The pure PbI2based device remarkably achieved a0.4%efficiency with a J sc of2.1mA/cm2and a V oc of564mV (Figure3(a)).Upon progressive conversion of the PbI2layer to MAPbI3,we observe two different regions(Figure4,Table I).In thefirst region,the expected behavior is observed;as more PbI2is converted to MAPbI3,the trend is toward higher photovoltaic efficiency,due both to J sc and V oc, until1.7%PbI2is reached.The increase in J sc is attributable,at least in part,to increasing absorption of light by the perovskite.We speculate that progressive elimination of PbI2,present as a layer between TiO2and the perovskite,also leads to higher net yields for electron injection into TiO2and therefore,higher J values.For a sufficiently thick PbI2spacer layer,electron injection would occur instepwise fashion,i.e.,perovskite→PbI2→TiO2.Finally,the photovoltage increase is attributable toFIG.4.Summary of J-V data vs.PbI2concentration of CH3NH3PbI3-based devices(Region1:0to15min dipping time, Region2:15min to2h dipping time).TABLE I.Photovoltaic performance of CH3NH3PbI3-based devices as a function of unconverted PbI2fraction.Dipping time PbI2concentration a J sc(mA/cm2)V oc(V)Fill factor(%)Efficiency(%) 0s100% 2.10.564320.45s9.5%10.10.93352 4.960s7.2%13.40.96052 6.72min 5.3%14.00.964557.45min 3.7%14.70.995578.315min 1.7%15.1 1.036629.730min0.8%13.60.968648.51h0.4%12.40.938657.62h0.3%9.70.84668 5.5a Determined from the Rietveld analysis of X-ray diffraction data.the positive shift in TiO2’s quasi-Fermi level as the population of photo-injected electrons is higher with increased concentration of MAPbI3.The second region yields a notably different trend;surprisingly,below a concentration of2% PbI2,J sc,V oc,and ultimatelyηdecrease.Considering that the light-harvesting efficiency would increase when the remaining2%PbI2is converted to MAPbI3(albeit to only a small degree),then the remnant PbI2must have some other role.We posit that remnant PbI2serves to inhibit detrimental electron-transfer processes(Figure5).Two such processes are back electron transfer from TiO2to holes in the valence band of the perovskite(charge-recombination)or to the holes in the HOMO of the HTM(charge-interception).This retardation of electron interception/recombination observation is reminiscent of the behavior of atomic layer deposited Al2O3/ZrO2layers that have been employed in dye-sensitized solar cells.15–18It is conceivable that the conversion of PbI2to MAPbI3occurs from the solution interface toward the TiO2/PbI2interface and thus would leave sandwiched between TiO2and MAPbI3a blocking layer of PbI2that inhibits charge-interception/recombination.For this hypothesis to be correct,it is crucial that the conduction-band-edge energy(E cb)of the PbI2be higher than the E cb of the TiO2.19–21 The work function of PbI2was measured by ultraviolet photoelectron spectroscopy(UPS)and was observed to be at6.35eV vs.vacuum level,which is0.9eV lower than the valence-band-edge energy(E vb)of MAPbI3(see Sec.S7of the supplementary material23for the work function of PbI2);the E cb(4.05eV)was calculated by subtracting the work function from the band gap(2.30eV).solar cell.FIG.6.Dark current of CH3NH3PbI3-based devices as a function of unconverted PbI2phase fraction.The E cb of PbI2is0.26eV higher than the E cb of TiO2and thus PbI2satisfies the conditions of a charge-recombination/interception barrier layer.In order to probe the hypothesis that PbI2acts as a charge-interception barrier,dark current measurements,in which electronsflow from TiO2to the HOMO of the HTM,were made.Consistent with our hypothesis,Figure6illustrates that the onset of the dark current occurs at lower potentials as the PbI2concentration decreases.In the absence of other effects,the increasing dark current with increasing fraction of perovskite(and decreasing fraction of PbI2)should result in progressively lower open-circuit photovoltages.Instead,the photocurrent density and the open-circuit photovoltage bothincrease,at least until to PbI2fraction reaches1.7%.As discussed above,thinning of a PbI2-basedFIG.7.Cross-sectional SEM images of CH3NH3PbI3film with different dipping time.sandwich layer should lead to higher net injection yields,but excessive thinning would diminish the effectiveness of PbI2as a barrier layer for back electron transfer reactions.Given the surprising role of remnant PbI2in these devices,we further probed the two-step conversion process by using scanning-electron microscopy(SEM)(Figure7).Two domains of lead-containing materials(PbI2and MAPbI3)are present.Thefirst domain is sited within the mesoporous TiO2network(area1)while the second grows on top of the network(area2).Area2initially contains 200nm crystals.As the dipping time is increased,the crystals show marked changes in size and morphology.The formation of bigger perovskite crystals is likely the result of the thermodynamically driven Ostwald ripening process,i.e.,smaller perovskite crystals dissolves and re-deposits onto larger perovskite crystals.22The rate of charge-interception,as measured via dark current,is proportional to the contact area between the perovskite and the HTM.Thus,the eventual formation of large, high-aspect-ratio crystals,as shown in Figure7,may well lead to increases in contact area and thereby contributes to the dark-current in Figure6.Regardless,we found that the formation of large perovskite crystals greatly decreased our success rate in constructing high-functioning,non-shorting solar cells.In summary,residual PbI2appears to play an important role in boosting overall efficiencies for CH3NH3PbI3-containing photovoltaics.PbI2’s role appears to be that of a TiO2-supported blocking layer,thereby slowing rates of electron(TiO2)/hole(perovskite)recombination,as well as decreasing rates of electron interception by the hole-transporting material.Optimal performance for energy conversion is observed when ca.98%of the initially present PbI2has been converted to the perovskite. Conversion to this extent requires about15min.Pushing beyond98%(and beyond15min of reaction time)diminishes cell performance and diminishes the success rate in constructing non-shorting cells.The latter problem is evidently a consequence of conversion of small and more-or-less uniformly packed perovskite crystallites to larger,poorly packed crystallites of varying shape and size.Finally,the essential,but previously unrecognized,role played by remnant PbI2 provides an additional explanation for why cells prepared dissolving and then depositing pre-formed CH3NH3PbI3generally under-perform those prepared via the intermediacy of PbI2.We thank Prof.Tobin Marks for use of the solar simulator and EQE measurement system. Electron microscopy was done at the Electron Probe Instrumentation Center(EPIC)at Northwestern University.Ultraviolet Photoemission Spectroscopy was done at the Keck Interdisciplinary SurfaceScience facility(Keck-II)at Northwestern University.This research was supported as part of theANSER Center,an Energy Frontier Research Center funded by the U.S Department of Energy, Office of Science,Office of Basic Energy Sciences,under Award No.DE-SC0001059.1R.Monastersky,Nature(London)497(7447),13(2013).2H.J.Snaith,J.Phys.Chem.Lett.4(21),3623(2013).3M.M.Lee,J.Teuscher,T.Miyasaka,T.N.Murakami,and H.J.Snaith,Science338(6107),643(2012).4J.Burschka,N.Pellet,S.J.Moon,R.Humphry-Baker,P.Gao,M.K.Nazeeruddin,and M.Gratzel,Nature(London) 499(7458),316(2013).5F.Hao,C.C.Stoumpos,D.H.Cao,R.P.H.Chang,and M.G.Kanatzidis,Nat.Photonics8(6),489(2014);F.Hao,C.C. Stoumpos,R.P.H.Chang,and M.G.Kanatzidis,J.Am.Chem.Soc.136,8094–8099(2014).6N.K.Noel,S.D.Stranks,A.Abate,C.Wehrenfennig,S.Guarnera,A.Haghighirad,A.Sadhanala,G.E.Eperon,M.B. Johnston,A.M.Petrozza,L.M.Herz,and H.J.Snaith,Energy Environ.Sci.7,3061(2014).7H.S.Kim,C.R.Lee,J.H.Im,K.B.Lee,T.Moehl,A.Marchioro,S.J.Moon,R.Humphry-Baker,J.H.Yum,J.E.Moser, M.Gratzel,and N.G.Park,Sci.Rep.2,591(2012).8D.Y.Liu and T.L.Kelly,Nat.Photonics8(2),133(2014).9C.C.Stoumpos,C.D.Malliakas,and M.G.Kanatzidis,Inorg.Chem.52(15),9019(2013).10J.H.Noh,S.H.Im,J.H.Heo,T.N.Mandal,and S.I.Seok,Nano Lett.13(4),1764(2013).11Diffuse reflectance measurements of MAPbI3films were converted to absorption spectra using the Kubelka-Munk equation,α/S=(1-R)2/2R,where R is the percentage of reflected light,andαand S are the absorption and scattering coefficients, respectively.The band gap values are the energy value at the intersection point of the absorption spectrum’s tangent line and the energy axis.12L.F.Gate,Appl.Opt.13(2),236(1974).13O.V oskoboynikov,C.P.Lee,and I.Tretyak,Phys.Rev.B63(16),165306(2001).14X.X.Xue,W.Ji,Z.Mao,H.J.Mao,Y.Wang,X.Wang,W.D.Ruan,B.Zhao,and J.R.Lombardi,J.Phys.Chem.C 116(15),8792(2012).15E.Palomares,J.N.Clifford,S.A.Haque,T.Lutz,and J.R.Durrant,J.Am.Chem.Soc.125(2),475(2003).16C.Prasittichai,J.R.Avila,O.K.Farha,and J.T.Hupp,J.Am.Chem.Soc.135(44),16328(2013).17A.K.Chandiran,M.K.Nazeeruddin,and M.Gratzel,Adv.Funct.Mater.24(11),1615(2014).18M.J.Katz,M.J.D.Vermeer,O.K.Farha,M.J.Pellin,and J.T.Hupp,Langmuir29(2),806(2013).19M.J.DeVries,M.J.Pellin,and J.T.Hupp,Langmuir26(11),9082(2010).20C.Prasittichai and J.T.Hupp,J.Phys.Chem.Lett.1(10),1611(2010).21F.Fabregat-Santiago,J.Garcia-Canadas,E.Palomares,J.N.Clifford,S.A.Haque,J.R.Durrant,G.Garcia-Belmonte, and J.Bisquert,J.Appl.Phys.96(11),6903(2004).22Alan D.McNaught and Andrew Wilkinson,IUPAC Compendium of Chemical Terminology(Blackwell Scientific Publica-tions,Oxford,1997).23See supplementary material at /10.1063/1.4895038for experimental details,absorption spectrum of bulk CH3NH3PbI3,fraction,size,absorption spectrum,work function of unconverted PbI2,and average photovoltaic perfor-mance.。
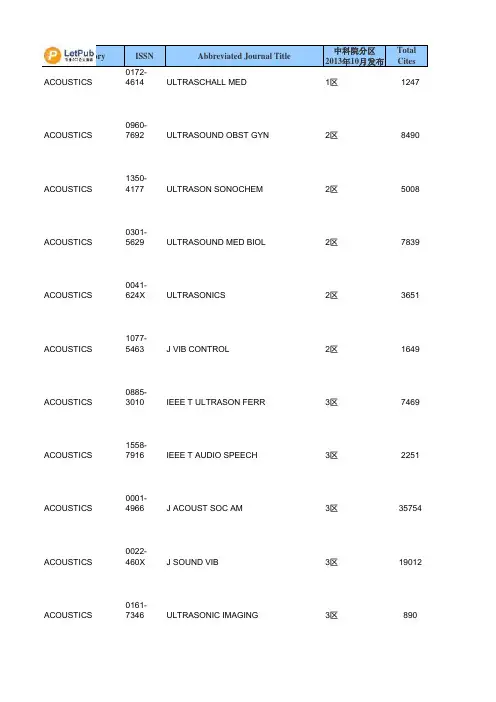
Subcategory ISSN Abbreviated Journal Title中科院分区2013年10月发TotalCitesACOUSTICS0960-7692ULTRASOUND OBST GYN2区 8490 ACOUSTICS1350-4177ULTRASON SONOCHEM2区 5008 ACOUSTICS0301-5629ULTRASOUND MED BIOL2区 7839 ACOUSTICS0041-624XULTRASONICS2区 3651 ACOUSTICS1077-5463J VIB CONTROL2区 1649 ACOUSTICS0885-3010IEEE T ULTRASON FERR3区 7469 ACOUSTICS1558-7916IEEE T AUDIO SPEECH3区 2251 ACOUSTICS0001-4966J ACOUST SOC AM3区 35754 ACOUSTICS0022-460XJ SOUND VIB3区 19012 ACOUSTICS0161-7346ULTRASONIC IMAGING3区 890 ACOUSTICS0165-2125WAVE MOTION3区 1367 ACOUSTICS0278-4297J ULTRAS MED3区 3907 ACOUSTICS0167-6393SPEECH COMMUN3区 1982 ACOUSTICS1048-9002J VIB ACOUST3区 1825 ACOUSTICS0003-682XAPPL ACOUST4区 1975 ACOUSTICS1549-4950J AUDIO ENG SOC4区 832 ACOUSTICS0137-5075ARCH ACOUST4区 242 ACOUSTICS1610-1928ACTA ACUST UNITED AC4区 1808 ACOUSTICS0091-2751J CLIN ULTRASOUND4区 1642 ACOUSTICS0031-8388PHONETICA4区 570 ACOUSTICS0218-396XJ COMPUT ACOUST4区 328 ACOUSTICS1687-4722EURASIP J AUDIO SPEE4区 59 ACOUSTICS1475-472XINT J AEROACOUST4区 190 ACOUSTICS1070-9622SHOCK VIB4区 320 ACOUSTICS1063-7710ACOUST PHYS+4区 600 ACOUSTICS1027-5851INT J ACOUST VIB4区 57 ACOUSTICS0736-2501NOISE CONTROL ENG J4区 273 ACOUSTICS0814-6039ACOUST AUST4区 49 ACOUSTICS0263-0923J LOW FREQ NOISE V A4区 79 ACOUSTICS1541-0161SOUND VIB4区 195 AGRICULTURAL ECON0306-9192FOOD POLICY2区 1736 AGRICULTURAL ECON0165-1587EUR REV AGRIC ECON2区 744 AGRICULTURAL ECON2040-5790APPL ECON PERSPECT P3区 101 AGRICULTURAL ECON0021-857XJ AGR ECON3区 829 AGRICULTURAL ECON1364-985XAUST J AGR RESOUR EC3区 473 AGRICULTURAL ECON0169-5150AGR ECON-BLACKWELL3区 1284 AGRICULTURAL ECON0002-9092AM J AGR ECON4区 4462 AGRICULTURAL ECON0008-3976CAN J AGR ECON4区 378 AGRICULTURAL ECON1068-5502J AGR RESOUR ECON4区 534 AGRICULTURAL ECON1756-137XCHINA AGR ECON REV4区 43 AGRICULTURAL ECON1559-2448INT FOOD AGRIBUS MAN4区 199 AGRICULTURAL ECON1699-6887ITEA-INF TEC ECON AG4区 54 AGRICULTURAL ECON0303-1853AGREKON4区 104 AGRICULTURAL ECON1808-2882CUST AGRONEGOCIO4区 3 AGRICULTURAL ENGIN0960-8524BIORESOURCE TECHNOL2区 44257 AGRICULTURAL ENGIN0961-9534BIOMASS BIOENERG2区 10805 AGRICULTURAL ENGIN0926-6690IND CROP PROD3区 4151 AGRICULTURAL ENGIN0144-8609AQUACULT ENG3区 1214 AGRICULTURAL ENGIN1537-5110BIOSYST ENG3区 2799 AGRICULTURAL ENGIN0733-9437J IRRIG DRAIN E-ASCE4区 2413 AGRICULTURAL ENGIN1611-2490PADDY WATER ENVIRON4区 2462151-0032T ASABE4区 6871 AGRICULTURAL ENGIN0100-6916ENG AGR-JABOTICABAL4区 568 AGRICULTURAL ENGIN1415-4366REV BRAS ENG AGR AMB4区 1022 AGRICULTURAL ENGIN0883-8542APPL ENG AGRIC4区 1234 AGRICULTURAL ENGIN0084-5841AMA-AGR MECH ASIA AF4区 90 AGRICULTURAL ENGINAGRICULTURE, DAIRY0999-193XGENET SEL EVOL1区 1539 AGRICULTURE, DAIRY0268-9146ANIM GENET1区 3284 AGRICULTURE, DAIRY0022-0302J DAIRY SCI2区 29407 AGRICULTURE, DAIRY0739-7240DOMEST ANIM ENDOCRIN2区 1927 AGRICULTURE, DAIRY0021-8812J ANIM SCI2区 23776 AGRICULTURE, DAIRY0378-4320ANIM REPROD SCI2区 5803 AGRICULTURE, DAIRY0916-8818J REPROD DEVELOP2区 1622 AGRICULTURE, DAIRY0931-2668J ANIM BREED GENET2区 1150 AGRICULTURE, DAIRY1751-7311ANIMAL2区 1894 AGRICULTURE, DAIRY0377-8401ANIM FEED SCI TECH2区 6366 AGRICULTURE, DAIRY0032-5791POULTRY SCI3区 13639 AGRICULTURE, DAIRY0168-1591APPL ANIM BEHAV SCI3区 5989 AGRICULTURE, DAIRY0936-6768REPROD DOMEST ANIM3区 2880 AGRICULTURE, DAIRY0022-0299J DAIRY RES3区 2641 AGRICULTURE, DAIRY0931-2439J ANIM PHYSIOL AN N3区 1202 AGRICULTURE, DAIRY1871-1413LIVEST SCI3区 2542 AGRICULTURE, DAIRY0043-9339WORLD POULTRY SCI J3区 1320 AGRICULTURE, DAIRY0007-1668BRIT POULTRY SCI3区 3285 AGRICULTURE, DAIRY0921-4488SMALL RUMINANT RES3区 4134 AGRICULTURE, DAIRY1745-039XARCH ANIM NUTR3区 629 AGRICULTURE, DAIRY0049-4747TROP ANIM HEALTH PRO3区 1749 AGRICULTURE, DAIRY1344-3941ANIM SCI J3区 737 AGRICULTURE, DAIRY0008-3984CAN J ANIM SCI3区 1829 AGRICULTURE, DAIRY1212-1819CZECH J ANIM SCI3区 613 AGRICULTURE, DAIRY0860-4037ANIM SCI PAP REP3区 306 AGRICULTURE, DAIRY1049-5398ANIM BIOTECHNOL3区 330 AGRICULTURE, DAIRY1056-6171J APPL POULTRY RES4区 1465 AGRICULTURE, DAIRY1594-4077ITAL J ANIM SCI4区 676 AGRICULTURE, DAIRY1230-1388J ANIM FEED SCI4区 560 AGRICULTURE, DAIRY0990-0632PROD ANIM4区 263 AGRICULTURE, DAIRY1346-7395J POULT SCI4区 308 AGRICULTURE, DAIRY1758-1559AVIAN BIOL RES4区 43 AGRICULTURE, DAIRY1011-2367ASIAN AUSTRAL J ANIM4区 1609 AGRICULTURE, DAIRY1257-5011WORLD RABBIT SCI4区 269 AGRICULTURE, DAIRY0906-4702ACTA AGR SCAND A-AN4区 427 AGRICULTURE, DAIRY0990-0632INRA PROD ANIM4区 126 AGRICULTURE, DAIRY0375-1589S AFR J ANIM SCI4区 544 AGRICULTURE, DAIRY0003-9438ARCH TIERZUCHT4区 456 AGRICULTURE, DAIRY1642-3402ANN ANIM SCI4区 135 AGRICULTURE, DAIRY0004-9433AUST J DAIRY TECHNOL4区 360 AGRICULTURE, DAIRY0044-5401ZUCHTUNGSKUNDE4区 167 AGRICULTURE, DAIRY0003-9098ARCH GEFLUGELKD4区 323 AGRICULTURE, DAIRY0429-2766FOURRAGES4区 251 AGRICULTURE, DAIRY0972-2963ANIM NUTR FEED TECHN4区 78 AGRICULTURE, DAIRY1516-635XBRAZ J POULTRY SCI4区 280 AGRICULTURE, DAIRY2007-1124REV MEX CIENC PECU4区 25 AGRICULTURE, DAIRY0026-704XMLJEKARSTVO4区 90 AGRICULTURE, DAIRY0120-0690REV COLOMB CIENC PEC4区 69 AGRICULTURE, DAIRY0122-0268REV MVZ CORDOBA4区 56AGRICULTURE, DAIRY0367-8318INDIAN J ANIM SCI4区 733 AGRICULTURE, DAIRY0971-2119J APPL ANIM RES4区 221 AGRICULTURE, DAIRY1124-4593LARGE ANIM REV4区 31 AGRICULTURE, DAIRY0125-6726BUFFALO BULL4区 43 AGRICULTURE, DAIRY0367-6722INDIAN J ANIM RES4区 33 AGRICULTURE, MULTI0021-8561J AGR FOOD CHEM1区 76046 AGRICULTURE, MULTI0021-8596J AGR SCI1区 3886 AGRICULTURE, MULTI0167-8809AGR ECOSYST ENVIRON2区 10733 AGRICULTURE, MULTI0308-521XAGR SYST2区 2915 AGRICULTURE, MULTI0003-4746ANN APPL BIOL2区 3678 AGRICULTURE, MULTI0168-1699COMPUT ELECTRON AGR2区 2535 AGRICULTURE, MULTI0022-5142J SCI FOOD AGR2区 12723 AGRICULTURE, MULTI1670-567XICELAND AGR SCI2区 65 AGRICULTURE, MULTI1385-2256PRECIS AGRIC2区 703 AGRICULTURE, MULTI1473-5903INT J AGR SUSTAIN2区 275 AGRICULTURE, MULTI1537-5110BIOSYST ENG2区 2799 AGRICULTURE, MULTI0889-048XAGR HUM VALUES3区 790 AGRICULTURE, MULTI1187-7863J AGR ENVIRON ETHIC3区 494 AGRICULTURE, MULTI1573-5214NJAS-WAGEN J LIFE SC3区 528 AGRICULTURE, MULTI0552-9034PAK J AGR SCI3区 325 AGRICULTURE, MULTI1836-0939ANIM PROD SCI3区 541 AGRICULTURE, MULTI1836-0947CROP PASTURE SCI3区 537 AGRICULTURE, MULTI1742-1705RENEW AGR FOOD SYST3区 356 AGRICULTURE, MULTI0028-8233NEW ZEAL J AGR RES3区 1235 AGRICULTURE, MULTI0008-0845CALIF AGR3区 592 AGRICULTURE, MULTI1560-8530INT J AGRIC BIOL3区 1292 AGRICULTURE, MULTI0103-9016SCI AGR3区 1258 AGRICULTURE, MULTI1459-6067AGR FOOD SCI3区 363 AGRICULTURE, MULTI1044-0046J SUSTAIN AGR3区 462 AGRICULTURE, MULTI0045-6888REV CIENC AGRON3区 418 AGRICULTURE, MULTI1680-7073J AGR SCI TECH-IRAN3区 282 AGRICULTURE, MULTI0100-204XPESQUI AGROPECU BRAS3区 3338 AGRICULTURE, MULTI1695-971XSPAN J AGRIC RES3区 555 AGRICULTURE, MULTI1018-7081J ANIM PLANT SCI4区 240 AGRICULTURE, MULTI1861-3829J PLANT DIS PROTECT4区 305 AGRICULTURE, MULTI1166-7699CAH AGRIC4区 300 AGRICULTURE, MULTI1392-3196ZEMDIRBYSTE4区 138 AGRICULTURE, MULTI0718-5839CHIL J AGR RES4区 171 AGRICULTURE, MULTI0030-7270OUTLOOK AGR4区 289 AGRICULTURE, MULTI1671-2927AGR SCI CHINA4区 754 AGRICULTURE, MULTI1744-6961GRASSL SCI4区 215 AGRICULTURE, MULTI0021-3551JARQ-JPN AGR RES Q4区 421 AGRICULTURE, MULTI1413-7054CIENC AGROTEC4区 1168 AGRICULTURE, MULTI0005-9080BER LANDWIRTSCH4区 55 AGRICULTURE, MULTI0458-6859LANDBAUFORSCHUNG-GER4区 21 AGRICULTURE, MULTI0791-6833IRISH J AGR FOOD RES4区 190 AGRICULTURE, MULTI0031-7454PHILIPP AGRIC SCI4区 145 AGRICULTURE, MULTI1405-3195AGROCIENCIA-MEXICO4区 333 AGRICULTURE, MULTI0718-1620CIENC INVESTIG AGRAR4区 155 AGRICULTURE, MULTI1676-546XSEMIN-CIENC AGRAR4区 327 AGRICULTURE, MULTI1516-3725BIOSCI J4区 248 AGRICULTURE, MULTI0023-6152J FAC AGR KYUSHU U4区 205 AGRICULTURE, MULTI1594-5685NEW MEDIT4区 37 AGRICULTURE, MULTI0019-5022INDIAN J AGR SCI4区 776AGRICULTURE, MULTI0370-4661REV FAC CIENC AGRAR4区 36 AGRICULTURE, MULTI1663-7852AGRARFORSCH SCHWEIZ+4区 34 AGRICULTURE, MULTI1310-0351BULG J AGRIC SCI4区 115 AGRICULTURE, MULTI0122-1450CUAD DESARRO RURAL4区 11 AGRICULTURE, MULTI0002-1482AGR HIST4区 193 AGRICULTURE, MULTI1612-9830J AGR RURAL DEV TROP4区 45 AGRICULTURE, MULTI1300-7580TARIM BILIM DERG4区 54 AGRICULTURE, MULTI2095-3119J INTEGR AGR4区 88 AGRONOMY0065-2113ADV AGRON1区 2753 AGRONOMY1757-1693GCB BIOENERGY1区 585 AGRONOMY0040-5752THEOR APPL GENET1区 20361 AGRONOMY1774-0746AGRON SUSTAIN DEV2区 992 AGRONOMY0168-1923AGR FOREST METEOROL2区 10024 AGRONOMY1380-3743MOL BREEDING2区 3453 AGRONOMY1161-0301EUR J AGRON2区 3312 AGRONOMY0032-0862PLANT PATHOL2区 4386 AGRONOMY0032-079XPLANT SOIL2区 19896 AGRONOMY1526-498XPEST MANAG SCI2区 4883 AGRONOMY0378-4290FIELD CROP RES2区 6943 AGRONOMY0926-6690IND CROP PROD2区 4151 AGRONOMY0925-5214POSTHARVEST BIOL TEC2区 6155 AGRONOMY1939-8425RICE2区 224 AGRONOMY0342-7188IRRIGATION SCI2区 1297 AGRONOMY0378-3774AGR WATER MANAGE3区 5581 AGRONOMY0931-2250J AGRON CROP SCI3区 1411 AGRONOMY0043-1737WEED RES3区 2410 AGRONOMY0043-1745WEED SCI3区 4523 AGRONOMY0014-2336EUPHYTICA3区 7762 AGRONOMY0929-1873EUR J PLANT PATHOL3区 3736 AGRONOMY0925-9864GENET RESOUR CROP EV3区 2277 AGRONOMY0142-5242GRASS FORAGE SCI3区 1331 AGRONOMY0002-1962AGRON J3区 9966 AGRONOMY0011-183XCROP SCI3区 14370 AGRONOMY1436-8730J PLANT NUTR SOIL SC3区 2267 AGRONOMY0167-4366AGROFOREST SYST3区 2319 AGRONOMY0261-2194CROP PROT3区 4245 AGRONOMY0179-9541PLANT BREEDING3区 2696 AGRONOMY1735-6814INT J PLANT PROD3区 178 AGRONOMY0890-037XWEED TECHNOL3区 2762 AGRONOMY1214-1178PLANT SOIL ENVIRON3区 789 AGRONOMY1099-209XAM J POTATO RES3区 719 AGRONOMY0014-4797EXP AGR3区 762 AGRONOMY1344-7610BREEDING SCI3区 947 AGRONOMY0236-8722INT AGROPHYS3区 533 AGRONOMY1611-2490PADDY WATER ENVIRON3区 246 AGRONOMY0038-0768SOIL SCI PLANT NUTR3区 1724 AGRONOMY1343-943XPLANT PROD SCI4区 563 AGRONOMY0031-9465PHYTOPATHOL MEDITERR4区 640 AGRONOMY1044-0046J SUSTAIN AGR4区 462 AGRONOMY1300-011XTURK J AGRIC FOR4区 637 AGRONOMY1444-6162WEED BIOL MANAG4区 297 AGRONOMY0008-4220CAN J PLANT SCI4区 2567 AGRONOMY0906-4710ACTA AGR SCAND B-S P4区 565 AGRONOMY0251-0952SEED SCI TECHNOL4区 1101AGRONOMY1531-0353IRRIG DRAIN4区 736 AGRONOMY0971-4693ALLELOPATHY J4区 398 AGRONOMY1166-7699CAH AGRIC4区 300 AGRONOMY0014-3065POTATO RES4区 677 AGRONOMY0718-5839CHIL J AGR RES4区 171 AGRONOMY0133-3720CEREAL RES COMMUN4区 646 AGRONOMY1301-1111TURK J FIELD CROPS4区 66 AGRONOMY1984-7033CROP BREED APPL BIOT4区 307 AGRONOMY1744-6961GRASSL SCI4区 215 AGRONOMY1936-5209J PLANT REGIST4区 238 AGRONOMY0114-0671NEW ZEAL J CROP HORT4区 493 AGRONOMY0010-3624COMMUN SOIL SCI PLAN4区 3685 AGRONOMY1212-1975CZECH J GENET PLANT4区 123 AGRONOMY0103-8478CIENC RURAL4区 2136 AGRONOMY0144-8765BIOL AGRIC HORTIC4区 265 AGRONOMY1370-6233BIOTECHNOL AGRON SOC4区 243 AGRONOMY0534-0012GENETIKA-BELGRADE4区 119 AGRONOMY0025-6153MAYDICA4区 498 AGRONOMY1807-8621ACTA SCI-AGRON4区 365 AGRONOMY0100-316XREV CAATINGA4区 279 AGRONOMY1527-3741J AM POMOL SOC4区 179 AGRONOMY1516-3725BIOSCI J4区 248 AGRONOMY0187-7380REV FITOTEC MEX4区 171 AGRONOMY0020-8841INT SUGAR J4区 281 AGRONOMY0367-4223GESUNDE PFLANZ4区 109 AGRONOMY1222-4227ROM AGRIC RES4区 52 AGRONOMY0971-2070RANGE MANAG AGROFOR4区 37 AGRONOMY0115-463XPHILIPP J CROP SCI4区 55 AGRONOMY0972-1665J AGROMETEOROL4区 109 AGRONOMY0378-7818REV FAC AGRON LUZ4区 92 AGRONOMY0972-3226RES CROP4区 91 AGRONOMY0250-5371LEGUME RES4区 97 ALLERGY0091-6749J ALLERGY CLIN IMMUN1区 36074 ALLERGY0105-4538ALLERGY2区 12173 ALLERGY1080-0549CLIN REV ALLERG IMMU2区 1685 ALLERGY0954-7894CLIN EXP ALLERGY2区 10054 ALLERGY1081-1206ANN ALLERG ASTHMA IM3区 5372 ALLERGY1528-4050CURR OPIN ALLERGY CL3区 2255 ALLERGY0905-6157PEDIAT ALLERG IMM-UK3区 3045 ALLERGY0105-1873CONTACT DERMATITIS3区 4899 ALLERGY1529-7322CURR ALLERGY ASTHM R3区 1204 ALLERGY2092-7355ALLERGY ASTHMA IMMUN3区 229 ALLERGY0889-8561IMMUNOL ALLERGY CLIN3区 1136 ALLERGY1018-2438INT ARCH ALLERGY IMM4区 4635 ALLERGY1088-5412ALLERGY ASTHMA PROC4区 1201 ALLERGY1018-9068J INVEST ALLERG CLIN4区 1375 ALLERGY0277-0903J ASTHMA4区 2551 ALLERGY0301-0546ALLERGOL IMMUNOPATH4区 582 ALLERGY0125-877XASIAN PAC J ALLERGY4区 369 ALLERGY1642-395XPOSTEP DERM ALERGOL4区 149 ALLERGY1735-1502IRAN J ALLERGY ASTHM4区 213 ALLERGY2151-321XPEDIAT ALLER IMM PUL4区 51 ALLERGY0344-5062ALLERGOLOGIE4区 210 ALLERGY1877-0320REV FR ALLERGOL4区 253ALLERGY1609-3607CURR ALLERGY CLIN IM4区 30 ANATOMY & MORPHO1863-2653BRAIN STRUCT FUNCT2区 1259 ANATOMY & MORPHO1662-5129FRONT NEUROANAT2区 579 ANATOMY & MORPHO0301-5556ADV ANAT EMBRYOL CEL2区 352 ANATOMY & MORPHO1058-8388DEV DYNAM3区 10728 ANATOMY & MORPHO0021-8782J ANAT3区 7342 ANATOMY & MORPHO1422-6405CELLS TISSUES ORGANS3区 1951 ANATOMY & MORPHO0940-9602ANN ANAT3区 1160 ANATOMY & MORPHO1062-3345APPL IMMUNOHISTO M M3区 1373 ANATOMY & MORPHO0362-2525J MORPHOL3区 4364 ANATOMY & MORPHO1059-910XMICROSC RES TECHNIQ4区 4641 ANATOMY & MORPHO0001-7272ACTA ZOOL-STOCKHOLM4区 783 ANATOMY & MORPHO1932-8486ANAT REC4区 7324 ANATOMY & MORPHO0897-3806CLIN ANAT4区 2046 ANATOMY & MORPHO0720-213XZOOMORPHOLOGY4区 707 ANATOMY & MORPHO0930-1038SURG RADIOL ANAT4区 1843 ANATOMY & MORPHO0340-2096ANAT HISTOL EMBRYOL4区 768 ANATOMY & MORPHO1447-6959ANAT SCI INT4区 317 ANATOMY & MORPHO0015-5659FOLIA MORPHOL4区 448 ANATOMY & MORPHO0717-9502INT J MORPHOL4区 373 ANATOMY & MORPHO0003-2778J ANAT SOC INDIA4区 130 ANDROLOGY0105-6263INT J ANDROL2区 3259 ANDROLOGY0196-3635J ANDROL3区 4717 ANDROLOGY1008-682XASIAN J ANDROL4区 1739 ANDROLOGY1939-6368SYST BIOL REPROD MED4区 257 ANDROLOGY0303-4569ANDROLOGIA4区 1463 ANDROLOGY1698-031XREV INT ANDROL4区 25 ANESTHESIOLOGY0304-3959PAIN1区 29370 ANESTHESIOLOGY0003-3022ANESTHESIOLOGY2区 22547 ANESTHESIOLOGY0007-0912BRIT J ANAESTH2区 12587 ANESTHESIOLOGY0003-2409ANAESTHESIA2区 6605 ANESTHESIOLOGY1098-7339REGION ANESTH PAIN M2区 2816 ANESTHESIOLOGY0003-2999ANESTH ANALG3区 20930 ANESTHESIOLOGY1090-3801EUR J PAIN3区 4369 ANESTHESIOLOGY0375-9393MINERVA ANESTESIOL3区 1715 ANESTHESIOLOGY0265-0215EUR J ANAESTH3区 2714 ANESTHESIOLOGY1530-7085PAIN PRACT3区 922 ANESTHESIOLOGY0749-8047CLIN J PAIN3区 4291 ANESTHESIOLOGY1155-5645PEDIATR ANESTH3区 3380 ANESTHESIOLOGY0952-7907CURR OPIN ANESTHESIO3区 1692 ANESTHESIOLOGY0001-5172ACTA ANAESTH SCAND3区 5582 ANESTHESIOLOGY0832-610XCAN J ANESTH4区 4123 ANESTHESIOLOGY0959-289XINT J OBSTET ANESTH4区 974 ANESTHESIOLOGY0898-4921J NEUROSURG ANESTH4区 914 ANESTHESIOLOGY1053-0770J CARDIOTHOR VASC AN4区 2386 ANESTHESIOLOGY0310-057XANAESTH INTENS CARE4区 2189 ANESTHESIOLOGY1471-2253BMC ANESTHESIOL4区 170 ANESTHESIOLOGY0952-8180J CLIN ANESTH4区 1843 ANESTHESIOLOGY0932-433XSCHMERZ4区 659 ANESTHESIOLOGY0913-8668J ANESTH4区 866 ANESTHESIOLOGY0003-2417ANAESTHESIST4区 1101 ANESTHESIOLOGY0750-7658ANN FR ANESTH4区 1106 ANESTHESIOLOGY1387-1307J CLIN MONIT COMPUT4区 595 ANESTHESIOLOGY0170-5334ANASTH INTENSIVMED4区 235ANESTHESIOLOGY0939-2661ANASTH INTENSIV NOTF4区 309 ANESTHESIOLOGY1011-288XDOULEUR ANALG4区 27 0066-4146ANNU REV ASTRON ASTR1区 8240 ASTRONOMY & ASTRO0067-0049ASTROPHYS J SUPPL S1区 24764 ASTRONOMY & ASTRO1614-4961LIVING REV SOL PHYS2区 498 ASTRONOMY & ASTRO0935-4956ASTRON ASTROPHYS REV2区 973 ASTRONOMY & ASTRO0084-6597ANNU REV EARTH PL SC2区 4653 ASTRONOMY & ASTRO0004-637XASTROPHYS J2区 191940 ASTRONOMY & ASTRO2041-8205ASTROPHYS J LETT2区 45993 ASTRONOMY & ASTRO1475-7516J COSMOL ASTROPART P2区 13656 ASTRONOMY & ASTRO0035-8711MON NOT R ASTRON SOC2区 96083 ASTRONOMY & ASTRO0038-6308SPACE SCI REV2区 7134 ASTRONOMY & ASTRO0004-6361ASTRON ASTROPHYS2区 101305 ASTRONOMY & ASTROASTRONOMY & ASTRO0004-6256ASTRON J3区 346680927-6505ASTROPART PHYS3区 3658 ASTRONOMY & ASTRO1550-7998PHYS REV D3区 134999 ASTRONOMY & ASTRO0004-6280PUBL ASTRON SOC PAC3区 9054 ASTRONOMY & ASTRO0264-9381CLASSICAL QUANT GRAV3区 15025 ASTRONOMY & ASTRO0038-0938SOL PHYS3区 9921 ASTRONOMY & ASTROASTRONOMY & ASTRO0019-1035ICARUS3区 157541323-3580PUBL ASTRON SOC AUST3区 925 ASTRONOMY & ASTRO0922-6435EXP ASTRON3区 581 ASTRONOMY & ASTROASTRONOMY & ASTRO1531-1074ASTROBIOLOGY3区 17300001-5237ACTA ASTRONOM3区 1148 ASTRONOMY & ASTRO0004-6264PUBL ASTRON SOC JPN3区 4997 ASTRONOMY & ASTRO0923-2958CELEST MECH DYN ASTR3区 1982 ASTRONOMY & ASTRO0032-0633PLANET SPACE SCI3区 6073 ASTRONOMY & ASTRO0004-640XASTROPHYS SPACE SCI3区 5706 ASTRONOMY & ASTROASTRONOMY & ASTRO0001-7701GEN RELAT GRAVIT3区 42471384-1076NEW ASTRON4区 1442 ASTRONOMY & ASTRO1387-6473NEW ASTRON REV4区 1024 ASTRONOMY & ASTROASTRONOMY & ASTRO1631-0705CR PHYS4区 12760992-7689ANN GEOPHYS-GERMANY4区 5348 ASTRONOMY & ASTRO1473-5504INT J ASTROBIOL4区 349 ASTRONOMY & ASTRO0004-6337ASTRON NACHR4区 1887 ASTRONOMY & ASTRO1539-4956SPACE WEATHER4区 469 ASTRONOMY & ASTRO1674-4527RES ASTRON ASTROPHYS4区 553 ASTRONOMY & ASTRO0185-1101REV MEX ASTRON ASTR4区 593 ASTRONOMY & ASTRO0309-1929GEOPHYS ASTRO FLUID4区 685 ASTRONOMY & ASTRO0273-1177ADV SPACE RES4区 7209 ASTRONOMY & ASTRO0218-2718INT J MOD PHYS D4区 2727 ASTRONOMY & ASTRO0048-6604RADIO SCI4区 3254 ASTRONOMY & ASTRO1063-7737ASTRON LETT+4区 992 ASTRONOMY & ASTRO1225-4614J KOREAN ASTRON SOC4区 158 ASTRONOMY & ASTRO0167-9295EARTH MOON PLANETS4区 645 ASTRONOMY & ASTRO0304-9523B ASTRON SOC INDIA4区 202 ASTRONOMY & ASTROASTRONOMY & ASTRO1063-7729ASTRON REP+4区 14151990-3413ASTROPHYS BULL4区 156 ASTRONOMY & ASTROASTRONOMY & ASTRO0038-0946SOLAR SYST RES+4区 3680202-2893GRAVIT COSMOL-RUSSIA4区 268 ASTRONOMY & ASTRO0029-7704OBSERVATORY4区 278 ASTRONOMY & ASTRO1392-0049BALT ASTRON4区 330 ASTRONOMY & ASTRO0571-7256ASTROPHYSICS+4区 424 ASTRONOMY & ASTRO0884-5913KINEMAT PHYS CELEST+4区 148 ASTRONOMY & ASTRO1366-8781ASTRON GEOPHYS4区 130 ASTRONOMY & ASTRO0250-6335J ASTROPHYS ASTRON4区 286 ASTRONOMY & ASTRO0010-9525COSMIC RES+4区 332 ASTRONOMY & ASTRO1335-1842CONTRIB ASTRON OBS S4区 62 ASTRONOMY & ASTROAUDIOLOGY & SPEEC0093-934XBRAIN LANG1区 5351 AUDIOLOGY & SPEEC0196-0202EAR HEARING2区 3754 AUDIOLOGY & SPEEC0378-5955HEARING RES2区 7746 AUDIOLOGY & SPEEC1058-0360AM J SPEECH-LANG PAT2区 1102 AUDIOLOGY & SPEEC1420-3030AUDIOL NEURO-OTOL3区 1407 AUDIOLOGY & SPEEC0094-730XJ FLUENCY DISORD3区 833 AUDIOLOGY & SPEEC1092-4388J SPEECH LANG HEAR R3区 4781 AUDIOLOGY & SPEEC1463-1741NOISE HEALTH3区 665 AUDIOLOGY & SPEEC0001-4966J ACOUST SOC AM3区 35754 AUDIOLOGY & SPEEC1499-2027INT J AUDIOL3区 1804 AUDIOLOGY & SPEEC1050-0545J AM ACAD AUDIOL4区 1418 AUDIOLOGY & SPEEC0021-9924J COMMUN DISORD4区 1298 AUDIOLOGY & SPEEC1368-2822INT J LANG COMM DIS4区 1027 AUDIOLOGY & SPEEC0743-4618AUGMENT ALTERN COMM4区 541 AUDIOLOGY & SPEEC1754-9507INT J SPEECH-LANG PA4区 287 AUDIOLOGY & SPEEC1084-7138TRENDS AMPLIF4区 407 AUDIOLOGY & SPEEC1021-7762FOLIA PHONIATR LOGO4区 848 AUDIOLOGY & SPEEC1059-0889AM J AUDIOL4区 326 AUDIOLOGY & SPEEC0023-8309LANG SPEECH4区 855 AUDIOLOGY & SPEEC0269-9206CLIN LINGUIST PHONET4区 798 AUDIOLOGY & SPEEC0031-8388PHONETICA4区 570 AUDIOLOGY & SPEEC1401-5439LOGOP PHONIATR VOCO4区 335 AUTOMATION & CONT0278-0046IEEE T IND ELECTRON1区 17404 AUTOMATION & CONT1532-4435J MACH LEARN RES1区 6024 AUTOMATION & CONT1551-3203IEEE T IND INFORM2区 969 AUTOMATION & CONT1083-4419IEEE T SYST MAN CY B2区 5821 AUTOMATION & CONT1083-4435IEEE-ASME T MECH2区 2878 AUTOMATION & CONT0005-1098AUTOMATICA2区 15500 AUTOMATION & CONT0018-9286IEEE T AUTOMAT CONTR2区 23664 AUTOMATION & CONT1070-9932IEEE ROBOT AUTOM MAG2区 1163 AUTOMATION & CONT0016-0032J FRANKLIN I2区 2276 AUTOMATION & CONT1066-033XIEEE CONTR SYST MAG2区 2254 AUTOMATION & CONT0169-7439CHEMOMETR INTELL LAB2区 4880 AUTOMATION & CONT1063-6536IEEE T CONTR SYST T3区 4147 AUTOMATION & CONT0886-9383J CHEMOMETR3区 2658 AUTOMATION & CONT1049-8923INT J ROBUST NONLIN3区 2213 AUTOMATION & CONT0959-1524J PROCESS CONTR3区 2881 AUTOMATION & CONT1751-8644IET CONTROL THEORY A3区 1967 AUTOMATION & CONT1751-570XNONLINEAR ANAL-HYBRI3区 535 AUTOMATION & CONT1545-5955IEEE T AUTOM SCI ENG3区 871 AUTOMATION & CONT0967-0661CONTROL ENG PRACT3区 3413 AUTOMATION & CONT0167-6911SYST CONTROL LETT3区 4239 AUTOMATION & CONT0952-1976ENG APPL ARTIF INTEL3区 2085 AUTOMATION & CONT1562-2479INT J FUZZY SYST3区 284 AUTOMATION & CONT1561-8625ASIAN J CONTROL3区 853 AUTOMATION & CONT0363-0129SIAM J CONTROL OPTIM3区 4590 AUTOMATION & CONT0020-7721INT J SYST SCI3区 1996 AUTOMATION & CONT0957-4158MECHATRONICS3区 1681 AUTOMATION & CONT1367-5788ANNU REV CONTROL3区 662 AUTOMATION & CONT1292-8119ESAIM CONTR OPTIM CA3区 608AUTOMATION & CONT0947-3580EUR J CONTROL4区 636 AUTOMATION & CONT0890-6327INT J ADAPT CONTROL4区 809 AUTOMATION & CONT0268-3768INT J ADV MANUF TECH4区 7187 AUTOMATION & CONT0921-8890ROBOT AUTON SYST4区 1807 AUTOMATION & CONT0143-2087OPTIM CONTR APPL MET4区 411 AUTOMATION & CONT0020-7179INT J CONTROL4区 4282 AUTOMATION & CONT1641-876XINT J AP MAT COM-POL4区 562 AUTOMATION & CONT1598-6446INT J CONTROL AUTOM4区 774 AUTOMATION & CONT1387-2532AUTON AGENT MULTI-AG4区 500 AUTOMATION & CONT0022-0434J DYN SYST-T ASME4区 2738 AUTOMATION & CONT0265-0754IMA J MATH CONTROL I4区 303 AUTOMATION & CONT0332-7353MODEL IDENT CONTROL4区 170 AUTOMATION & CONT0924-6703DISCRETE EVENT DYN S4区 292 AUTOMATION & CONT0959-6518P I MECH ENG I-J SYS4区 567 AUTOMATION & CONT1392-124XINF TECHNOL CONTROL4区 125 AUTOMATION & CONT0142-3312T I MEAS CONTROL4区 272 AUTOMATION & CONT0144-5154ASSEMBLY AUTOM4区 252 AUTOMATION & CONT1220-1766STUD INFORM CONTROL4区 145 AUTOMATION & CONT0826-8185INT J ROBOT AUTOM4区 199 AUTOMATION & CONT1079-2724J DYN CONTROL SYST4区 239 AUTOMATION & CONT1841-9836INT J COMPUT COMMUN4区 175 AUTOMATION & CONT0932-4194MATH CONTROL SIGNAL4区 515 AUTOMATION & CONT1004-4132J SYST ENG ELECTRON4区 357 AUTOMATION & CONT1697-7912REV IBEROAM AUTOM IN4区 50 AUTOMATION & CONT0005-1144AUTOMATIKA4区 48 AUTOMATION & CONT0020-2940MEAS CONTROL-UK4区 183 AUTOMATION & CONT0178-2312AT-AUTOM4区 160 AUTOMATION & CONT1454-8658CONTROL ENG APPL INF4区 40 AUTOMATION & CONT0005-1179AUTOMAT REM CONTR+4区 812 AUTOMATION & CONT1064-2315J AUTOMAT INFORM SCI4区 41 BEHAVIORAL SCIENCE0140-525XBEHAV BRAIN SCI1区 64021364-6613TRENDS COGN SCI1区 15717 BEHAVIORAL SCIENCE0149-7634NEUROSCI BIOBEHAV R2区 12968 BEHAVIORAL SCIENCEBEHAVIORAL SCIENCE0010-9452CORTEX2区 52651662-5153FRONT BEHAV NEUROSCI2区 989 BEHAVIORAL SCIENCE1939-3792AUTISM RES2区 700 BEHAVIORAL SCIENCE1090-5138EVOL HUM BEHAV2区 2536 BEHAVIORAL SCIENCE1530-7026COGN AFFECT BEHAV NE2区 2402 BEHAVIORAL SCIENCE0018-506XHORM BEHAV2区 7968 BEHAVIORAL SCIENCE1601-1848GENES BRAIN BEHAV3区 2912 BEHAVIORAL SCIENCE0028-3932NEUROPSYCHOLOGIA3区 20370 BEHAVIORAL SCIENCE0301-0511BIOL PSYCHOL3区 5593 BEHAVIORAL SCIENCE0166-4328BEHAV BRAIN RES3区 19204 BEHAVIORAL SCIENCE1074-7427NEUROBIOL LEARN MEM3区 4648 BEHAVIORAL SCIENCE1025-3890STRESS3区 1631 BEHAVIORAL SCIENCEBEHAVIORAL SCIENCE0379-864XCHEM SENSES3区 37981045-2249BEHAV ECOL3区 7392 BEHAVIORAL SCIENCE0065-3454ADV STUD BEHAV3区 1119 BEHAVIORAL SCIENCEBEHAVIORAL SCIENCE0031-9384PHYSIOL BEHAV3区 170030003-3472ANIM BEHAV3区 21716 BEHAVIORAL SCIENCE0006-8977BRAIN BEHAV EVOLUT3区 2315 BEHAVIORAL SCIENCE1744-9081BEHAV BRAIN FUNCT3区 1055 BEHAVIORAL SCIENCE0340-5443BEHAV ECOL SOCIOBIOL3区 10205 BEHAVIORAL SCIENCE1435-9448ANIM COGN3区 1820 BEHAVIORAL SCIENCE0735-7044BEHAV NEUROSCI4区 8038 BEHAVIORAL SCIENCE0091-3057PHARMACOL BIOCHEM BE4区 12961 BEHAVIORAL SCIENCE0001-8244BEHAV GENET4区 3356 BEHAVIORAL SCIENCE0195-6663APPETITE4区 6200 BEHAVIORAL SCIENCE0097-7403J EXP PSYCHOL ANIM B4区 2311 BEHAVIORAL SCIENCE0955-8810BEHAV PHARMACOL4区 2896 BEHAVIORAL SCIENCE0096-140XAGGRESSIVE BEHAV4区 2025 BEHAVIORAL SCIENCE0179-1613ETHOLOGY4区 3596 BEHAVIORAL SCIENCE0735-7036J COMP PSYCHOL4区 2517 BEHAVIORAL SCIENCE1543-4494LEARN BEHAV4区 654 BEHAVIORAL SCIENCEBEHAVIORAL SCIENCE0340-7594J COMP PHYSIOL A4区 50301525-5050EPILEPSY BEHAV4区 4765 BEHAVIORAL SCIENCE0196-206XJ DEV BEHAV PEDIATR4区 2669 BEHAVIORAL SCIENCEBEHAVIORAL SCIENCE1095-0680J ECT4区 9040005-7959BEHAVIOUR4区 4666 BEHAVIORAL SCIENCE1558-7878J VET BEHAV4区 303 BEHAVIORAL SCIENCE0376-6357BEHAV PROCESS4区 2669 BEHAVIORAL SCIENCE0168-1591APPL ANIM BEHAV SCI4区 5989 BEHAVIORAL SCIENCE1543-3633COGN BEHAV NEUROL4区 634 BEHAVIORAL SCIENCE0018-7208HUM FACTORS4区 2872 BEHAVIORAL SCIENCE0873-9749ACTA ETHOL4区 240 BEHAVIORAL SCIENCE0394-9370ETHOL ECOL EVOL4区 631 BEHAVIORAL SCIENCE0022-5002J EXP ANAL BEHAV4区 2483 BEHAVIORAL SCIENCE0896-4289BEHAV MED4区 503 BEHAVIORAL SCIENCE0289-0771J ETHOL4区 600 BEHAVIORAL SCIENCEBIOCHEMICAL RESEA1548-7091NAT METHODS1区 19241 BIOCHEMICAL RESEA0907-4449ACTA CRYSTALLOGR D1区 12453 BIOCHEMICAL RESEA1754-2189NAT PROTOC1区 16922 BIOCHEMICAL RESEA0958-1669CURR OPIN BIOTECH2区 9501 BIOCHEMICAL RESEA1535-9476MOL CELL PROTEOMICS2区 14201 BIOCHEMICAL RESEA1473-0197LAB CHIP2区 16485 BIOCHEMICAL RESEA1367-4803BIOINFORMATICS2区 51753 BIOCHEMICAL RESEA0735-9640PLANT MOL BIOL REP2区 2985 BIOCHEMICAL RESEA1467-5463BRIEF BIOINFORM2区 3073 BIOCHEMICAL RESEA1535-3893J PROTEOME RES2区 17943 BIOCHEMICAL RESEA1553-7358PLOS COMPUT BIOL2区 11758 BIOCHEMICAL RESEA0021-9673J CHROMATOGR A2区 63419 BIOCHEMICAL RESEA1043-1802BIOCONJUGATE CHEM2区 13900 BIOCHEMICAL RESEA1615-9853PROTEOMICS2区 16435 BIOCHEMICAL RESEA1874-3919J PROTEOMICS3区 3175 BIOCHEMICAL RESEA1478-9450EXPERT REV PROTEOMIC3区 1343 BIOCHEMICAL RESEA1552-4922CYTOM PART A3区 2910 BIOCHEMICAL RESEA1618-2642ANAL BIOANAL CHEM3区 21971 BIOCHEMICAL RESEA1467-3037CURR ISSUES MOL BIOL3区 534 BIOCHEMICAL RESEA1046-2023METHODS3区 11612 BIOCHEMICAL RESEA1860-6768BIOTECHNOL J3区 2238 BIOCHEMICAL RESEA1535-3508MOL IMAGING3区 1090 BIOCHEMICAL RESEA0173-0835ELECTROPHORESIS3区 16985 BIOCHEMICAL RESEA1757-6180BIOANALYSIS3区 1318 BIOCHEMICAL RESEA1076-5174J MASS SPECTROM3区 5573 BIOCHEMICAL RESEA2156-7085BIOMED OPT EXPRESS3区 1552 BIOCHEMICAL RESEA1864-063XJ BIOPHOTONICS3区 1049 BIOCHEMICAL RESEA1471-2105BMC BIOINFORMATICS3区 17337 BIOCHEMICAL RESEA1862-8346PROTEOM CLIN APPL3区 1160BIOCHEMICAL RESEA1083-3668J BIOMED OPT3区 9022 BIOCHEMICAL RESEA0962-8819TRANSGENIC RES3区 2675 BIOCHEMICAL RESEA0003-2697ANAL BIOCHEM3区 39746 BIOCHEMICAL RESEA1752-7155J BREATH RES3区 445 BIOCHEMICAL RESEA0951-4198RAPID COMMUN MASS SP3区 13285 BIOCHEMICAL RESEA1570-0232J CHROMATOGR B3区 20776 BIOCHEMICAL RESEA0958-0344PHYTOCHEM ANALYSIS3区 1895 BIOCHEMICAL RESEA1477-5956PROTEOME SCI3区 740 BIOCHEMICAL RESEA0736-6205BIOTECHNIQUES4区 7614 BIOCHEMICAL RESEA1093-3263J MOL GRAPH MODEL4区 5010 BIOCHEMICAL RESEA1090-7807J MAGN RESON4区 10596 BIOCHEMICAL RESEA0022-1759J IMMUNOL METHODS4区 11972 BIOCHEMICAL RESEA1087-0571J BIOMOL SCREEN4区 2357 BIOCHEMICAL RESEA0167-7012J MICROBIOL METH4区 7548 BIOCHEMICAL RESEA0165-0270J NEUROSCI METH4区 11483 BIOCHEMICAL RESEA1574-8936CURR BIOINFORM4区 233 BIOCHEMICAL RESEA0076-6879METHOD ENZYMOL4区 25177 BIOCHEMICAL RESEA1386-2073COMB CHEM HIGH T SCR4区 1453 BIOCHEMICAL RESEA0269-3879BIOMED CHROMATOGR4区 2861 BIOCHEMICAL RESEA0166-0934J VIROL METHODS4区 6884 BIOCHEMICAL RESEA1540-658XASSAY DRUG DEV TECHN4区 835 BIOCHEMICAL RESEA0890-8508MOL CELL PROBE4区 1695 BIOCHEMICAL RESEA1053-0509J FLUORESC4区 2801 BIOCHEMICAL RESEA1871-6784NEW BIOTECHNOL4区 765 BIOCHEMICAL RESEA1045-1056BIOLOGICALS4区 1044 BIOCHEMICAL RESEA1545-5963IEEE ACM T COMPUT BI4区 1027 BIOCHEMICAL RESEA1748-7188ALGORITHM MOL BIOL4区 247 BIOCHEMICAL RESEA1066-5277J COMPUT BIOL4区 2743 BIOCHEMICAL RESEA2211-0682JALA-J LAB AUTOM4区 379 BIOCHEMICAL RESEA0009-5893CHROMATOGRAPHIA4区 5250 BIOCHEMICAL RESEA1046-5928PROTEIN EXPRES PURIF4区 4668 BIOCHEMICAL RESEA1536-1241IEEE T NANOBIOSCI4区 663 BIOCHEMICAL RESEA0362-4803J LABELLED COMPD RAD4区 1544 BIOCHEMICAL RESEA1751-8741IET NANOBIOTECHNOL4区 118 BIOCHEMICAL RESEA1480-9222BIOL PROCED ONLINE4区 331 BIOCHEMICAL RESEA1570-1646CURR PROTEOMICS4区 218 BIOCHEMICAL RESEA1976-0280BIOCHIP J4区 159 BIOCHEMICAL RESEA0021-9665J CHROMATOGR SCI4区 1775 BIOCHEMICAL RESEA1532-1819J IMMUNOASS IMMUNOCH4区 219 BIOCHEMICAL RESEA1082-6076J LIQ CHROMATOGR R T4区 2292 BIOCHEMICAL RESEA1744-3091ACTA CRYSTALLOGR F4区 1097 BIOCHEMICAL RESEA0712-4813SPECTROSC-INT J4区 340 BIOCHEMICAL RESEA0580-9517METHOD MICROBIOL4区 682 BIOCHEMICAL RESEA1082-6068PREP BIOCHEM BIOTECH4区 331 BIOCHEMICAL RESEA1554-0014HYBRIDOMA4区 517 BIOCHEMICAL RESEA2161-5063ACS SYNTH BIOL4区 51 0092-8674CELL1区 178762 BIOCHEMISTRY & MOL0066-4154ANNU REV BIOCHEM1区 19420 BIOCHEMISTRY & MOL1078-8956NAT MED1区 57350 BIOCHEMISTRY & MOL1097-2765MOL CELL1区 47818 BIOCHEMISTRY & MOL1359-4184MOL PSYCHIATR1区 12686 BIOCHEMISTRY & MOLBIOCHEMISTRY & MOL1088-9051GENOME RES1区 288560907-4449ACTA CRYSTALLOGR D1区 12453 BIOCHEMISTRY & MOL0968-0004TRENDS BIOCHEM SCI1区 15642 BIOCHEMISTRY & MOL。
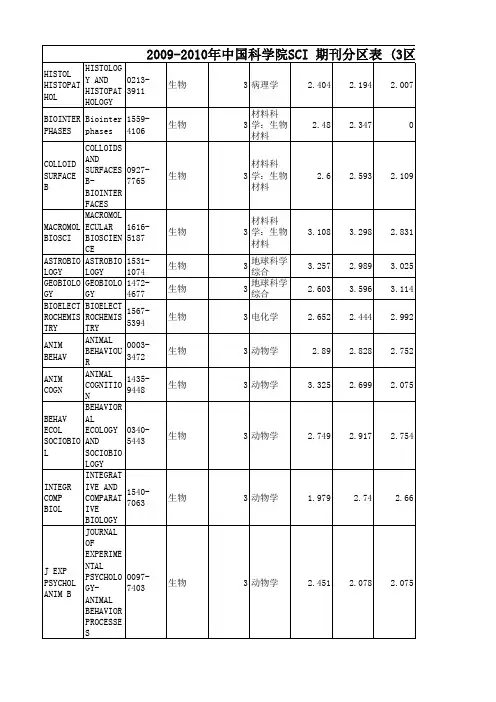

Experimental Vibrational Zero-Point Energies:Diatomic MoleculesKarl K.Irikura a…Physical and Chemical Properties Division,National Institute of Standards and Technology,Gaithersburg,Maryland20899-8380͑Received23August2006;revised manuscript received18December2006;accepted22December2006;published online18April2007͒Vibrational zero-point energies͑ZPEs͒,as determined from published spectroscopicconstants,are derived for85diatomic molecules.Standard uncertainties are also pro-vided,including estimated contributions from bias as well as the statistical uncertaintiespropagated from those reported in the spectroscopy literature.This compilation will behelpful for validating theoretical procedures for predicting ZPEs,which is a necessarystep in the ab initio prediction of molecular energetics.©2007by the U.S.Secretary ofCommerce on behalf of the United States.All rights reserved..͓DOI:10.1063/1.2436891͔Key words:molecular energetics;uncertainty;vibrational spectroscopy;zero-point energy.CONTENTS1.Introduction (389)2.Definition of Zero-Point Energy (389)3.Selection of Molecules (390)4.Contributions to Uncertainties (390)5.Analyses for Individual Molecules (394)6.Results and Discussion (396)7.Acknowledgments (396)8.References (396)List of Tables1.Spectroscopic constants for selected diatomicmolecules (391)2.Derivatives of ZPE with respect tospectroscopic constants,c (392)3.Estimated standard uncertainties associatedwith some diatomic ZPEs͑cm−1units͒asdetermined using the full͓Eq.͑6͔͒,diagonal͓Eq.͑7͔͒,and pessimistic͓Eq.͑8͔͒approximations (393)4.Truncation bias with respect to sextic diatomicvibrational model (394)5.Experimental vibrational zero-point energies͑cm−1͒for selected diatomic molecules (395)1.IntroductionOne of the most popular uses of computational quantum chemistry models is to predict molecular energetics,which is required for applications such as thermochemistry and reac-tion kinetics.As a result of steady progress in electronic structure theory and computational efficiency,the precision of ab initio molecular energetics has improved by a factor of about10every10years.With the advent of basis-set ex-trapolation methods and highly correlated theories,the ener-getics of small molecules can now be computed to high pre-cision almost routinely.1As the uncertainties in electronic energy fall away,other sources of uncertainty become in-creasingly noticeable.Vibrational zero-point energy͑ZPE͒has emerged as one of the principal remaining sources of uncertainty in calculations of molecular energetics.In careful work,it is necessary to go beyond the harmonic approxima-tion to obtain a reliable ZPE.2–5However,the most common practice is simply to scale the computed harmonic ZPE by a multiplicative correction factor.The empirical scaling factor carries uncertainty.We have recently quantified the uncer-tainties associated with scaling factors for fundamental vi-brational frequencies.6The uncertainties associated with the experimental vibrational frequencies were required in the analysis,although in the end their contribution was small enough to be neglected.Similar work is underway to provide scaling factors,with their associated uncertainties,appropri-ate for routine predictions of ZPE.Unfortunately,we have been unable to locate any recent compilations of experimen-tally derived ZPEs,or compilations of any age that include uncertainties.Moreover,existing compilations include few data for polyatomic molecules.We are now working tofill this gap by providing benchmark,experimental ZPEs along with the associated uncertainties.The present list,restricted to diatomic molecules,is not intended to be exhaustive.Nev-ertheless,it is the largest such list yet assembled,thefirst to employ spectroscopic data more recent than those compiled by Huber and Herzberg,7and thefirst to include uncertainties.2.Definition of Zero-Point EnergyIn quantum chemistry,the Born-Oppenheimer approxima-tion͑BOA͒is almost8always accepted.Thus,the most com-mon definition of the molecular ZPE is the energy difference between the vibrational ground state and the lowest point on the Born-Oppenheimer potential energy surface.Unfortu-a͒karl.irikura@©2007by the U.S.Secretary of Commerce on behalf of the United States.All rights reserved..0047-2689/2007/36…2…/389/9/$42.00J.Phys.Chem.Ref.Data,Vol.36,No.2,2007389nately,this definition is not convenient for experimentalZPEs,as the BOA is never adopted by real molecules.Ex-perimental spectra are generally analyzed,however,as if theBOA were followed by real molecules;explicit Born-Oppenheimer corrections are only made when simulta-neouslyfitting different isotopologs to the same effective po-tential.Thus,in the present compilation,the experimentalZPE is defined as the difference between the molecularground state and the lowest point on its isotope-specific ef-fective potential.The small difference between this definitionand that used by quantum chemists must be absorbed by theempirical scaling factor that is typically applied to theoreti-cally determined ZPEs.The ZPE cannot be measured directly since no moleculecan be observed below its ground state.Instead,the term“experimental ZPE”describes a value that is usually͑but notnecessarily9͒derived by combining experimental spectro-scopic constants with standard theoretical or empirical mod-els for anharmonic oscillators.Thus,“experimental”ZPEvalues are actually hybrids of experiment and theoreticalmodeling.Most available experimental ZPEs are for diatomic mol-ecules,because far fewer spectroscopic constants are neededfor diatomic molecules than for polyatomic molecules.Al-though thefield of molecular spectroscopy is home tocrowds of molecular constants,among nonspecialists themost common expression for the vibrational energy levels ofa diatomic molecule,relative to the minimum on the poten-tial energy curve,isG͑v͒=e͑v+12͒−e x e͑v+12͒2.͑1͒In Eq.͑1͒,e ande x e are the harmonic frequency and the first anharmonicity constant,respectively,and v is the vibra-tional quantum number,which can assume nonnegative inte-ger values.10Note that the symbole x e represents a single constant,not a product.Thus,the most popular expression for diatomic ZPE is,to second order in͑v+12͒,ZPE=G͑0͒=12e−14e x e.͑2͒This expression is derived by extrapolating Eq.͑1͒to v i=−1 2,which corresponds to the lowest point on the effectivepotential,to a good approximation.11Contributions from higher-order anharmonicities are generally negligible͑e.g., 0.1cm−1for H2,0.07cm−1for OH,and0.0013cm−1for CO͒.Unfortunately,as has been pointed out recently,2–5the popular expression is incorrect.In addition to the linear and quadratic terms in Eq.͑1͒, there is a constant term that is usually overlooked.This was demonstrated by Dunham in his classic power-series analysis.12The resulting energy levels,to second order,are given byG͑v͒=Y00+Y10͑v+12͒+Y20͑v+12͒2,͑3͒where Y10Ϸe and Y20Ϸ−e x e to good approximations.The constant Y00does not influence the line positions͑i.e.,energy intervals͒in a spectrum but contributes to the ZPE.In this paper we include Y00and even the third-order term͑second anharmonicity constant͒when available.Thus,ZPEs of di-atomic molecules are taken to beZPE=G͑0͒=Y00+12e−14e x e+18e y e,͑4͒where Y30Ϸe y e.To a good approximation,Y00can be ex-pressed in terms of conventional spectroscopic constants as13Y00ϷB e4+␣ee12B e+␣e2e2144B e3−e x e4.͑5͒The Dunham constants that correspond to the conventional rotational constants here are Y01ϷB e and Y11Ϸ−␣e.The value of Y00is largest for H2͑Y00=8.9cm−1͒,smaller for hydrides such as OH͑Y00=3.0cm−1͒,and less than one wavenumber for nonhydrides such as CO͑Y00=0.2cm−1͒.3.Selection of MoleculesAn initial list of molecules and associated ground-state spectroscopic constants was culled from the classic compila-tion by Huber and Herzberg7and from the NIST Diatomic Spectral Database.14For compatibility with the NIST Com-putational Chemistry Comparison and Benchmark Database,15only molecules composed of elements lighter than argon͑i.e.,atomic number ZϽ18͒were included.More recent values of constants were taken from the spectroscopic literature as available.Since our goal is a list of reliable ZPEs,diatomic molecules were excluded if their ZPEs obvi-ously͑upon cursory analysis͒had standard uncertainties of about0.5cm−1or more.Ourfinal list of diatomic molecules, with their spectroscopic constants,is provided in Table1.͑In Table1,and throughout this paper,H refers specifically to protium and D to deuterium.͒Standard uncertainties are listed in the spectroscopic style.For example,the quantity 12.345±0.067would be written12.345͑67͒.When the ground state is classically degenerate͑usually2⌸͒,averaged constants are used,as reported in the experimental sources cited.This choice was made for convenient comparison with conventional,non-relativistic quantum chemistry calcula-tions.For Cl2+,only separate constants for2⌸3/2and1⌸3/2 were reported.74.Contributions to UncertaintiesThe spectroscopic constants have both statistical uncer-tainty͑i.e.,uncertainty from random effects͒and bias͑i.e., systematic error͒.As we are interested in the ZPE,it is nec-essary to determine how the uncertainties in the constants contribute to the uncertainty in the corresponding ZPE.For this purpose,we accept the common linearized propagation of uncertainties16given byy2Ϸ͚iii͑ץy/ץx i͒2+2͚iϽjij͑ץy/ץx i͒͑ץy/ץx j͒,͑6͒for a quantity y=f͑x1,x2,...,x i,...͒,wherey2is the esti-mated variance of y andij=͗͑x i−x¯i͒͑x j−x¯j͒͘is the element of the covariance matrix that corresponds to the pair of vari-390K.K.IRIKURA J.Phys.Chem.Ref.Data,Vol.36,No.2,2007EXPERIMENTAL VIBRATIONAL ZERO-POINT ENERGIES391 T ABLE1.Spectroscopic constants for selected diatomic molecules.Standard uncertainties͑1͒are between parentheses and refer to the least significant digits. Values are in wavenumber units͑cm−1͒.Moleculeee x ee y e B e␣e ReferenceFirst-row elements onlyH24401.213͑18͒121.336͑18͒0.8129͑18͒60.8530͑18͒ 3.0622͑18͒7HD3813.15͑18͒91.65͑9͒0.723͑9͒45.655͑9͒ 1.986͑9͒7D23115.50͑9͒61.82͑9͒0.562͑9͒30.4436͑18͒ 1.0786͑9͒7Second-row elementsBeH2061.235͑15͒37.327͑22͒0.084͑17͒10.31992͑5͒0.3084͑2͒22BeD1529.986͑11͒20.557͑12͒0.035͑7͒ 5.68830͑3͒0.1261͑1͒22Be18O1457.09͑22͒11.311͑74͒0.0143͑83͒ 1.5847͑5͒0.01784͑15͒33BF1402.15865͑26͒11.82106͑15͒0.051595͑35͒ 1.51674399͑21͒0.01904848͑22͒23BH2366.7296͑16͒49.33983͑99͒0.362͑10͒12.025755͑45͒0.421565͑22͒34BO1885.286͑41͒11.694͑11͒−0.00952͑83͒ 1.781110͑31͒0.016516͑17͒35C21855.0663͑63͒13.6007͑54͒−0.116͑2͒ 1.820053͑11͒0.0179143͑44͒36C2−1781.189͑18͒11.6717͑48͒0.009981͑28͒ 1.74666͑32͒0.01651͑46͒37CF1307.93͑37͒11.08͑12͒0.093͑15͒ 1.41626͑48͒0.01844͑17͒38CH2860.7508͑98͒64.4387͑85͒0.3634͑27͒14.45988͑20͒0.53654͑33͒39CD2101.05193͑55͒34.72785͑58͒0.14147͑10͒7.8079823͑55͒0.212240͑11͒40CN2068.648͑11͒13.0971͑68͒−0.0124͑17͒ 1.89978316͑67͒0.0173720͑12͒41CO2169.75589͑8͒13.28803͑2͒0.0104109͑14͒ 1.9316023͑7͒0.01750513͑14͒42CO+2214.127͑35͒15.094͑21͒−0.0117͑34͒ 1.976941͑39͒0.018943͑34͒43and44F2916.929͑10͒11.3221͑10͒−0.10572͑67͒0.889294͑11͒0.0125952͑24͒45HF4138.3850͑7͒89.9432͑7͒0.92449͑31͒20.953712͑2͒0.7933704͑65͒46Li2351.4066͑10͒ 2.58324͑41͒−0.00583͑7͒0.6725297͑50͒0.0070461͑16͒47and48 LiF910.57272͑10͒8.207956͑46͒0.569166͑82͒ 1.34525715͑57͒0.02028749͑19͒49LiH1405.49805͑76͒23.1679͑7͒0.17093͑28͒7.5137315͑9͒0.2163911͑24͒50LiD1054.93973͑32͒13.05777͑21͒0.075478͑50͒ 4.23308131͑46͒0.09149428͑84͒50LiO814.62͑15͒7.78͑15͒NA 1.21282948͑11͒0.0178990͑25͒27N22358.57͑9͒14.324͑9͒−0.00226͑9͒ 1.998241͑18͒0.017318͑9͒7N2+2207.0115͑60͒16.0616͑23͒−0.04289͑25͒ 1.93176͑9͒0.01881͑9͒vib,51rot7 NF1141.37͑9͒8.99͑9͒NA 1.205679͑53͒0.014889͑53͒vib,7rot52 NH3282.72͑10͒79.04͑8͒0.367͑23͒16.66792͑6͒0.65038͑17͒53ND2399.126͑30͒42.106͑21͒0.1203͑54͒8.90867͑15͒0.25457͑19͒54NO1904.1346͑18͒14.08836͑89͒0.01005͑20͒ 1.7048885͑21͒0.0175416͑14͒55NO+2376.72͑11͒16.255͑18͒−0.01562͑92͒ 1.997195͑89͒0.018790͑69͒56O21580.161͑9͒11.95127͑9͒0.0458489͑9͒ 1.44562͑9͒0.0159305͑9͒57O2+1905.892͑82͒16.489͑13͒0.02057͑90͒ 1.689824͑91͒0.019363͑37͒58FO1053.0138͑12͒9.9194͑13͒−0.06096͑59͒ 1.05870763͑81͒0.0132951͑23͒59OH3737.761͑18͒84.8813͑18͒0.5409͑18͒18.9108͑18͒0.7242͑9͒7OD+2271.80͑9͒44.235͑9͒0.4267͑18͒8.9116͑18͒0.2896͑9͒7Third-row elementsAlCl481.77466͑20͒ 2.101811͑88͒0.006638͑15͒0.243930066͑12͒0.001611082͑12͒60AlF802.32447͑11͒ 4.849915͑44͒0.0195738͑68͒0.552480208͑65͒0.004984261͑44͒23AlH1682.37474͑31͒29.05098͑29͒0.24762͑12͒ 6.3937842͑17͒0.1870527͑15͒61AlD1211.77402͑15͒15.06477͑11͒0.09244͑4͒ 3.3183929͑8͒0.0698773͑4͒61AlO979.4852͑50͒7.0121͑32͒−0.00206͑56͒0.6413856͑54͒0.0057796͑8͒62AlS617.1169͑33͒ 3.3310͑19͒−0.00924͑32͒0.2800368͑33͒0.00178225͑55͒63BCl840.29472͑63͒ 5.49170͑33͒0.02995͑7͒0.684282͑12͒0.0068124͑14͒17BeS997.94͑9͒ 6.137͑9͒NA0.79059͑9͒0.00664͑9͒7BS1179.91͑3͒ 6.25͑3͒−0.0083͑58͒0.79478͑5͒0.00578͑4͒64CCl876.89749͑69͒ 5.44698͑54͒0.02607͑15͒0.697137͑34͒0.00685277͑45͒25Cl2559.751͑20͒ 2.69427͑20͒−0.003325͑2͒0.24415͑20͒0.001516͑20͒65Cl2+645.61͑9͒ 3.015͑9͒0.007͑9͒0.26950͑18͒0.00164͑9͒7͑⍀=1/2͒644.77͑9͒ 2.988͑9͒NA0.2697͑9͒0.00167͑9͒7ClF783.4534͑24͒ 4.9487͑6͒−0.0176͑1͒0.5164805͑31͒0.0043385͑8͒66ClO853.64268͑13͒ 5.51828͑6͒−0.01256͑30͒0.62345797͑4͒0.0059357͑1͒28CP1239.79924͑8͒ 6.833769͑46͒−0.001377͑7͒0.79886775͑8͒0.00596933͑19͒67CS1285.15464͑10͒ 6.502605͑53͒0.003887͑9͒0.82004356͑4͒0.00591835͑5͒68HCl2990.9248͑15͒52.8000͑15͒0.21803͑55͒10.5933002͑13͒0.3069985͑41͒69J.Phys.Chem.Ref.Data,Vol.36,No.2,2007ables x i and x j.For the problem at hand,y is the ZPE and the x i are generic spectroscopic constants.The required deriva-tives,summarized in Table2,are straightforward but do not appear to have been compiled previously.Typically,only the diagonal elements,ii,of the covari-ance matrix͑i.e.,the variances of thefitting constants͒are reported and available to us.Omitting the off-diagonal con-tributions in Eq.͑6͒yields the estimatey2Ϸ͚ii2͑ץy/ץx i͒2,͑7͒where the standard uncertainties of the x i arei=ͱii.This diagonal approximation introduces a bias in the estimated uncertainty,which will be different for different molecules. To estimate the magnitude of the bias,we consider the situ-ation for BCl,for which the covariance matrix has been published.17Using the derivatives in Table2and the param-eter values and statistical uncertainties from Table1,the standard uncertainty͑i.e.,the square root of the variance͒for the ZPE is0.00065cm−1.When only the diagonal terms are included,the resulting standard uncertainty is only slightly smaller,0.00057cm−1.Given only the variances,the most pessimistic scenario for the correlation coefficients r ij͑−1Յr ijϵij/ijՅ1͒leads to the upper boundT ABLE1.Spectroscopic constants for selected diatomic molecules.Standard uncertainties͑1͒are between parentheses and refer to the least significant digits. Values are in wavenumber units͑cm−1͒.—ContinuedMoleculeee x ee y e B e␣e ReferenceDCl2145.1326͑11͒27.1593͑7͒0.07993͑20͒ 5.4487838͑6͒0.1132345͑15͒69HCl+2673.69͑9͒52.537͑9͒NA9.95661͑18͒0.32716͑18͒7LiCl642.95453͑93͒ 4.47253͑40͒0.020118͑49͒0.70652247͑20͒0.00801019͑32͒70NaLi256.5412͑19͒ 1.62271͑96͒−0.00495͑22͒0.3758620͑89͒0.0031465͑15͒30Mg251.121͑18͒ 1.645͑9͒0.01624͑18͒0.09287͑9͒0.003776͑18͒7MgH1492.7763͑36͒29.847͑4͒−0.3048͑20͒ 5.8255229͑41͒0.177298͑14͒71MgD1077.2976͑26͒15.521͑2͒−0.1184͑7͒ 3.0343436͑21͒0.066607͑5͒71MgO785.2183͑6͒ 5.1327͑3͒0.01649͑7͒0.5748414͑3͒0.0053223͑3͒72MgS528.74͑9͒ 2.704͑9͒NA0.26797͑9͒0.00176͑9͒7Na2159.08548͑44͒0.70866͑29͒−0.004632͑77͒0.15473537͑29͒0.00086375͑11͒29NaCl364.6842͑4͒ 1.7761͑2͒0.005937͑35͒0.21806302͑8͒0.00162479͑6͒73NaF535.65805͑21͒ 3.57523͑13͒0.018453͑34͒0.43690153͑7͒0.00455918͑7͒74NaH1171.968͑12͒19.703͑10͒0.175͑2͒ 4.90327͑13͒0.1370͑4͒75NCl827.95767͑75͒ 5.30015͑61͒−0.00480͑19͒0.649767390͑85͒0.00641432͑20͒76P2780.77͑9͒ 2.835͑18͒−0.00462͑9͒0.30362͑9͒0.00149͑9͒7P2+672.20͑9͒ 2.74͑9͒NA0.27600͑18͒0.00151͑9͒7PF846.75͑9͒ 4.489͑9͒0.019͑9͒0.5667427͑35͒0.00456͑9͒7but B e77 PH2363.774͑36͒43.907͑27͒0.1059͑73͒8.53904͑19͒0.25339͑28͒54PN1336.948͑20͒ 6.8958͑57͒−0.00605͑48͒0.7864844͑28͒0.0055337͑36͒78PO1233.34͑9͒ 6.56͑9͒NA0.733223657͑22͒0.005466162͑50͒vib,7rot79S2725.7102͑97͒ 2.8582͑25͒NA0.29539516͑30͒0.00159754͑59͒80SF837.6418͑5͒ 4.46953͑18͒NA0.555173͑4͒0.004459͑10͒81SH2696.2475͑58͒48.7420͑28͒0.1124͑6͒9.600247͑51͒0.27990͑10͒82Si2510.98͑9͒ 2.02͑9͒NA0.2390͑9͒0.00135͑18͒7SiCl535.59͑2͒ 2.1757͑50͒0.00604͑36͒0.256103͑14͒0.0015817͑72͒83SiF837.32507͑22͒ 4.83419͑9͒0.019807͑16͒0.58125735͑21͒0.00503859͑39͒84SiH2042.5229͑8͒36.0552͑5͒0.1254͑1͒7.503898͑30͒0.21814͑2͒85SiH+2157.17͑9͒34.24͑9͒NA7.6603͑9͒0.2096͑9͒7SiN1151.284͑43͒ 6.455͑21͒−0.0069͑20͒0.730927͑15͒0.005685͑30͒86bute y e87 SiO1241.54388͑7͒ 5.97437͑2͒0.006090͑3͒0.72675206͑2͒0.00503784͑1͒88SiS749.64559͑7͒ 2.58623͑4͒0.001048͑9͒0.303527856͑6͒0.001473130͑3͒88SO1150.7913͑10͒ 6.4096͑5͒0.01306͑11͒0.72082210͑2͒0.00575080͑2͒vib,89rot90T ABLE2.Derivatives of ZPE with respect to spectroscopic constants,c.cץ͑ZPE͒/ץcZPE=e2−e x e2+e y e8+B e4+␣ee12B e+␣e2e2144B e3e1/2+B ee s͑2s+1͒,where sϵ␣ee12B ee x e−1/2e y e1/8B e1/4−s͑3s+1͒␣e B e␣e s͑2s+1͒392K.K.IRIKURA J.Phys.Chem.Ref.Data,Vol.36,No.2,2007yՅ͚ii͉ץy/ץx i͉,͑8͒oryՅ0.00098cm−1for the BCl example.This is50% larger than the result from Eq.͑6͒and appears inferior to the diagonal approximation.These various estimates are summa-rized in Table3,along with analogous estimates for three other diatomic molecules,based upon unpublishedfitting data generously provided by Dr.F.J.Lovas.Based upon the data for these four molecules,the diagonal approximation appears reasonable and in this paper we use Eq.͑7͒to esti-mate the statistical contribution to the standard uncertainty of the diatomic ZPEs.Most of the standard uncertainties presented in Table1are from the experimental papers in which the constants were reported.In some cases,uncertainties were reported but not described;they have been assumed to represent standard un-certainties͑1͒.When uncertainties have been described as 95%confidence intervals,they have been divided by2to estimate standard uncertainties.Other special cases are de-scribed in Sec.5.The uncertainties associated with spectroscopic constants only reflect the statistical uncertainties resulting fromfitting the observed line positions to a model Hamiltonian.In this context,a Hamiltonian is a physically motivatedfitting func-tion involving selected spectroscopic constants and various quantum numbers.In some cases,the uncertainties were propagated from v-dependentfitting constants or from esti-mated͓type B͑Ref.18͔͒uncertainties in line positions.In addition to the statistical uncertainties,there are sources of bias͑“systematic error”͒which may dominate the com-bined uncertainties.True bias cannot be known,since that requires knowledge of true values.When ancillary informa-tion is available about the possible values of bias,we can make a correction for bias,as recommended in the Guide to the Expression of Uncertainty in Measurement,published by the International Organization for Standardization.19There is uncertainty associated with the correction for bias,corre-sponding to the uncertainty of the ancillary information.If we have no information,we choose the value of the correc-tion to be zero,but there is still an uncertainty associated with this͑null͒value.This uncertainty is combined with the statistical uncertainties to obtain the combined uncertainty. One source of bias derives from the truncation in Taylor-series expansions such as Eq.͑1͒.Such model-dependent uncertainties are seldom discussed.20–22Thefitted values of low-order constants are affected when higher-order constants are included in thefitting procedure.Further,our ZPE com-putation͓Eq.͑4͔͒includes terms only through third order. For diatomic molecules,we estimate the uncertainty due to truncation using the simplified analytical model described in the following paragraph.Assume,for this approximate model,that the diatomic vi-brational energy levels are perfectly described by the sextic polynomialG͑v͒=͚i=06b i͑v+12͒i,͑9͒where v is the quantum number,G͑v͒is the energy of the associated level,and the b i are constants.The exact zero-point energy within this model isZPE=b0+12b1+14b2+18b3+116b4+132b5+164b6.͑10͒Also suppose that n transition frequencies,y j=G͑j͒−G͑0͒, are observed and arefitted to the empirical expressionG͑v͒Ϸ͚i=0na i͑v+12͒i,͑11͒where a i=Y i0or the equivalent conventional constant and n Յ6.This is an exactfit because we have assumed,for math-ematical convenience,that the number of free parameters is equal to the number of data.Then the apparent zero-point energy isZPE app=a0+12a1+14a2+18a3.͑12͒ZPE app depends upon the number,n,offitted constants even though the higher-order constants are not explicit in Eq.͑12͒. The difference͑ZPE app−ZPE͒,determined from Eqs.͑10͒and͑12͒,is the truncation bias in this simple model;its ab-solute value is an estimate for the uncertainty arising from truncation of the spectroscopic Hamiltonian.The biases in the cubic approximation͑12͒,for different orders n of the fitting polynomial͑11͒,are listed in the second column of Table4.The value for n=2uses the more severely truncatedquadratic expression ZPE app=a0+12a1+14a2instead of Eq.͑12͒,because a3is undefined.The differences͑a0−b0͒, which appear in the expressions for estimated bias,require evaluation.If we identify a0and b0with Y00,we can use Eq.͑5͒to estimate that͑a0−b0͒Ϸ١Y00·͑a−b͒=␣e12B eͩ1+␣e6B e2eͪ͑a1−b1͒−14͑a2−b2͒.͑13͒The expressions for͑a1−b1͒and for͑a2−b2͒are given in the last two columns of Table4.To obtain numerical values for the uncertainty arising from truncation for a particular di-atomic molecule,we determine n based upon the order of the experimentalfit,choose b iϷY i0,and combine Eq.͑13͒with the expressions in Table4.However,frequently the higherT ABLE3.Estimated standard uncertainties associated with some diatomicZPEs͑cm−1units͒as determined using the full͓Eq.͑6͔͒,diagonal͓Eq.͑7͔͒,and pessimistic͓Eq.͑8͔͒approximations.Molecule Full Diagonal a PessimisticBCl0.000650.00057͑−12%͒0.00098͑+51%͒CS0.000440.00045͑+2%͒0.00050͑+14%͒SiO0.000330.00052͑+58%͒0.00082͑+148%͒SiS0.000360.00037͑+3%͒0.00041͑+14%͒a Percentage deviation from“full”value given between parentheses.EXPERIMENTAL VIBRATIONAL ZERO-POINT ENERGIES393J.Phys.Chem.Ref.Data,Vol.36,No.2,2007Y i0are unknown.As typically͉Y i+1,0͉Ͻ͉Y i0͉,we estimate the missing Y i0crudely by geometric extrapolation from the two highest measured Y i0.As pointed out by a referee,Y i0and Y i+1,0usually differ in sign.Thus,an alternating sign is as-sumed in the present work.However,for about one-third of the data at hand,the sign does not change,i.e.,Y i0Y i−1,0Ͼ0͑for iϾ3͒.To test the sensitivity of the results to the choice of sign,we consider a positive sign in the extrapolation.This changes the magnitude of͑ZPE app−ZPE͒by a mean factor of 1.8͑standard deviationϭ1.5͒.Thus,we multiply the trunca-tion bias values by1.8to reflect the uncertainty of the ex-trapolation.Among the remaining sources of uncertainty,the most im-portant is probably that Dunham’s canonical treatment is it-self an approximation that leads to bias.In particular,any perturbations from electronically or vibrationally excited states will displace some rovibrational levels,skewing the values of thefitting constants.These effects are specific to individual molecules.We do not attempt to quantify the as-sociated uncertainties.However,they are probably small for the ground states of diatomic molecules.Dunham’s analysis is also for non-degenerate electronic states͑1⌺͒and must be considered more approximate in other cases.To illustrate the current procedure,consider BF as an ex-ample.Constants from Table1are substituted into Eq.͑5͒to obtain Y00=0.3111cm−1.Applying Eq.͑4͒yields ZPE =698.4416cm−1,as listed in Table5.The associated uncer-tainty is computed by propagating the uncertainties in the spectroscopic constants,estimating the uncertainty associ-ated with the͑null͒correction for truncation bias,and com-bining these two quantities to obtain a combined standard uncertainty.To propagate uncertainties,substitute values from Table1into the formulas in Table2to obtain the partial derivativesץ͑ZPE͒/ץ͑e͒=0.50307,ץ͑ZPE͒/ץ͑e x e͒=−0.5,ץ͑ZPE͒/ץ͑e y e͒=0.125,ץ͑ZPE͒/ץ͑B e͒=−3.5257,and ץ͑ZPE͒/ץ͑␣e͒=226.11.The intermediate quantity s =0.96750͑Table2͒.The standard uncertainties for the spec-troscopic constants,from Table1,are combined with thesederivatives according to Eq.͑7͒to obtain the estimated sta-tistical contribution to the variance of the ZPE.Taking thesquare root gives u statϷ0.000159cm−1,as listed in Table5͑rounded to two digits͒.To estimate the uncertainty associ-ated with the͑null correction for͒truncation bias,check theliterature͑Zhang et al.23͒tofind the available Dunham con-stants Y n0.In this case,Y40=0.0003464cm−1and no higher constants are available.Thus,the appropriate row in Table4 is that for n=4.Values of b j in Table4are approximated as b jϷY j0.The missing constants are estimated by geometric extrapolation with sign alternation as Y i+1,0Ϸ−͉Y i,02/Y i−1,0͉͑Y i,0/͉Y i,0͉͒,so Y50Ϸ−2.33ϫ10−6cm−1and Y60Ϸ1.56ϫ10−8cm−1.Then from the third column of Table 4,͑a1−b1͒=0.000225cm−1and from the fourth column of Table4,͑a2−b2͒=−0.000255cm−1.Substituting these val-ues into Eq.͑13͒then gives͑a0−b0͒=6.435ϫ10−5cm−1.Fi-nally the expression in the second column of Table4evalu-ates to͑ZPE app−ZPE͒=0.000107cm−1.Multiplying by1.8 to account for sensitivity to guessed Y i0values,and taking the absolute value,u truncϷ0.000193cm−1.Thefinal,esti-mated,combined standard uncertainty is then=͑u stat2 +u trunc2͒1/2=0.000250cm−1,which is rounded to a single digit in Table5.5.Analyses for Individual MoleculesIn their classic compilation,Huber and Herzberg͑H&H͒did not attempt to estimate the uncertainties associated withthe reported spectroscopic constants.7Instead,they providedthe following guidance.“…we hope that the number of dig-its quoted may serve as a very rough indication of the esti-mated order of magnitude of the error,generally±9units ofthe last decimal place.Where the last digit is given as asubscript,we expect that the uncertainty may considerablyT ABLE4.Truncation bias with respect to sextic diatomic vibrational model͓Eqs.͑9͒–͑13͔͒.The order of thefitting polynomial is n. n ZPE app−ZPE͑a1−b1͒͑a2−b2͒60005͑a0−b0͒−116b4−132b5+6794b648818b6−1213916b64͑a0−b0͒−116b4−48516b5−600516b6−168916b5−1290b64754b5+2206116b63͑a0−b0͒+10516b4+1052b5+892532b622b4+271116b5+17652b6−432b4−150b5−1190916b62͑a0−b0͒−158b3−13516b4−43516b5−247532b6−234b3−24b4−119916b5−210b692b3+292b4+1654b5+177116b6394K.K.IRIKURA J.Phys.Chem.Ref.Data,Vol.36,No.2,2007。
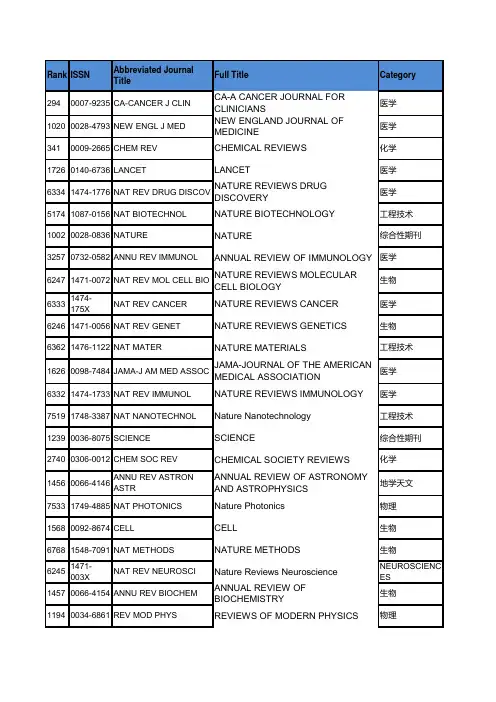
Rank ISSN Abbreviated JournalTitleFull Title Category2940007-9235CA-CANCER J CLIN CA-A CANCER JOURNAL FOR CLINICIAN医学10200028-4793NEW ENGL J MED NEW ENGLAND JOURNAL OF MEDICINE医学3410009-2665CHEM REV CHEMICAL REVIEWS化学17260140-6736LANCET LANCET医学63341474-1776NAT REV DRUG DISCOV NATURE REVIEWS DRUG DISCOVERY医学51741087-0156NAT BIOTECHNOL NATURE BIOTECHNOLOGY工程技术10020028-0836NATURE NATURE综合性期刊32570732-0582ANNU REV IMMUNOL ANNUAL REVIEW OF IMMUNOLOGY医学62471471-0072NAT REV MOL CELL BIO NATURE REVIEWS MOLECULAR CELL B生物63331474-175X NAT REV CANCER NATURE REVIEWS CANCER医学62461471-0056NAT REV GENET NATURE REVIEWS GENETICS生物63621476-1122NAT MATER NATURE MATERIALS工程技术16260098-7484JAMA-J AM MED ASSOC JAMA-JOURNAL OF THE AMERICAN ME医学63321474-1733NAT REV IMMUNOL NATURE REVIEWS IMMUNOLOGY医学75191748-3387NAT NANOTECHNOL Nature Nanotechnology工程技术12390036-8075SCIENCE SCIENCE综合性期刊27400306-0012CHEM SOC REV CHEMICAL SOCIETY REVIEWS化学14560066-4146ANNU REV ASTRON ASTR ANNUAL REVIEW OF ASTRONOMY AND地学天文75331749-4885NAT PHOTONICS Nature Photonics物理15680092-8674CELL CELL生物67681548-7091NAT METHODS NATURE METHODS生物62451471-003X NAT REV NEUROSCI Nature Reviews Neuroscience NEUROSCIENCES 14570066-4154ANNU REV BIOCHEM ANNUAL REVIEW OF BIOCHEMISTRY生物11940034-6861REV MOD PHYS REVIEWS OF MODERN PHYSICS物理48751061-4036NAT GENET NATURE GENETICS生物15060079-6425PROG MATER SCI PROGRESS IN MATERIALS SCIENCE工程技术51051078-8956NAT MED NATURE MEDICINE医学11140031-9333PHYSIOL REV PHYSIOLOGICAL REVIEWS医学15100079-6700PROG POLYM SCI PROGRESS IN POLYMER SCIENCE化学76261755-4330NAT CHEM Nature Chemistry化学62371470-2045LANCET ONCOL LANCET ONCOLOGY医学74001740-1526NAT REV MICROBIOL NATURE REVIEWS MICROBIOLOGY生物66151535-6108CANCER CELL CANCER CELL医学67091543-5008ANNU REV PLANT BIOL ANNUAL REVIEW OF PLANT BIOLOGY生物63191473-3099LANCET INFECT DIS LANCET INFECTIOUS DISEASES医学150001-4842ACCOUNTS CHEM RES ACCOUNTS OF CHEMICAL RESEARCH化学80331934-5909CELL STEM CELL Cell Stem Cell生物57941364-6613TRENDS COGN SCI TRENDS IN COGNITIVE SCIENCES医学63351474-4422LANCET NEUROL LANCET NEUROLOGY医学14660066-4308ANNU REV PSYCHOL ANNUAL REVIEW OF PSYCHOLOGY医学50621074-7613IMMUNITY IMMUNITY医学19000163-769X ENDOCR REV ENDOCRINE REVIEWS医学580001-8732ADV PHYS ADVANCES IN PHYSICS物理17250140-525X BEHAV BRAIN SCI BEHAVIORAL AND BRAIN SCIENCES医学76141754-5692ENERG ENVIRON SCI Energy & Environmental Science化学74791745-2473NAT PHYS Nature Physics物理29820370-1573PHYS REP PHYSICS REPORTS-REVIEW SECTION 物理65501529-2908NAT IMMUNOL NATURE IMMUNOLOGY医学61991465-7392NAT CELL BIOL NATURE CELL BIOLOGY生物84372159-8274CANCER DISCOV Cancer discovery ONCOLOGY 18180147-006X ANNU REV NEUROSCI ANNUAL REVIEW OF NEUROSCIENCE医学60511433-8351LIVING REV RELATIV Living Reviews in Relativity物理28850360-1285PROG ENERG COMBUST PROGRESS IN ENERGY AND COMBUST工程技术医学68241553-4006ANNU REV PATHOL-MECH Annual Review of Pathology-Mechanisms o14640066-4278ANNU REV PHYSIOL ANNUAL REVIEW OF PHYSIOLOGY医学32580732-183X J CLIN ONCOL JOURNAL OF CLINICAL ONCOLOGY医学29200362-1642ANNU REV PHARMACOL ANNUAL REVIEW OF PHARMACOLOGY 医学1520003-4819ANN INTERN MED ANNALS OF INTERNAL MEDICINE医学地学天文39930935-4956ASTRON ASTROPHYS REV ASTRONOMY AND ASTROPHYSICS REV71031614-4961LIVING REV SOL PHYS LIVING REVIEWS IN SOLAR PHYSICS ASTRONOMY & AS 67881550-4131CELL METAB Cell Metabolism生物39970935-9648ADV MATER ADVANCED MATERIALS工程技术76391756-1833BMJ-BRIT MED J BMJ-British Medical Journal MEDICINE, GENER 36650893-8512CLIN MICROBIOL REV CLINICAL MICROBIOLOGY REVIEWS医学1830003-9926ARCH INTERN MED ARCHIVES OF INTERNAL MEDICINE医学10940031-6997PHARMACOL REV PHARMACOLOGICAL REVIEWS医学680002-5100ALDRICHIM ACTA ALDRICHIMICA ACTA化学11890034-4885REP PROG PHYS REPORTS ON PROGRESS IN PHYSICS物理生物26600301-5556ADV ANAT EMBRYOL CEL ADVANCES IN ANATOMY EMBRYOLOGY化学14630066-426X ANNU REV PHYS CHEM ANNUAL REVIEW OF PHYSICAL CHEMIS5210016-5085GASTROENTEROLOGY GASTROENTEROLOGY医学51251081-0706ANNU REV CELL DEV BI ANNUAL REVIEW OF CELL AND DEVELO生物医学32960735-1097J AM COLL CARDIOL JOURNAL OF THE AMERICAN COLLEGE20440169-5347TRENDS ECOL EVOL TRENDS IN ECOLOGY & EVOLUTION生物71041614-6832ADV ENERGY MATER Advanced Energy Materials CHEMISTRY, PHYS 52791097-6256NAT NEUROSCI NATURE NEUROSCIENCE医学化学59481389-5567J PHOTOCH PHOTOBIO C JOURNAL OF PHOTOCHEMISTRY AND P81491946-6234SCI TRANSL MED Science Translational Medicine CELL BIOLOGY医学64501520-765X EUR HEART J SUPPL EUROPEAN HEART JOURNAL SUPPLEM14600066-4197ANNU REV GENET ANNUAL REVIEW OF GENETICS生物39160927-796X MAT SCI ENG R MATERIALS SCIENCE & ENGINEERING 工程技术80491936-122X ANNU REV BIOPHYS Annual Review of Biophysics生物76891759-4758NAT REV NEUROL Nature Reviews Neurology医学21640195-668X EUR HEART J EUROPEAN HEART JOURNAL医学37130896-6273NEURON NEURON医学20380169-409X ADV DRUG DELIVER REV ADVANCED DRUG DELIVERY REVIEWS医学75131748-0132NANO TODAY Nano Today工程技术86088755-1209REV GEOPHYS REVIEWS OF GEOPHYSICS地学81551947-5454ANNU REV CONDEN MA P Annual Review of Condensed Matter Physi PHYSICS, CONDEN 19620167-5729SURF SCI REP SURFACE SCIENCE REPORTS化学11550033-2909PSYCHOL BULL PSYCHOLOGICAL BULLETIN医学5530017-5749GUT GUT医学51931088-9051GENOME RES GENOME RESEARCH生物生物52311092-2172MICROBIOL MOL BIOL R MICROBIOLOGY AND MOLECULAR BIOL83072047-7538LIGHT-SCI APPL Light-Science & Applications OPTICS 76811758-678X NAT CLIM CHANGE Nature climate change ENVIRONMENTAL 57581359-4184MOL PSYCHIATR MOLECULAR PSYCHIATRY医学1820003-990X ARCH GEN PSYCHIAT ARCHIVES OF GENERAL PSYCHIATRY医学3590009-7322CIRCULATION CIRCULATION医学67721549-1676PLOS MED PLOS MEDICINE MEDICINE, GENER 49041063-5157SYST BIOL SYSTEMATIC BIOLOGY生物81041941-1405ANNU REV MAR SCI Annual Review of Marine Science地学73441723-8617WORLD PSYCHIATRY World Psychiatry医学64881523-9829ANNU REV BIOMED ENG ANNUAL REVIEW OF BIOMEDICAL ENG工程技术76901759-4774NAT REV CLIN ONCOL Nature Reviews Clinical Oncology医学58401369-7021MATER TODAY Materials Today工程技术52771097-2765MOL CELL MOLECULAR CELL生物26720302-2838EUR UROL EUROPEAN UROLOGY医学14580066-4170ANNU REV ENTOMOL ANNUAL REVIEW OF ENTOMOLOGY生物65641530-6984NANO LETT NANO LETTERS工程技术19380166-2236TRENDS NEUROSCI TRENDS IN NEUROSCIENCES医学67381545-9993NAT STRUCT MOL BIOL NATURE STRUCTURAL & MOLECULAR BBIOCHEMISTRY & 76941759-5029NAT REV ENDOCRINOL Nature Reviews Endocrinology医学19350166-0616STUD MYCOL STUDIES IN MYCOLOGY生物20150168-6445FEMS MICROBIOL REV FEMS MICROBIOLOGY REVIEWS生物医学7260021-9738J CLIN INVEST JOURNAL OF CLINICAL INVESTIGATION84752168-6106JAMA INTERN MED JAMA Internal Medicine MEDICINE, GENER医学50501073-449X AM J RESP CRIT CARE AMERICAN JOURNAL OF RESPIRATORY68091552-4450NAT CHEM BIOL Nature Chemical Biology生物57731360-1385TRENDS PLANT SCI TRENDS IN PLANT SCIENCE生物14610066-4219ANNU REV MED ANNUAL REVIEW OF MEDICINE医学80481936-0851ACS NANO ACS Nano工程技术医学67671548-5943ANNU REV CLIN PSYCHO ANNUAL REVIEW OF CLINICAL PSYCHO76951759-5045NAT REV GASTRO HEPAT Nature Reviews Gastroenterology & Hepat医学10000027-8874JNCI-J NATL CANCER I JNCI-Journal of the National Cancer Institu ONCOLOGY医学7570022-1007J EXP MED JOURNAL OF EXPERIMENTAL MEDICINE44551001-0602CELL RES CELL RESEARCH生物68181552-5260ALZHEIMERS DEMENT Alzheimers & Dementia医学79881931-3128CELL HOST MICROBE Cell Host & Microbe生物1050002-953X AM J PSYCHIAT AMERICAN JOURNAL OF PSYCHIATRY医学3970010-8545COORDIN CHEM REV COORDINATION CHEMISTRY REVIEWS化学14620066-4227ANNU REV MICROBIOL ANNUAL REVIEW OF MICROBIOLOGY生物770002-7863J AM CHEM SOC JOURNAL OF THE AMERICAN CHEMICA化学84802168-622X JAMA PSYCHIAT JAMA Psychiatry PSYCHIATRY 43000962-8924TRENDS CELL BIOL TRENDS IN CELL BIOLOGY生物19870167-7799TRENDS BIOTECHNOL TRENDS IN BIOTECHNOLOGY工程技术76871759-0876WIRES COMPUT MOL SCI W IREs Computational Molecular Science CHEMISTRY, MUL 65801531-7331ANNU REV MATER RES ANNUAL REVIEW OF MATERIALS RESE工程技术1150003-0007B AM METEOROL SOC BULLETIN OF THE AMERICAN METEOR地学71151616-301X ADV FUNCT MATER ADVANCED FUNCTIONAL MATERIALS工程技术68331554-8627AUTOPHAGY Autophagy生物75871752-0894NAT GEOSCI Nature Geoscience地学医学19310165-6147TRENDS PHARMACOL SCI TRENDS IN PHARMACOLOGICAL SCIEN15560091-6749J ALLERGY CLIN IMMUN JOURNAL OF ALLERGY AND CLINICAL I医学82642041-1723NAT COMMUN Nature communications Biological Science Disciplines 20200168-8278J HEPATOL JOURNAL OF HEPATOLOGY医学化学60491433-7851ANGEW CHEM INT EDIT ANGEWANDTE CHEMIE-INTERNATIONA19030163-8998ANNU REV NUCL PART S ANNUAL REVIEW OF NUCLEAR AND PA物理43630968-0004TRENDS BIOCHEM SCI TRENDS IN BIOCHEMICAL SCIENCES生物14690067-0049ASTROPHYS J SUPPL S ASTROPHYSICAL JOURNAL SUPPLEME地学天文物理14590066-4189ANNU REV FLUID MECH ANNUAL REVIEW OF FLUID MECHANICS24880270-9139HEPATOLOGY HEPATOLOGY医学3600009-7330CIRC RES CIRCULATION RESEARCH医学940002-9297AM J HUM GENET AMERICAN JOURNAL OF HUMAN GENE生物74601744-4292MOL SYST BIOL Molecular Systems Biology生物61981465-6906GENOME BIOL GENOME BIOLOGY BIOTECHNOLOGY 36240890-9369GENE DEV GENES & DEVELOPMENT生物370001-6322ACTA NEUROPATHOL ACTA NEUROPATHOLOGICA医学930002-9270AM J GASTROENTEROL AMERICAN JOURNAL OF GASTROENTE医学61551461-023X ECOL LETT ECOLOGY LETTERS环境科学67111543-592X ANNU REV ECOL EVOL S ANNUAL REVIEW OF ECOLOGY EVOLU生物2530006-4971BLOOD BLOOD医学84252157-1724KIDNEY INT SUPPL Kidney International Supplements UROLOGY & NEPHROLOGY 23760261-4189EMBO J EMBO JOURNAL生物35670887-6924LEUKEMIA LEUKEMIA医学62881471-4906TRENDS IMMUNOL TRENDS IN IMMUNOLOGY医学医学1580003-4967ANN RHEUM DIS ANNALS OF THE RHEUMATIC DISEASES85252211-2855NANO ENERGY Nano Energy CHEMISTRY, PHYS 2400006-3223BIOL PSYCHIAT BIOLOGICAL PSYCHIATRY医学16200098-2997MOL ASPECTS MED MOLECULAR ASPECTS OF MEDICINE医学57341355-4786HUM REPROD UPDATE HUMAN REPRODUCTION UPDATE医学16720105-2896IMMUNOL REV IMMUNOLOGICAL REVIEWS医学79641884-4049NPG ASIA MATER NPG Asia Materials MATERIALS SCIEN 81361943-8206ADV OPT PHOTONICS Advances in Optics and Photonics OPTICS24090265-0568NAT PROD REP NATURAL PRODUCT REPORTS化学85472214-109X LANCET GLOB HEALTH Lancet Global Health PUBLIC, ENVIRON 19010163-7827PROG LIPID RES PROGRESS IN LIPID RESEARCH生物28830360-0564ADV CATAL ADVANCES IN CATALYSIS化学26360301-0082PROG NEUROBIOL PROGRESS IN NEUROBIOLOGY医学29480364-5134ANN NEUROL ANNALS OF NEUROLOGY医学20260168-9525TRENDS GENET TRENDS IN GENETICS生物76911759-4790NAT REV RHEUMATOL Nature Reviews Rheumatology医学7130021-9525J CELL BIOLJOURNAL OF CELL BIOLOGY 生物18980163-7258PHARMACOL THERAPEUT P HARMACOLOGY & THERAPEUTICS 医学66051534-5807DEV CELL DEVELOPMENTAL CELL 生物9990027-8424P NATL ACAD SCI USA PROCEEDINGS OF THE NATIONAL ACA 综合性期刊76111754-2189NAT PROTOC Nature Protocols 生物61911464-7931BIOL REV BIOLOGICAL REVIEWS 生物85402213-2600LANCET RESP MED Lancet Respiratory Medicine RESPIRATORY SYSTEM 14650066-4286ANNU REV PHYTOPATHOL ANNUAL REVIEW OF PHYTOPATHOLOG 生物62141467-5463BRIEF BIOINFORM BRIEFINGS IN BIOINFORMATICS 生物33610742-3098J PINEAL RES JOURNAL OF PINEAL RESEARCH 医学42810960-9822CURR BIOL CURRENT BIOLOGY 生物84242157-1422CSH PERSPECT MED Cold Spring Harbor perspectives in medicin MEDICINE, RESEA 62891471-4914TRENDS MOL MED TRENDS IN MOLECULAR MEDICINE 医学47001043-2760TRENDS ENDOCRIN MET TRENDS IN ENDOCRINOLOGY AND MET 医学47361046-6673J AM SOC NEPHROL JOURNAL OF THE AMERICAN SOCIETY 医学67321545-7885PLOS BIOL PLoS Biology BIOCHEMISTRY &46501040-4651PLANT CELL PLANT CELL 生物47171044-579X SEMIN CANCER BIOL SEMINARS IN CANCER BIOLOGY 医学3220008-5472CANCER RES CANCER RESEARCH 医学83162050-084X ELIFE eLife BIOLOGY 84112155-5435ACS CATAL ACS catalysis CHEMISTRY, PHYSICAL 75601751-7362ISME J ISME Journal 环境科学82692041-6520CHEM SCI Chemical science CHEMISTRY, MULT 2610006-8950BRAIN BRAIN 医学11620033-3190PSYCHOTHER PSYCHOSOM PSYCHOTHERAPY AND PSYCHOSOMAT 医学43460966-842X TRENDS MICROBIOL TRENDS IN MICROBIOLOGY 生物85462213-8587LANCET DIABETES ENDO Lancet Diabetes & Endocrinology ENDOCRINOLOGY 76931759-5002NAT REV CARDIOL Nature Reviews Cardiology 医学26130300-5771INT J EPIDEMIOL INTERNATIONAL JOURNAL OF EPIDEMI 医学58301368-7646DRUG RESIST UPDATE DRUG RESISTANCE UPDATES 医学27240305-1048NUCLEIC ACIDS RES NUCLEIC ACIDS RESEARCH 生物33250737-4038MOL BIOL EVOL MOLECULAR BIOLOGY AND EVOLUTION 生物62311469-221X EMBO REP EMBO REPORTS 生物84392160-3308PHYS REV X Physical Review X PHYSICS, MULTIDISCIPLINAR 32930734-9750BIOTECHNOL ADV BIOTECHNOLOGY ADVANCES 工程技术65391527-8204ANNU REV GENOM HUM G A NNUAL REVIEW OF GENOMICS AND H 生物48421058-4838CLIN INFECT DIS CLINICAL INFECTIOUS DISEASES 医学80501936-1327ANNU REV ANAL CHEM Annual Review of Analytical Chemistry CHEMISTRY, ANAL 18530149-7634NEUROSCI BIOBEHAV R NEUROSCIENCE AND BIOBEHAVIORAL 医学49101063-6706IEEE T FUZZY SYST IEEE TRANSACTIONS ON FUZZY SYSTE 工程技术56991350-9462PROG RETIN EYE RES PROGRESS IN RETINAL AND EYE RESE 医学50971078-0432CLIN CANCER RES CLINICAL CANCER RESEARCH 医学81271943-0264CSH PERSPECT BIOL Cold Spring Harbor perspectives in biology CELL BIOLOGY 81541947-5438ANNU REV CHEM BIOMOL Annual review of chemical and biomolecula CHEMISTRY, APPL 47411047-3211CEREB CORTEX CEREBRAL CORTEX 医学76581757-4676EMBO MOL MED EMBO Molecular Medicine 医学15250084-6597ANNU REV EARTH PL SC ANNUAL REVIEW OF EARTH AND PLAN地学天文85372212-6864PHYS DARK UNIVERSE Physics of the Dark Universe ASTRONOMY & ASTROPHYSI 15260085-2538KIDNEY INT KIDNEY INTERNATIONAL医学17610142-9612BIOMATERIALS BIOMATERIALS工程技术76961759-5061NAT REV NEPHROL Nature Reviews Nephrology医学51771087-0792SLEEP MED REV SLEEP MEDICINE REVIEWS医学41100950-6608INT MATER REV INTERNATIONAL MATERIALS REVIEWS工程技术化学18720161-4940CATAL REV CATALYSIS REVIEWS-SCIENCE AND EN41800955-0674CURR OPIN CELL BIOL CURRENT OPINION IN CELL BIOLOGY生物41120950-9232ONCOGENE ONCOGENE医学31840586-7614SCHIZOPHRENIA BULL SCHIZOPHRENIA BULLETIN医学21910198-6325MED RES REV MEDICINAL RESEARCH REVIEWS医学18490149-5992DIABETES CARE DIABETES CARE医学37140896-8411J AUTOIMMUN JOURNAL OF AUTOIMMUNITY医学71011613-6810SMALL SMALL工程技术21950199-9885ANNU REV NUTR ANNUAL REVIEW OF NUTRITION医学85242211-1247CELL REP Cell reports CELL BIOLOGY37210897-4756CHEM MATER CHEMISTRY OF MATERIALS工程技术1570003-4932ANN SURG ANNALS OF SURGERY医学13180040-6376THORAX THORAX医学10150028-3878NEUROLOGY NEUROLOGY医学62101467-2960FISH FISH FISH AND FISHERIES农林科学56481342-937X GONDWANA RES GONDWANA RESEARCH地学56981350-9047CELL DEATH DIFFER CELL DEATH AND DIFFERENTIATION生物85532223-7755GEOCHEM PERSPECT Geochemical Perspectives GEOCHEMISTRY & 4220012-1797DIABETES DIABETES医学57201354-1013GLOBAL CHANGE BIOL GLOBAL CHANGE BIOLOGY环境科学77831838-7640THERANOSTICS THERANOSTICS MEDICINE, RESEA 61781463-9262GREEN CHEM GREEN CHEMISTRY化学78381863-8880LASER PHOTONICS REV Laser & Photonics Reviews物理62171467-7881OBES REV Obesity Reviews医学2760007-1250BRIT J PSYCHIAT BRITISH JOURNAL OF PSYCHIATRY医学74121741-7007BMC BIOL BMC BIOLOGY生物环境科学15570091-6765ENVIRON HEALTH PERSP ENVIRONMENTAL HEALTH PERSPECTIV11570033-295X PSYCHOL REV PSYCHOLOGICAL REVIEW医学69511568-9972AUTOIMMUN REV AUTOIMMUNITY REVIEWS医学3680009-9147CLIN CHEM CLINICAL CHEMISTRY医学医学3730009-9236CLIN PHARMACOL THER CLINICAL PHARMACOLOGY & THERAPE67011542-3565CLIN GASTROENTEROL H Clinical Gastroenterology and Hepatology医学4390012-8252EARTH-SCI REV EARTH-SCIENCE REVIEWS地学58361369-5266CURR OPIN PLANT BIOL CURRENT OPINION IN PLANT BIOLOGY生物医学27100304-419X BBA-REV CANCER BIOCHIMICA ET BIOPHYSICA ACTA-REV生物11730033-5835Q REV BIOPHYS QUARTERLY REVIEWS OF BIOPHYSICS化学560001-8686ADV COLLOID INTERFAC ADVANCES IN COLLOID AND INTERFAC1990004-3591ARTHRITIS RHEUM-US ARTHRITIS & RHEUMATISM-ARTHRITIS医学78321863-2297SEMIN IMMUNOPATHOL Seminars in Immunopathology医学46731040-9238CRIT REV BIOCHEM MOL CRITICAL REVIEWS IN BIOCHEMISTRY 生物25510277-7037MASS SPECTROM REV MASS SPECTROMETRY REVIEWS物理20130168-3659J CONTROL RELEASE JOURNAL OF CONTROLLED RELEASE医学10210028-646X NEW PHYTOL NEW PHYTOLOGIST生物78421864-5631CHEMSUSCHEM ChemSusChem化学37510903-1936EUR RESPIR J EUROPEAN RESPIRATORY JOURNAL医学27360305-7372CANCER TREAT REV CANCER TREATMENT REVIEWS医学48921062-7995PROG PHOTOVOLTAICS PROGRESS IN PHOTOVOLTAICS工程技术生物42350959-437X CURR OPIN GENET DEV CURRENT OPINION IN GENETICS & DEV68281553-7366PLOS PATHOG PLoS Pathogens生物68291553-7390PLOS GENET PLoS Genetics生物11050031-9007PHYS REV LETT PHYSICAL REVIEW LETTERS物理65491529-1839OPTOMETRY OPTOMETRY OPHTHALMOLOGY4270012-3692CHEST CHEST医学41370952-7915CURR OPIN IMMUNOL CURRENT OPINION IN IMMUNOLOGY医学69851571-0645PHYS LIFE REV Physics of Life Reviews生物81631948-7185J PHYS CHEM LETT The journal of physical chemistry letters CHEMISTRY, PHYS 83212050-7488J MATER CHEM A Journal of Materials Chemistry A CHEMISTRY, PHYS环境科学66791540-9295FRONT ECOL ENVIRON FRONTIERS IN ECOLOGY AND THE ENV环境科学38970926-3373APPL CATAL B-ENVIRON APPLIED CATALYSIS B-ENVIRONMENTA8310022-3417J PATHOL JOURNAL OF PATHOLOGY医学1840003-9942ARCH NEUROL-CHICAGO ARCHIVES OF NEUROLOGY医学64781523-0864ANTIOXID REDOX SIGN ANTIOXIDANTS & REDOX SIGNALING生物82562040-3364NANOSCALE Nanoscale CHEMISTRY, MUL80181933-0219MUCOSAL IMMUNOL Mucosal Immunology医学75341749-5016SOC COGN AFFECT NEUR Social Cognitive and Affective Neuroscienc医学42930962-4929ACTA NUMER ACTA NUMERICA MATHEMATICS80611936-8798JACC-CARDIOVASC INTE JACC. Cardiovascular interventions CARDIAC & CARDI 52851098-3600GENET MED GENETICS IN MEDICINE医学1190003-0090B AM MUS NAT HIST BULLETIN OF THE AMERICAN MUSEUM环境科学84892190-5991J CACHEXIA SARCOPENI Journal of Cachexia Sarcopenia and Muscl MEDICINE, GENER 84762168-6149JAMA NEUROL JAMA Neurology CLINICAL NEUROLOGY医学36210890-8567J AM ACAD CHILD PSY JOURNAL OF THE AMERICAN ACADEMY74131741-7015BMC MED BMC Medicine医学76241755-263X CONSERV LETT Conservation Letters BIODIVERSITY CONSERVATIO物理15070079-6565PROG NUCL MAG RES SP PROGRESS IN NUCLEAR MAGNETIC RE19850167-7659CANCER METAST REV CANCER AND METASTASIS REVIEWS医学医学7860022-202X J INVEST DERMATOL JOURNAL OF INVESTIGATIVE DERMATO28540342-4642INTENS CARE MED INTENSIVE CARE MEDICINE医学42370959-440X CURR OPIN STRUC BIOL CURRENT OPINION IN STRUCTURAL BI生物80601936-878X JACC-CARDIOVASC IMAG JACC. Cardiovascular imaging CARDIAC & CARD33330738-8551CRIT REV BIOTECHNOL CRITICAL REVIEWS IN BIOTECHNOLOG工程技术84792168-6203JAMA PEDIATR JAMA Pediatrics PEDIATRICS42210958-1669CURR OPIN BIOTECH CURRENT OPINION IN BIOTECHNOLOG工程技术74501743-8977PART FIBRE TOXICOL Particle and fibre toxicology TOXICOLOGY生物42960962-8436PHILOS T R SOC B PHILOSOPHICAL TRANSACTIONS OF THNEUROPSYCHOPHARMACOLOGY医学36560893-133X NEUROPSYCHOPHARMACO38530923-7534ANN ONCOL ANNALS OF ONCOLOGY医学15500091-3022FRONT NEUROENDOCRIN FRONTIERS IN NEUROENDOCRINOLOG医学17840144-235X INT REV PHYS CHEM INTERNATIONAL REVIEWS IN PHYSICA化学82261998-0124NANO RES Nano Research化学化学14450065-3055ADV ORGANOMET CHEM ADVANCES IN ORGANOMETALLIC CHEM4410012-9615ECOL MONOGR ECOLOGICAL MONOGRAPHS环境科学17300140-7791PLANT CELL ENVIRON PLANT CELL AND ENVIRONMENT生物7120021-9517J CATAL JOURNAL OF CATALYSIS工程技术11830033-8419RADIOLOGY RADIOLOGY医学11250032-0889PLANT PHYSIOL PLANT PHYSIOLOGY生物50551073-8584NEUROSCIENTIST NEUROSCIENTIST医学57701359-7345CHEM COMMUN CHEMICAL COMMUNICATIONS化学35080884-0431J BONE MINER RES JOURNAL OF BONE AND MINERAL RES医学生物58211367-5931CURR OPIN CHEM BIOL CURRENT OPINION IN CHEMICAL BIOLO8170022-3050J NEUROL NEUROSUR PS JOURNAL OF NEUROLOGY NEUROSUR医学COMPUTER SCIEN 68301553-877X IEEE COMMUN SURV TUT IEEE COMMUNICATIONS SURVEYS AND83942150-7511MBIO mBio MICROBIOLOGY 72461674-2788J MOL CELL BIOL Journal of molecular cell biology CELL BIOLOGY 890002-9165AM J CLIN NUTR AMERICAN JOURNAL OF CLINICAL NUT医学52751096-7176METAB ENG METABOLIC ENGINEERING工程技术14380065-2660ADV GENET ADVANCES IN GENETICS生物51221080-6040EMERG INFECT DIS EMERGING INFECTIOUS DISEASES医学81421944-8244ACS APPL MATER INTER ACS Applied Materials & Interfaces工程技术57651359-6446DRUG DISCOV TODAY DRUG DISCOVERY TODAY医学4230012-186X DIABETOLOGIA DIABETOLOGIA医学21520193-936X EPIDEMIOL REV EPIDEMIOLOGIC REVIEWS医学医学47911053-2498J HEART LUNG TRANSPL JOURNAL OF HEART AND LUNG TRANS50601074-5521CHEM BIOL CHEMISTRY & BIOLOGY生物42360959-4388CURR OPIN NEUROBIOL CURRENT OPINION IN NEUROBIOLOGY医学化学15120079-6786PROG SOLID STATE CH PROGRESS IN SOLID STATE CHEMISTR66181535-9476MOL CELL PROTEOMICS MOLECULAR & CELLULAR PROTEOMIC生物75431750-1326MOL NEURODEGENER Molecular Neurodegeneration医学82652041-210X METHODS ECOL EVOL Methods in ecology and evolution ECOLOGY环境科学62071466-822X GLOBAL ECOL BIOGEOGR GLOBAL ECOLOGY AND BIOGEOGRAPH59381388-9842EUR J HEART FAIL EUROPEAN JOURNAL OF HEART FAILU医学49671066-5099STEM CELLS STEM CELLS医学17070129-0657INT J NEURAL SYST International Journal of Neural Systems工程技术25530278-0046IEEE T IND ELECTRON IEEE TRANSACTIONS ON INDUSTRIAL E工程技术42900962-1083MOL ECOL MOLECULAR ECOLOGY生物21560194-911X HYPERTENSION HYPERTENSION医学19340165-9936TRAC-TREND ANAL CHEM TRAC-TRENDS IN ANALYTICAL CHEMIS化学18990163-7525ANNU REV PUBL HEALTH ANNUAL REVIEW OF PUBLIC HEALTH医学41020950-1991DEVELOPMENT DEVELOPMENT生物医学7220021-9630J CHILD PSYCHOL PSYC JOURNAL OF CHILD PSYCHOLOGY AND物理46661040-8436CRIT REV SOLID STATE CRITICAL REVIEWS IN SOLID STATE AN81451945-4589AGING-US Aging CELL BIOLOGY 59271387-6473NEW ASTRON REV NEW ASTRONOMY REVIEWS地学天文74431743-5390NANOTOXICOLOGY Nanotoxicology医学41970956-5663BIOSENS BIOELECTRON BIOSENSORS & BIOELECTRONICS工程技术11870034-4257REMOTE SENS ENVIRON REMOTE SENSING OF ENVIRONMENT环境科学43220964-6906HUM MOL GENET HUMAN MOLECULAR GENETICS生物64871523-7060ORG LETT ORGANIC LETTERS化学61691462-8902DIABETES OBES METAB DIABETES OBESITY & METABOLISM医学81681949-2553ONCOTARGET Oncotarget Oncogenes 47971053-8119NEUROIMAGE NEUROIMAGE医学24860270-6474J NEUROSCI JOURNAL OF NEUROSCIENCE医学63411474-9718AGING CELL AGING CELL生物72451674-2052MOL PLANT Molecular Plant生物8020022-2593J MED GENET JOURNAL OF MEDICAL GENETICS医学79091874-9399BBA-GENE REGUL MECH Biochimica et Biophysica Acta-Gene Regul生物15320090-3493CRIT CARE MED CRITICAL CARE MEDICINE医学81051941-1413ANNU REV FOOD SCI T Annual review of food science and technoloFOOD SCIENCE & 12760038-6308SPACE SCI REV SPACE SCIENCE REVIEWS地学天文81441945-0877SCI SIGNAL Science Signaling BIOCHEMISTRY & 26820303-4240REV PHYSIOL BIOCH P REVIEWS OF PHYSIOLOGY BIOCHEMIS医学84482162-4011ONCOIMMUNOLOGY OncoImmunology ONCOLOGY生物51631084-9521SEMIN CELL DEV BIOL SEMINARS IN CELL & DEVELOPMENTAL9450025-6196MAYO CLIN PROC MAYO CLINIC PROCEEDINGS医学工程技术57541359-0286CURR OPIN SOLID ST M CURRENT OPINION IN SOLID STATE & M79001873-9946J CROHNS COLITIS Journal of Crohns & Colitis医学64971525-0016MOL THER MOLECULAR THERAPY医学14460065-308X ADV PARASIT ADVANCES IN PARASITOLOGY医学68991560-2745FUNGAL DIVERS FUNGAL DIVERSITY生物51441083-4419IEEE T SYST MAN CY B IEEE TRANSACTIONS ON SYSTEMS MA工程技术81121941-7640CIRC-CARDIOVASC INTE Circulation: Cardiovascular Interventions CARDIAC & CARDI 30630378-7753J POWER SOURCES JOURNAL OF POWER SOURCES工程技术生物33830748-3007CLADISTICS CLADISTICS-THE INTERNATIONAL JOUR生物58771383-5742MUTAT RES-REV MUTAT MUTATION RESEARCH-REVIEWS IN MU7250021-972X J CLIN ENDOCR METAB JOURNAL OF CLINICAL ENDOCRINOLO医学62901471-4922TRENDS PARASITOL TRENDS IN PARASITOLOGY医学61641462-2912ENVIRON MICROBIOL ENVIRONMENTAL MICROBIOLOGY环境科学3240008-6223CARBON CARBON工程技术47121044-3983EPIDEMIOLOGY EPIDEMIOLOGY医学36600893-3952MODERN PATHOL MODERN PATHOLOGY医学18730161-5505J NUCL MED JOURNAL OF NUCLEAR MEDICINE医学68751558-3724POLYM REV Polymer Reviews工程技术67831549-9634NANOMED-NANOTECHNOL Nanomedicine-Nanotechnology Biology an工程技术18760161-6420OPHTHALMOLOGY OPHTHALMOLOGY医学29840370-2693PHYS LETT B PHYSICS LETTERS B物理63151473-0197LAB CHIP LAB ON A CHIP工程技术46281029-8479J HIGH ENERGY PHYS JOURNAL OF HIGH ENERGY PHYSICS PHYSICS, PARTICL10420029-6643NUTR REV NUTRITION REVIEWS医学79501879-6257CURR OPIN VIROL Current Opinion in Virology VIROLOGY 41750954-6820J INTERN MED JOURNAL OF INTERNAL MEDICINE医学76151754-6834BIOTECHNOL BIOFUELS Biotechnology for Biofuels工程技术医学62321469-493X COCHRANE DB SYST REV Cochrane Database of Systematic Reviews36740894-1491GLIA GLIA医学16730105-4538ALLERGY ALLERGY医学74331742-7061ACTA BIOMATER Acta Biomaterialia工程技术46641040-841X CRIT REV MICROBIOL CRITICAL REVIEWS IN MICROBIOLOGY生物76631757-7004WIRES RNA WIRES RNA CELL BIOLOGY工程技术35420885-8993IEEE T POWER ELECTR IEEE TRANSACTIONS ON POWER ELEC51121079-5642ARTERIOSCL THROM VAS ARTERIOSCLEROSIS THROMBOSIS AN医学7830022-1899J INFECT DIS JOURNAL OF INFECTIOUS DISEASES医学2090004-637X ASTROPHYS J ASTROPHYSICAL JOURNAL地学天文28390340-5761ARCH TOXICOL ARCHIVES OF TOXICOLOGY医学42700960-7412PLANT J PLANT JOURNAL生物49631065-9471HUM BRAIN MAPP HUMAN BRAIN MAPPING医学5280016-6731GENETICS GENETICS生物14410065-2776ADV IMMUNOL ADVANCES IN IMMUNOLOGY医学34470820-3946CAN MED ASSOC J CANADIAN MEDICAL ASSOCIATION JOU医学3260008-6363CARDIOVASC RES CARDIOVASCULAR RESEARCH医学11560033-2917PSYCHOL MED PSYCHOLOGICAL MEDICINE医学43190964-4563TOB CONTROL TOBACCO CONTROL医学57881364-0321RENEW SUST ENERG REV R ENEWABLE & SUSTAINABLE ENERGY工程技术医学25140272-6386AM J KIDNEY DIS AMERICAN JOURNAL OF KIDNEY DISEA58371369-5274CURR OPIN MICROBIOL CURRENT OPINION IN MICROBIOLOGY生物环境科学67121543-5938ANNU REV ENV RESOUR ANNUAL REVIEW OF ENVIRONMENT AN81081941-3289CIRC-HEART FAIL Circulation-Heart Failure医学35880889-1591BRAIN BEHAV IMMUN BRAIN BEHAVIOR AND IMMUNITY医学1350003-3022ANESTHESIOLOGY ANESTHESIOLOGY医学47501049-250X ADV ATOM MOL OPT PHY ADVANCES IN ATOMIC MOLECULAR AN物理医学53951180-4882J PSYCHIATR NEUROSCI JOURNAL OF PSYCHIATRY & NEUROSC47961053-5888IEEE SIGNAL PROC MAG IEEE SIGNAL PROCESSING MAGAZINE工程技术57551359-0294CURR OPIN COLLOID IN CURRENT OPINION IN COLLOID & INTE化学31020390-6078HAEMATOLOGICA HAEMATOLOGICA HEMATOLOGY地学天文63551475-7516J COSMOL ASTROPART P JOURNAL OF COSMOLOGY AND ASTRO生物60021420-682X CELL MOL LIFE SCI CMLS-Cellular and Molecular Life Sciences9170024-9297MACROMOLECULES MACROMOLECULES工程技术84962192-2640ADV HEALTHC MATER Advanced healthcare materials工程技术82952046-2441OPEN BIOL Open biology BIOCHEMISTRY &工程技术18870162-8828IEEE T PATTERN ANAL IEEE TRANSACTIONS ON PATTERN ANA54041198-743X CLIN MICROBIOL INFEC CLINICAL MICROBIOLOGY AND INFECT医学84402161-1653ACS MACRO LETT ACS Macro Letters化学62151467-7644PLANT BIOTECHNOL J PLANT BIOTECHNOLOGY JOURNAL生物62121467-3037CURR ISSUES MOL BIOL CURRENT ISSUES IN MOLECULAR BIOL生物65071525-7797BIOMACROMOLECULES BIOMACROMOLECULES化学36340891-5849FREE RADICAL BIO MED FREE RADICAL BIOLOGY AND MEDICIN医学40690947-6539CHEM-EUR J CHEMISTRY-A EUROPEAN JOURNAL化学50391072-4710ARCH PEDIAT ADOL MED ARCHIVES OF PEDIATRICS & ADOLESC医学医学24640269-2813ALIMENT PHARM THER ALIMENTARY PHARMACOLOGY & THER9590025-7974MEDICINE MEDICINE医学12840039-2499STROKE STROKE医学69011560-7917EUROSURVEILLANCE EUROSURVEILLANCE INFECTIOUS DISEASES 66541538-7933J THROMB HAEMOST JOURNAL OF THROMBOSIS AND HAEM医学84262157-6564STEM CELL TRANSL MED Stem cells translational medicine CELL & TISSUE ENGINEERING 15130079-6816PROG SURF SCI PROGRESS IN SURFACE SCIENCE工程技术84982192-8606NANOPHOTONICS-BERLIN N anophotonics NANOSCIENCE &66161535-7163MOL CANCER THER MOLECULAR CANCER THERAPEUTICS医学医学70591600-6135AM J TRANSPLANT AMERICAN JOURNAL OF TRANSPLANTA35210885-3185MOVEMENT DISORD MOVEMENT DISORDERS医学35010883-7694MRS BULL MRS BULLETIN工程技术26690301-9268PRECAMBRIAN RES PRECAMBRIAN RESEARCH地学71071615-4150ADV SYNTH CATAL ADVANCED SYNTHESIS & CATALYSIS化学42180957-9672CURR OPIN LIPIDOL CURRENT OPINION IN LIPIDOLOGY医学81131941-7705CIRC-CARDIOVASC QUAL Circulation: Cardiovascular Quality and Ou CARDIAC & CARDI1320003-2700ANAL CHEM ANALYTICAL CHEMISTRY化学26970304-3835CANCER LETT CANCER LETTERS医学84312158-3188TRANSL PSYCHIAT Translational Psychiatry PSYCHIATRY43740969-2126STRUCTURE STRUCTURE生物78331863-2653BRAIN STRUCT FUNCT Brain Structure & Function医学27410306-2619APPL ENERG APPLIED ENERGY工程技术450001-690X ACTA PSYCHIAT SCAND ACTA PSYCHIATRICA SCANDINAVICA医学17750143-5221CLIN SCI CLINICAL SCIENCE医学57371355-6037HEART HEART医学82852045-2322SCI REP-UK Scientific reports Natural Science Disciplines 47861052-9276REV MED VIROL REVIEWS IN MEDICAL VIROLOGY医学34040749-6419INT J PLASTICITY INTERNATIONAL JOURNAL OF PLASTIC工程技术24600268-960X BLOOD REV BLOOD REVIEWS医学64761522-8517NEURO-ONCOLOGY NEURO-ONCOLOGY医学18600160-4120ENVIRON INT ENVIRONMENT INTERNATIONAL环境科学24800269-9370AIDS AIDS医学2780007-1323BRIT J SURG BRITISH JOURNAL OF SURGERY医学13480043-1354WATER RES WATER RESEARCH环境科学57241354-3784EXPERT OPIN INV DRUG EXPERT OPINION ON INVESTIGATIONA医学7550022-0957J EXP BOT JOURNAL OF EXPERIMENTAL BOTANY生物7450022-0477J ECOL JOURNAL OF ECOLOGY环境科学77031759-9954POLYM CHEM-UK POLYMER CHEMISTRY POLYMER SCIENCE 82141994-0416CRYOSPHERE Cryosphere GEOGRAPHY, PHY 18630160-6689J CLIN PSYCHIAT JOURNAL OF CLINICAL PSYCHIATRY医学67821549-9618J CHEM THEORY COMPUT J ournal of Chemical Theory and Computat化学65401527-8999CHEM REC CHEMICAL RECORD化学61951465-542X BREAST CANCER RES BREAST CANCER RESEARCH ONCOLOGY。

NAT NANOTECHNOL Nature Nanotechnology工程技术127.270 NAT BIOTECHNOL NATURE BIOTECHNOLOGY工程技术123.268 ENERGY EDUC SCI TECH Energy Education Science and Technology工程技术131.677 PROG MATER SCI PROGRESS IN MATERIALS SCIENCE工程技术118.216 MAT SCI ENG R MATERIALS SCIENCE & ENGINEERING R-RE工程技术114.951 NANO TODAY Nano Today工程技术115.355 PROG ENERG COMBUST PROGRESS IN ENERGY AND COMBUSTION工程技术114.220 NANO LETT NANO LETTERS工程技术113.198 ANNU REV BIOMED ENG ANNUAL REVIEW OF BIOMEDICAL ENGINEE工程技术112.214 ADV MATER ADVANCED MATERIALS工程技术113.877 ANNU REV MATER RES ANNUAL REVIEW OF MATERIALS RESEARC工程技术113.073 ACS NANO ACS NANO工程技术111.421 PROG SURF SCI PROGRESS IN SURFACE SCIENCE工程技术18.636 TRENDS BIOTECHNOL TRENDS IN BIOTECHNOLOGY工程技术19.148 ADV FUNCT MATER ADVANCED FUNCTIONAL MATERIALS工程技术110.179 BIOTECHNOL ADV BIOTECHNOLOGY ADVANCES工程技术19.646 CURR OPIN BIOTECH CURRENT OPINION IN BIOTECHNOLOGY工程技术17.711 MATER TODAY Materials Today工程技术1 5.565 BIOMATERIALS BIOMATERIALS工程技术17.404 ANNU REV CHEM BIOMOL Annual Review of Chemical and Biomolecular E工程技术17.294 SMALL SMALL工程技术18.349 PROG CRYST GROWTH CH PROGRESS IN CRYSTAL GROWTH AND CHA工程技术1 5.750 ACM COMPUT SURV ACM COMPUTING SURVEYS工程技术1 4.529 POLYM REV Polymer Reviews工程技术1 6.281 CHEM MATER CHEMISTRY OF MATERIALS工程技术17.286 LAB CHIP LAB ON A CHIP工程技术1 5.670 INT MATER REV INTERNATIONAL MATERIALS REVIEWS工程技术1 6.962 NANOMED-NANOTECHNOL Nanomedicine-Nanotechnology Biology and Me工程技术1 6.692 PROG PHOTOVOLTAICS PROGRESS IN PHOTOVOLTAICS工程技术1 5.789 P IEEE PROCEEDINGS OF THE IEEE工程技术1 6.810 J CATAL JOURNAL OF CATALYSIS工程技术1 6.002 NPG ASIA MATER NPG Asia Materials工程技术1 5.533 BIOSENS BIOELECTRON BIOSENSORS & BIOELECTRONICS工程技术1 5.602 PROG QUANT ELECTRON PROGRESS IN QUANTUM ELECTRONICS工程技术17.000 MRS BULL MRS BULLETIN工程技术1 4.950 J MATER CHEM JOURNAL OF MATERIALS CHEMISTRY工程技术1 5.968 METAB ENG METABOLIC ENGINEERING工程技术1 5.614 RENEW SUST ENERG REV RENEWABLE & SUSTAINABLE ENERGY REV工程技术1 6.018 CRIT REV BIOTECHNOL CRITICAL REVIEWS IN BIOTECHNOLOGY工程技术1 6.472 NANOSCALE Nanoscale工程技术1 5.914 IEEE COMMUN SURV TUT IEEE Communications Surveys and Tutorials工程技术1 6.311 IEEE SIGNAL PROC MAG IEEE SIGNAL PROCESSING MAGAZINE工程技术1 4.066 CARBON CARBON工程技术1 5.378 IEEE T PATTERN ANAL IEEE TRANSACTIONS ON PATTERN ANALYS工程技术1 4.908 MACROMOLECULES MACROMOLECULES工程技术1 5.167 INT J PLASTICITY INTERNATIONAL JOURNAL OF PLASTICITY工程技术1 4.603 BIOTECHNOL BIOFUELS Biotechnology for Biofuels工程技术1 6.088 MIS QUART MIS QUARTERLY工程技术1 4.447 SIAM J IMAGING SCI SIAM Journal on Imaging Sciences工程技术1 4.656 SOFT MATTER Soft Matter工程技术1 4.390 ACTA BIOMATER Acta Biomaterialia工程技术1 4.865 BIORESOURCE TECHNOL BIORESOURCE TECHNOLOGY工程技术1 4.980 ADV APPL MECH ADVANCES IN APPLIED MECHANICS工程技术1 5.000 ELECTROCHEM COMMUN ELECTROCHEMISTRY COMMUNICATIONS工程技术1 4.859MOL NUTR FOOD RES MOLECULAR NUTRITION & FOOD RESEARC工程技术1 4.301 IEEE T IND ELECTRON IEEE TRANSACTIONS ON INDUSTRIAL ELEC工程技术1 5.160 MACROMOL RAPID COMM MACROMOLECULAR RAPID COMMUNICATIO工程技术1 4.596 SOL ENERG MAT SOL C SOLAR ENERGY MATERIALS AND SOLAR CE工程技术1 4.542 J POWER SOURCES JOURNAL OF POWER SOURCES工程技术1 4.951 CRIT REV FOOD SCI CRITICAL REVIEWS IN FOOD SCIENCE AND工程技术1 4.789 CURR OPIN SOLID ST M CURRENT OPINION IN SOLID STATE & MATE工程技术1 4.233 INT J COMPUT VISION INTERNATIONAL JOURNAL OF COMPUTER 工程技术1 3.741 IEEE T EVOLUT COMPUT IEEE TRANSACTIONS ON EVOLUTIONARY C工程技术1 3.341 INT J HYDROGEN ENERG INTERNATIONAL JOURNAL OF HYDROGEN 工程技术1 4.054 J HAZARD MATER JOURNAL OF HAZARDOUS MATERIALS工程技术1 4.173 MED IMAGE ANAL MEDICAL IMAGE ANALYSIS工程技术1 4.424 HUM-COMPUT INTERACT HUMAN-COMPUTER INTERACTION工程技术1 1.476 MICROB CELL FACT Microbial Cell Factories工程技术1 3.552 INT J NEURAL SYST International Journal of Neural Systems工程技术1 4.284 TRENDS FOOD SCI TECH TRENDS IN FOOD SCIENCE & TECHNOLOGY工程技术1 3.672 IEEE J SEL AREA COMM IEEE JOURNAL ON SELECTED AREAS IN CO工程技术1 3.413 ORG ELECTRON ORGANIC ELECTRONICS工程技术1 4.047 ACTA MATER ACTA MATERIALIA工程技术1 3.755 APPL ENERG APPLIED ENERGY工程技术1 5.106 ACS APPL MATER INTER ACS Applied Materials & Interfaces工程技术1 4.525 BIOTECHNOL BIOENG BIOTECHNOLOGY AND BIOENGINEERING工程技术1 3.946 IEEE T POWER ELECTR IEEE TRANSACTIONS ON POWER ELECTRO工程技术1 4.650 BIOMASS BIOENERG BIOMASS & BIOENERGY工程技术1 3.646 ELECTROCHIM ACTA ELECTROCHIMICA ACTA工程技术1 3.832 ANNU REV FOOD SCI T Annual Review of Food Science and Technolog工程技术1 3.600 NANOTECHNOLOGY NANOTECHNOLOGY工程技术1 3.979 ACM T GRAPHIC ACM TRANSACTIONS ON GRAPHICS工程技术1 3.489 J MEMBRANE SCI JOURNAL OF MEMBRANE SCIENCE工程技术1 3.850 SENSOR ACTUAT B-CHEM SENSORS AND ACTUATORS B-CHEMICAL工程技术1 3.898 APPL SPECTROSC REV APPLIED SPECTROSCOPY REVIEWS工程技术2 3.385 IEEE J SEL TOP QUANT IEEE JOURNAL OF SELECTED TOPICS IN QU工程技术2 3.780 IEEE T FUZZY SYST IEEE TRANSACTIONS ON FUZZY SYSTEMS工程技术2 4.260 FOOD CHEM FOOD CHEMISTRY工程技术2 3.655 PLASMONICS Plasmonics 工程技术2 2.989 J NEURAL ENG Journal of Neural Engineering工程技术2 3.837 MICROFLUID NANOFLUID Microfluidics and Nanofluidics工程技术2 3.371 ENERGY ENERGY工程技术2 3.487 FUEL FUEL工程技术2 3.248 INT J NONLIN SCI NUM INTERNATIONAL JOURNAL OF NONLINEAR 工程技术2 1.484 FOOD MICROBIOL FOOD MICROBIOLOGY工程技术2 3.283 BIOMED MICRODEVICES BIOMEDICAL MICRODEVICES工程技术2 3.032 APPL MICROBIOL BIOT APPLIED MICROBIOLOGY AND BIOTECHNO工程技术2 3.425 CURR PROTEOMICS Current Proteomics工程技术2 3.179 BIOMECH MODEL MECHAN Biomechanics and Modeling in Mechanobiology工程技术2 3.192 IEEE J SOLID-ST CIRC IEEE JOURNAL OF SOLID-STATE CIRCUITS工程技术2 3.226 INT J FOOD MICROBIOL INTERNATIONAL JOURNAL OF FOOD MICRO工程技术2 3.327 INT J NANOMED International Journal of Nanomedicine 工程技术2 3.130 CHEM ENG J CHEMICAL ENGINEERING JOURNAL工程技术2 3.461 SCI TECHNOL ADV MAT SCIENCE AND TECHNOLOGY OF ADVANCED工程技术2 3.513 FOOD HYDROCOLLOID FOOD HYDROCOLLOIDS工程技术2 3.473 CORROS SCI CORROSION SCIENCE工程技术2 3.734 MICROSC MICROANAL MICROSCOPY AND MICROANALYSIS工程技术2 3.007 J MECH BEHAV BIOMED Journal of the Mechanical Behavior of Biomedic工程技术2 2.814 COMBUST FLAME COMBUSTION AND FLAME工程技术2 3.585 INT J ROBOT RES INTERNATIONAL JOURNAL OF ROBOTICS R工程技术2 3.107 ENVIRON MODELL SOFTW ENVIRONMENTAL MODELLING & SOFTWAR工程技术2 3.114IEEE COMMUN MAG IEEE COMMUNICATIONS MAGAZINE工程技术2 3.785 FUEL CELLS Fuel Cells工程技术2 3.149 J NANOPART RES JOURNAL OF NANOPARTICLE RESEARCH工程技术2 3.287 MAR BIOTECHNOL MARINE BIOTECHNOLOGY工程技术2 3.430 J STAT SOFTW Journal of Statistical Software工程技术2 4.010 INFORM SCIENCES INFORMATION SCIENCES工程技术2 2.833 DENT MATER DENTAL MATERIALS工程技术2 3.135 COMPOS SCI TECHNOL COMPOSITES SCIENCE AND TECHNOLOGY工程技术2 3.144 J BIOTECHNOL JOURNAL OF BIOTECHNOLOGY工程技术2 3.045 IEEE COMPUT INTELL M IEEE Computational Intelligence Magazine工程技术2 3.368 J CIV ENG MANAG Journal of Civil Engineering and Management工程技术2 2.171 IEEE T IMAGE PROCESS IEEE TRANSACTIONS ON IMAGE PROCESSI工程技术2 3.042 IEEE T SYST MAN CY B IEEE TRANSACTIONS ON SYSTEMS MAN AN工程技术2 3.080 INT J ELECTROCHEM SC International Journal of Electrochemical Science工程技术2 3.729 P COMBUST INST PROCEEDINGS OF THE COMBUSTION INST工程技术2 3.633 DYES PIGMENTS DYES AND PIGMENTS工程技术2 3.126 IEEE IND ELECTRON M IEEE Industrial Electronics Magazine工程技术2 5.000 SEP PURIF TECHNOL SEPARATION AND PURIFICATION TECHNOL工程技术2 2.921 CELLULOSE CELLULOSE工程技术2 3.600 GOLD BULL GOLD BULLETIN工程技术2 3.517 COMPUT-AIDED CIV INF COMPUTER-AIDED CIVIL AND INFRASTRUC工程技术2 3.382 J BIOMED MATER RES A JOURNAL OF BIOMEDICAL MATERIALS RES工程技术2 2.625 J SUPERCRIT FLUID JOURNAL OF SUPERCRITICAL FLUIDS工程技术2 2.860 IEEE T NEURAL NETWOR IEEE TRANSACTIONS ON NEURAL NETWOR工程技术2 2.952 SCRIPTA MATER SCRIPTA MATERIALIA工程技术2 2.699 IBM J RES DEV IBM JOURNAL OF RESEARCH AND DEVELO工程技术20.723 J ACM JOURNAL OF THE ACM工程技术2 2.353 J BIOACT COMPAT POL JOURNAL OF BIOACTIVE AND COMPATIBLE工程技术2 2.953 J MACH LEARN RES JOURNAL OF MACHINE LEARNING RESEAR工程技术2 2.561 VLDB J VLDB JOURNAL工程技术2 1.564 NANOSCALE RES LETT Nanoscale Research Letters工程技术2 2.726 IEEE ELECTR DEVICE L IEEE ELECTRON DEVICE LETTERS工程技术2 2.849 IEEE T INFORM THEORY IEEE TRANSACTIONS ON INFORMATION TH工程技术2 3.009 FUEL PROCESS TECHNOL FUEL PROCESSING TECHNOLOGY工程技术2 2.945 INNOV FOOD SCI EMERG Innovative Food Science & Emerging Technolo工程技术2 3.030 J NEUROENG REHABIL Journal of NeuroEngineering and Rehabilitation工程技术2 3.264 PROG AEROSP SCI PROGRESS IN AEROSPACE SCIENCES工程技术2 3.000 BIOFABRICATION Biofabrication工程技术2 3.480 IEEE T SOFTWARE ENG IEEE TRANSACTIONS ON SOFTWARE ENGI工程技术2 1.980 FOOD RES INT FOOD RESEARCH INTERNATIONAL工程技术2 3.150 SCI ADV MATER Science of Advanced Materials工程技术2 3.308 ADV BIOCHEM ENG BIOT ADVANCES IN BIOCHEMICAL ENGINEERING工程技术2 1.644 BMC BIOTECHNOL BMC BIOTECHNOLOGY工程技术2 2.349 FOOD CONTROL FOOD CONTROL工程技术2 2.656 ANNU REV INFORM SCI ANNUAL REVIEW OF INFORMATION SCIENC工程技术2 2.955 IEEE INTELL SYST IEEE INTELLIGENT SYSTEMS工程技术2 2.154 J FIELD ROBOT Journal of Field Robotics工程技术2 2.244 ARTIF INTELL ARTIFICIAL INTELLIGENCE工程技术2 2.252 IEEE T INTELL TRANSP IEEE TRANSACTIONS ON INTELLIGENT TRA工程技术2 3.452 RENEW ENERG RENEWABLE ENERGY工程技术2 2.978 IEEE-ASME T MECH IEEE-ASME TRANSACTIONS ON MECHATRO工程技术2 2.865 BIOTECHNIQUES BIOTECHNIQUES工程技术2 2.669 MABS-AUSTIN mAbs工程技术2 3.174 COMPR REV FOOD SCI F COMPREHENSIVE REVIEWS IN FOOD SCIEN工程技术2 3.724 FOOD CHEM TOXICOL FOOD AND CHEMICAL TOXICOLOGY工程技术2 2.999 IEEE T ROBOT IEEE Transactions on Robotics工程技术2 2.536 AUTOMATICA AUTOMATICA工程技术2 2.829IEEE INTERNET COMPUT IEEE INTERNET COMPUTING工程技术2 2.000 INTEGR COMPUT-AID E INTEGRATED COMPUTER-AIDED ENGINEER工程技术2 3.451 IEEE T GEOSCI REMOTE IEEE TRANSACTIONS ON GEOSCIENCE AND工程技术2 2.895 BIOCHEM ENG J BIOCHEMICAL ENGINEERING JOURNAL工程技术2 2.645 PATTERN RECOGN PATTERN RECOGNITION工程技术2 2.292 IEEE MICRO IEEE MICRO工程技术2 1.783 EARTHQ SPECTRA EARTHQUAKE SPECTRA工程技术20.894 J WEB SEMANT Journal of Web Semantics工程技术2 1.302 IEEE T SIGNAL PROCES IEEE TRANSACTIONS ON SIGNAL PROCESS工程技术2 2.628 REACT FUNCT POLYM REACTIVE & FUNCTIONAL POLYMERS工程技术2 2.479 ENERG FUEL ENERGY & FUELS工程技术2 2.721 COMPOS PART A-APPL S COMPOSITES PART A-APPLIED SCIENCE AN工程技术2 2.695 IEEE T ENERGY CONVER IEEE TRANSACTIONS ON ENERGY CONVER工程技术2 2.272 ISPRS J PHOTOGRAMM ISPRS JOURNAL OF PHOTOGRAMMETRY A工程技术2 2.885 IEEE POWER ENERGY M IEEE Power & Energy Magazine工程技术2 2.408 CEMENT CONCRETE RES CEMENT AND CONCRETE RESEARCH工程技术2 2.781 IEEE T MOBILE COMPUT IEEE TRANSACTIONS ON MOBILE COMPUTI工程技术2 2.283 COMPUT EDUC COMPUTERS & EDUCATION工程技术2 2.621 ENTERP INF SYST-UK Enterprise Information Systems工程技术2 3.684 J INF TECHNOL JOURNAL OF INFORMATION TECHNOLOGY工程技术2 2.321 J ELECTROCHEM SOC JOURNAL OF THE ELECTROCHEMICAL SOC工程技术2 2.590 J LIGHTWAVE TECHNOL JOURNAL OF LIGHTWAVE TECHNOLOGY工程技术2 2.784 J CEREAL SCI JOURNAL OF CEREAL SCIENCE工程技术2 2.073 TRANSPORT RES B-METH TRANSPORTATION RESEARCH PART B-MET工程技术2 2.856 J CHEM THERMODYN JOURNAL OF CHEMICAL THERMODYNAMIC工程技术2 2.422 ARCH COMPUT METHOD E ARCHIVES OF COMPUTATIONAL METHODS工程技术2 2.767 ANN BIOMED ENG ANNALS OF BIOMEDICAL ENGINEERING工程技术2 2.368 APPL SOFT COMPUT APPLIED SOFT COMPUTING工程技术2 2.612 INFORM MANAGE-AMSTER INFORMATION & MANAGEMENT工程技术2 2.214 J INSTRUM Journal of Instrumentation工程技术2 1.869 MAT SCI ENG C-MATER Materials Science & Engineering C-Materials fo工程技术2 2.686 IEEE CONTR SYST MAG IEEE CONTROL SYSTEMS MAGAZINE工程技术2 2.491 COMPUT INTELL-US COMPUTATIONAL INTELLIGENCE工程技术20.971 EXPERT SYST APPL EXPERT SYSTEMS WITH APPLICATIONS工程技术2 2.203 IEEE T ELECTRON DEV IEEE TRANSACTIONS ON ELECTRON DEVIC工程技术2 2.318 J EUR CERAM SOC JOURNAL OF THE EUROPEAN CERAMIC SO工程技术2 2.353 IEEE PHOTONICS J IEEE Photonics Journal工程技术2 2.320 INT DAIRY J INTERNATIONAL DAIRY JOURNAL工程技术2 2.401 PLANT FOOD HUM NUTR PLANT FOODS FOR HUMAN NUTRITION工程技术2 2.505 IEEE T POWER SYST IEEE TRANSACTIONS ON POWER SYSTEMS工程技术2 2.678 LWT-FOOD SCI TECHNOL LEBENSMITTEL-WISSENSCHAFT UND-TECH工程技术2 2.545 J IND MICROBIOL BIOT JOURNAL OF INDUSTRIAL MICROBIOLOGY 工程技术2 2.735 CHEM ENG SCI CHEMICAL ENGINEERING SCIENCE工程技术2 2.431 IEEE T AUTON MENT DE IEEE Transactions on Autonomous Mental Deve工程技术2 2.310 BIOTECHNOL PROGR BIOTECHNOLOGY PROGRESS工程技术2 2.340 J FOOD ENG JOURNAL OF FOOD ENGINEERING工程技术2 2.414 AUST J GRAPE WINE R AUSTRALIAN JOURNAL OF GRAPE AND WIN工程技术2 2.467 MEAT SCI MEAT SCIENCE工程技术2 2.275 IEEE PERVAS COMPUT IEEE PERVASIVE COMPUTING工程技术2 1.554 USER MODEL USER-ADAP USER MODELING AND USER-ADAPTED INTE工程技术2 1.400 EVOL COMPUT EVOLUTIONARY COMPUTATION 工程技术2 1.061 FOOD QUAL PREFER FOOD QUALITY AND PREFERENCE工程技术2 1.824 IEEE WIREL COMMUN IEEE WIRELESS COMMUNICATIONS工程技术2 2.575 IEEE J-STSP IEEE Journal of Selected Topics in Signal Proce工程技术2 2.880 SOL ENERGY SOLAR ENERGY工程技术2 2.475 IEEE T WIREL COMMUN IEEE TRANSACTIONS ON WIRELESS COMM工程技术2 2.586 COMMUN ACM COMMUNICATIONS OF THE ACM工程技术2 1.919BIOINTERPHASES Biointerphases 工程技术2 2.208 IEEE T AUTOMAT CONTR IEEE TRANSACTIONS ON AUTOMATIC CONT工程技术2 2.110 MATER CHEM PHYS MATERIALS CHEMISTRY AND PHYSICS工程技术2 2.234 ULTRAMICROSCOPY ULTRAMICROSCOPY工程技术2 2.471 J MATER SCI-MATER M JOURNAL OF MATERIALS SCIENCE-MATERI工程技术2 2.316 BIOMED MATER Biomedical Materials工程技术2 2.158 FUTURE GENER COMP SY FUTURE GENERATION COMPUTER SYSTEM工程技术2 1.978 J STRATEGIC INF SYST JOURNAL OF STRATEGIC INFORMATION SY工程技术2 1.457 J ALLOY COMPD JOURNAL OF ALLOYS AND COMPOUNDS工程技术2 2.289 PHOTONIC NANOSTRUCT Photonics and Nanostructures-Fundamentals an工程技术2 1.681 J BIOMED MATER RES B JOURNAL OF BIOMEDICAL MATERIALS RES工程技术2 2.147 COMPUT METHOD APPL M COMPUTER METHODS IN APPLIED MECHAN工程技术2 2.651 INT J COAL GEOL INTERNATIONAL JOURNAL OF COAL GEOLO工程技术2 2.542 J AM SOC INF SCI TEC JOURNAL OF THE AMERICAN SOCIETY FOR工程技术2 2.081 IEEE T VIS COMPUT GR IEEE TRANSACTIONS ON VISUALIZATION A工程技术2 2.215 DESALINATION DESALINATION工程技术2 2.590 J FOOD COMPOS ANAL JOURNAL OF FOOD COMPOSITION AND AN工程技术2 2.079 DECIS SUPPORT SYST DECISION SUPPORT SYSTEMS工程技术2 1.687 KNOWL INF SYST KNOWLEDGE AND INFORMATION SYSTEMS工程技术2 2.225 ARTIF LIFE ARTIFICIAL LIFE工程技术2 2.282 J AM CERAM SOC JOURNAL OF THE AMERICAN CERAMIC SOC工程技术2 2.272 J MICROMECH MICROENG JOURNAL OF MICROMECHANICS AND MICR工程技术2 2.105 IEEE ACM T NETWORK IEEE-ACM TRANSACTIONS ON NETWORKIN工程技术2 2.033 MATER LETT MATERIALS LETTERS工程技术2 2.307 NEURAL COMPUT NEURAL COMPUTATION工程技术2 1.884 INT J SEMANT WEB INF International Journal on Semantic Web and Info工程技术2 2.308 BUILD ENVIRON BUILDING AND ENVIRONMENT工程技术2 2.400 IEEE J QUANTUM ELECT IEEE JOURNAL OF QUANTUM ELECTRONIC工程技术2 1.879 IEEE NETWORK IEEE NETWORK工程技术2 2.239 COMPOS STRUCT COMPOSITE STRUCTURES工程技术2 2.240 IEEE ROBOT AUTOM MAG IEEE ROBOTICS & AUTOMATION MAGAZINE工程技术2 1.985 CHEMOMETR INTELL LAB CHEMOMETRICS AND INTELLIGENT LABOR工程技术2 1.920 INT J HEAT MASS TRAN INTERNATIONAL JOURNAL OF HEAT AND M工程技术2 2.407 FLUID PHASE EQUILIBR FLUID PHASE EQUILIBRIA工程技术2 2.139 IEEE T SYST MAN CY A IEEE TRANSACTIONS ON SYSTEMS MAN AN工程技术2 2.123 AICHE J AICHE JOURNAL工程技术2 2.261 J BIOMATER APPL JOURNAL OF BIOMATERIALS APPLICATION工程技术2 2.082 ENERG CONVERS MANAGE ENERGY CONVERSION AND MANAGEMENT工程技术2 2.216 IEEE T IND INFORM IEEE Transactions on Industrial Informatics工程技术2 2.990 IEEE T BIO-MED ENG IEEE TRANSACTIONS ON BIOMEDICAL ENG工程技术2 2.278 J CRYPTOL JOURNAL OF CRYPTOLOGY工程技术2 1.250 INTERMETALLICS INTERMETALLICS工程技术2 1.649 COMPUT CHEM ENG COMPUTERS & CHEMICAL ENGINEERING工程技术2 2.320 IEEE T CIRC SYST VID IEEE TRANSACTIONS ON CIRCUITS AND SY工程技术2 1.649 J SOLID STATE ELECTR JOURNAL OF SOLID STATE ELECTROCHEM工程技术2 2.131 J MANAGE INFORM SYST JOURNAL OF MANAGEMENT INFORMATION工程技术2 1.423 J MICROELECTROMECH S JOURNAL OF MICROELECTROMECHANICAL工程技术2 2.098 IEEE T SYST MAN CY C IEEE TRANSACTIONS ON SYSTEMS MAN AN工程技术2 2.009 J ASSOC INF SYST Journal of the Association for Information Syste工程技术2 1.667 MATER RES BULL MATERIALS RESEARCH BULLETIN工程技术2 2.105 FOOD ADDIT CONTAM A FOOD ADDITIVES AND CONTAMINANTS工程技术2 1.765 J ENG EDUC JOURNAL OF ENGINEERING EDUCATION工程技术2 1.569 BALT J ROAD BRIDGE E Baltic Journal of Road and Bridge Engineering工程技术2 1.610 INT J ELEC POWER INTERNATIONAL JOURNAL OF ELECTRICAL工程技术2 2.247 IND ENG CHEM RES INDUSTRIAL & ENGINEERING CHEMISTRY R工程技术2 2.237 EUR PHYS J E EUROPEAN PHYSICAL JOURNAL E工程技术2 1.944 INT J MACH TOOL MANU INTERNATIONAL JOURNAL OF MACHINE TO工程技术2 2.169J BIOMAT SCI-POLYM E JOURNAL OF BIOMATERIALS SCIENCE-POL工程技术2 1.691 NEURAL NETWORKS NEURAL NETWORKS工程技术2 2.182 J CHEM TECHNOL BIOT JOURNAL OF CHEMICAL TECHNOLOGY AND工程技术2 2.168 ACM T SENSOR NETWORK ACM Transactions on Sensor Networks 工程技术2 1.808 HYDROMETALLURGY HYDROMETALLURGY工程技术2 2.027 ENERG BUILDINGS ENERGY AND BUILDINGS工程技术2 2.386 MAT SCI ENG A-STRUCT MATERIALS SCIENCE AND ENGINEERING A工程技术2 2.003 IEEE PHOTONIC TECH L IEEE PHOTONICS TECHNOLOGY LETTERS工程技术2 2.191 IEEE ENG MED BIOL IEEE ENGINEERING IN MEDICINE AND BIOL工程技术2 2.057 INT J ADHES ADHES INTERNATIONAL JOURNAL OF ADHESION A工程技术2 2.170 INT J NUMER METH ENG INTERNATIONAL JOURNAL FOR NUMERICA工程技术2 2.009 FUNCT MATER LETT Functional Materials Letters工程技术20.724 IEEE T MICROW THEORY IEEE TRANSACTIONS ON MICROWAVE THE工程技术2 1.853 SMART MATER STRUCT SMART MATERIALS & STRUCTURES工程技术2 2.089 STRUCT SAF STRUCTURAL SAFETY工程技术2 1.867 INT J ENERG RES INTERNATIONAL JOURNAL OF ENERGY RES工程技术2 2.122 COMPUT LINGUIST COMPUTATIONAL LINGUISTICS工程技术20.721 J DISP TECHNOL Journal of Display Technology 工程技术2 2.280 BIODEGRADATION BIODEGRADATION工程技术2 2.017 IEEE T ANTENN PROPAG IEEE TRANSACTIONS ON ANTENNAS AND P工程技术2 2.151 MECH MATER MECHANICS OF MATERIALS工程技术2 1.769 J IND ENG CHEM JOURNAL OF INDUSTRIAL AND ENGINEERIN工程技术2 1.977 IEEE T NANOTECHNOL IEEE TRANSACTIONS ON NANOTECHNOLO工程技术2 2.292 ELECTROCHEM SOLID ST ELECTROCHEMICAL AND SOLID STATE LET工程技术2 1.995 APPL THERM ENG APPLIED THERMAL ENGINEERING工程技术2 2.064 SURF COAT TECH SURFACE & COATINGS TECHNOLOGY工程技术2 1.867 IEEE T KNOWL DATA EN IEEE TRANSACTIONS ON KNOWLEDGE AND工程技术2 1.657 CEMENT CONCRETE COMP CEMENT & CONCRETE COMPOSITES工程技术3 2.421 COAST ENG COASTAL ENGINEERING工程技术3 1.757 IET NANOBIOTECHNOL IET Nanobiotechnology工程技术3 1.833 DATA MIN KNOWL DISC DATA MINING AND KNOWLEDGE DISCOVER工程技术3 1.545 INT J APPROX REASON INTERNATIONAL JOURNAL OF APPROXIMAT工程技术3 1.948 POWDER TECHNOL POWDER TECHNOLOGY工程技术3 2.080 BIOPROC BIOSYST ENG BIOPROCESS AND BIOSYSTEMS ENGINEER工程技术3 1.809 STRUCT HEALTH MONIT STRUCTURAL HEALTH MONITORING-AN INT工程技术3 1.500 TRANSPORT RES A-POL TRANSPORTATION RESEARCH PART A-POL工程技术3 2.354 MECH SYST SIGNAL PR MECHANICAL SYSTEMS AND SIGNAL PROC工程技术3 1.824 MACROMOL MATER ENG MACROMOLECULAR MATERIALS AND ENGI工程技术3 1.986 J FUNCT FOODS Journal of Functional Foods工程技术3 2.446 J FOOD PROTECT JOURNAL OF FOOD PROTECTION工程技术3 1.937 COMPUT OPER RES COMPUTERS & OPERATIONS RESEARCH工程技术3 1.720 SYNTHETIC MET SYNTHETIC METALS工程技术3 1.829 ACM T WEB ACM Transactions on the Web 工程技术30.871 J PROCESS CONTR JOURNAL OF PROCESS CONTROL工程技术3 1.696 INT J THERM SCI INTERNATIONAL JOURNAL OF THERMAL SC工程技术3 2.142 ADSORPTION ADSORPTION-JOURNAL OF THE INTERNATI工程技术3 2.000 DIAM RELAT MATER DIAMOND AND RELATED MATERIALS工程技术3 1.913 TRANSPORT RES E-LOG TRANSPORTATION RESEARCH PART E-LOG工程技术3 1.648 THIN SOLID FILMS THIN SOLID FILMS工程技术3 1.890 COMPUT VIS IMAGE UND COMPUTER VISION AND IMAGE UNDERSTA工程技术3 1.340 IEEE T MULTIMEDIA IEEE TRANSACTIONS ON MULTIMEDIA工程技术3 1.935 ARTIF ORGANS ARTIFICIAL ORGANS工程技术3 2.000 APPL SURF SCI APPLIED SURFACE SCIENCE工程技术3 2.103 PROG ORG COAT PROGRESS IN ORGANIC COATINGS工程技术3 1.977 IEEE T INF FOREN SEC IEEE Transactions on Information Forensics an工程技术3 1.340 COMPUTER COMPUTER工程技术3 1.470 ACM T MATH SOFTWARE ACM TRANSACTIONS ON MATHEMATICAL S工程技术3 1.922J CHEM ENG DATA JOURNAL OF CHEMICAL AND ENGINEERING工程技术3 1.693 IEEE T BIOMED CIRC S IEEE Transactions on Biomedical Circuits and S工程技术3 2.032 ACM T COMPUT SYST ACM TRANSACTIONS ON COMPUTER SYST工程技术3 1.188 ENERG J ENERGY JOURNAL工程技术3 2.198 IEEE ACM T COMPUT BI IEEE-ACM Transactions on Computational Biolo工程技术3 1.543 FOOD SECUR Food Security工程技术3 1.970 PLASMA CHEM PLASMA P PLASMA CHEMISTRY AND PLASMA PROCES工程技术3 1.602 OPT MATER OPTICAL MATERIALS工程技术3 2.023 CHEM VAPOR DEPOS CHEMICAL VAPOR DEPOSITION工程技术3 1.796 SENSOR ACTUAT A-PHYS SENSORS AND ACTUATORS A-PHYSICAL工程技术3 1.802 MATER DESIGN MATERIALS & DESIGN工程技术3 2.200 IEEE MICROW WIREL CO IEEE MICROWAVE AND WIRELESS COMPON工程技术3 1.717 IEEE T BROADCAST IEEE TRANSACTIONS ON BROADCASTING工程技术3 1.703 CHEM ENG PROCESS CHEMICAL ENGINEERING AND PROCESSIN工程技术3 1.924 INT J CIRC THEOR APP INTERNATIONAL JOURNAL OF CIRCUIT THE工程技术3 1.625 TRANSPORT RES C-EMER TRANSPORTATION RESEARCH PART C-EME工程技术3 1.957 J FRANKLIN I JOURNAL OF THE FRANKLIN INSTITUTE-EN工程技术3 2.724 J MATER SCI JOURNAL OF MATERIALS SCIENCE工程技术3 2.015 SENSORS-BASEL SENSORS工程技术3 1.739 IET RENEW POWER GEN IET Renewable Power Generation工程技术3 1.742 POLYM ADVAN TECHNOL POLYMERS FOR ADVANCED TECHNOLOGIE工程技术3 2.007 KNOWL-BASED SYST KNOWLEDGE-BASED SYSTEMS工程技术3 2.422 INFORM FUSION Information Fusion工程技术3 1.467 POLYM TEST POLYMER TESTING工程技术3 1.608 WEAR WEAR工程技术3 1.872 J NUCL MATER JOURNAL OF NUCLEAR MATERIALS工程技术3 2.052 EMPIR SOFTW ENG EMPIRICAL SOFTWARE ENGINEERING工程技术3 1.854 APPL OPTICS APPLIED OPTICS工程技术3 1.748 AAPG BULL AAPG BULLETIN工程技术3 1.831 APPL BIOCHEM BIOTECH APPLIED BIOCHEMISTRY AND BIOTECHNOL工程技术3 1.943 FOOD NUTR BULL FOOD AND NUTRITION BULLETIN工程技术3 1.922 INT J HEAT FLUID FL INTERNATIONAL JOURNAL OF HEAT AND FL工程技术3 1.927 SIAM J COMPUT SIAM JOURNAL ON COMPUTING工程技术3 1.288 MACH LEARN MACHINE LEARNING工程技术3 1.587 COMPOS PART B-ENG COMPOSITES PART B-ENGINEERING工程技术3 1.731 FOOD BIOPHYS Food Biophysics工程技术3 2.187 MED ENG PHYS MEDICAL ENGINEERING & PHYSICS工程技术3 1.623 GPS SOLUT GPS SOLUTIONS工程技术3 1.667 INT J DAMAGE MECH INTERNATIONAL JOURNAL OF DAMAGE ME工程技术3 1.928 BIOINSPIR BIOMIM Bioinspiration & Biomimetics工程技术3 1.952 J AM OIL CHEM SOC JOURNAL OF THE AMERICAN OIL CHEMISTS工程技术3 1.773 MACROMOL RES MACROMOLECULAR RESEARCH工程技术3 1.153 EXP FLUIDS EXPERIMENTS IN FLUIDS工程技术3 1.735 INT J HUM-COMPUT ST INTERNATIONAL JOURNAL OF HUMAN-COM工程技术3 1.171 CURR NANOSCI Current Nanoscience工程技术3 1.776 J MICROSC-OXFORD JOURNAL OF MICROSCOPY-OXFORD工程技术3 1.631 ANNU REV CONTROL ANNUAL REVIEWS IN CONTROL工程技术3 1.319 BIOTECHNOL LETT BIOTECHNOLOGY LETTERS工程技术3 1.683 IEEE T INF TECHNOL B IEEE TRANSACTIONS ON INFORMATION TE工程技术3 1.676 METROLOGIA METROLOGIA工程技术3 1.750 IEEE SOFTWARE IEEE SOFTWARE工程技术3 1.508 IEEE T CONTR SYST T IEEE TRANSACTIONS ON CONTROL SYSTE工程技术3 1.766 EUR J LIPID SCI TECH EUROPEAN JOURNAL OF LIPID SCIENCE AN工程技术3 1.733 MACROMOL REACT ENG Macromolecular Reaction Engineering 工程技术3 1.848 COMPUT STRUCT COMPUTERS & STRUCTURES工程技术3 1.874 MATERIALS Materials工程技术3 1.677 DIG J NANOMATER BIOS Digest Journal of Nanomaterials and Biostructur工程技术3 1.200MODEL SIMUL MATER SC MODELLING AND SIMULATION IN MATERIAL工程技术3 2.298 J DATABASE MANAGE JOURNAL OF DATABASE MANAGEMENT工程技术30.875 AD HOC NETW Ad Hoc Networks工程技术3 2.110 CIRP ANN-MANUF TECHN CIRP ANNALS-MANUFACTURING TECHNOLO工程技术3 1.708 J THERM SPRAY TECHN JOURNAL OF THERMAL SPRAY TECHNOLOG工程技术3 1.812 IEEE T DEVICE MAT RE IEEE TRANSACTIONS ON DEVICE AND MAT工程技术3 1.543 ACM T SOFTW ENG METH ACM TRANSACTIONS ON SOFTWARE ENGIN工程技术3 1.269 J FOOD SCI JOURNAL OF FOOD SCIENCE工程技术3 1.658 APPL PHYS A-MATER APPLIED PHYSICS A-MATERIALS SCIENCE &工程技术3 1.630 IEEE T CIRCUITS-I IEEE TRANSACTIONS ON CIRCUITS AND SY工程技术3 1.970 J MICROENCAPSUL JOURNAL OF MICROENCAPSULATION工程技术3 1.553 APPL SPECTROSC APPLIED SPECTROSCOPY工程技术3 1.663 IEEE T ULTRASON FERR IEEE TRANSACTIONS ON ULTRASONICS FE工程技术3 1.694 INT J ROBUST NONLIN INTERNATIONAL JOURNAL OF ROBUST AND工程技术3 1.554 INT J FATIGUE INTERNATIONAL JOURNAL OF FATIGUE工程技术3 1.546 BIOL CYBERN BIOLOGICAL CYBERNETICS工程技术3 1.586 IEEE T AUDIO SPEECH IEEE Transactions on Audio Speech and Langu工程技术3 1.498 J APPL ELECTROCHEM JOURNAL OF APPLIED ELECTROCHEMISTR工程技术3 1.745 IEEE COMPUT GRAPH IEEE COMPUTER GRAPHICS AND APPLICAT工程技术3 1.411 J AUTOM REASONING JOURNAL OF AUTOMATED REASONING工程技术30.714 CERAM INT CERAMICS INTERNATIONAL工程技术3 1.751 LETT APPL MICROBIOL LETTERS IN APPLIED MICROBIOLOGY工程技术3 1.622 IEEE T VEH TECHNOL IEEE TRANSACTIONS ON VEHICULAR TECH工程技术3 1.921 J TAIWAN INST CHEM E Journal of the Taiwan Institute of Chemical Eng工程技术3 2.110 DATA KNOWL ENG DATA & KNOWLEDGE ENGINEERING工程技术3 1.422 INT J REFRACT MET H INTERNATIONAL JOURNAL OF REFRACTOR工程技术3 1.693 IEEE T COMP INTEL AI IEEE Transactions on Computational Intelligenc工程技术3 1.617 MACROMOL THEOR SIMUL MACROMOLECULAR THEORY AND SIMULAT工程技术3 1.709 CONTROL ENG PRACT CONTROL ENGINEERING PRACTICE工程技术3 1.481 NANOSC MICROSC THERM Nanoscale and Microscale Thermophysical Eng工程技术3 1.032 METALL MATER TRANS A METALLURGICAL AND MATERIALS TRANSA工程技术3 1.545 AUTON AGENT MULTI-AG AUTONOMOUS AGENTS AND MULTI-AGENT工程技术3 1.213 WIND ENERGY WIND ENERGY工程技术3 1.768 TRIBOL LETT TRIBOLOGY LETTERS工程技术3 1.582 J ARTIF INTELL RES JOURNAL OF ARTIFICIAL INTELLIGENCE RE工程技术3 1.143 TRIBOL INT TRIBOLOGY INTERNATIONAL工程技术3 1.553 MICRON MICRON工程技术3 1.527 DRY TECHNOL DRYING TECHNOLOGY工程技术3 2.084 COMPUT GRAPH FORUM COMPUTER GRAPHICS FORUM工程技术3 1.636 INT J REFRIG INTERNATIONAL JOURNAL OF REFRIGERAT工程技术3 1.817 NUMER HEAT TR A-APPL NUMERICAL HEAT TRANSFER PART A-APPL工程技术3 2.492 IEEE MICROW MAG IEEE MICROWAVE MAGAZINE工程技术3 2.111 IMAGE VISION COMPUT IMAGE AND VISION COMPUTING工程技术3 1.723 J MATER PROCESS TECH JOURNAL OF MATERIALS PROCESSING TEC工程技术3 1.783 AUTON ROBOT AUTONOMOUS ROBOTS工程技术3 1.500 INFORM SYST INFORMATION SYSTEMS工程技术3 1.198 MATER SCI ENG B-ADV Materials Science and Engineering B-Advanced工程技术3 1.518 WOOD SCI TECHNOL WOOD SCIENCE AND TECHNOLOGY工程技术3 1.727 J INTEL MAT SYST STR JOURNAL OF INTELLIGENT MATERIAL SYST工程技术3 1.953 MOB INF SYST Mobile Information Systems 工程技术3 2.432 J NANOPHOTONICS Journal of Nanophotonics工程技术3 1.570 SOFT COMPUT SOFT COMPUTING工程技术3 1.880 CHEM ENG RES DES CHEMICAL ENGINEERING RESEARCH & DES工程技术3 1.968 IEEE T PARALL DISTR IEEE TRANSACTIONS ON PARALLEL AND DI工程技术3 1.402 ELECTRON MATER LETT Electronic Materials Letters工程技术3 1.819 SOFT MATER SOFT MATERIALS工程技术3 1.154 IEEE T NANOBIOSCI IEEE TRANSACTIONS ON NANOBIOSCIENCE工程技术3 1.280J OPT COMMUN NETW Journal of Optical Communications and Networ工程技术3 1.872 ADV ENG MATER ADVANCED ENGINEERING MATERIALS工程技术3 1.185 INT COMMUN HEAT MASS INTERNATIONAL COMMUNICATIONS IN HEA工程技术3 1.892 OPT LASER ENG OPTICS AND LASERS IN ENGINEERING工程技术3 1.838 COMPUT IND COMPUTERS IN INDUSTRY工程技术3 1.529 ENG LIFE SCI ENGINEERING IN LIFE SCIENCES工程技术3 1.925 CONSTR BUILD MATER CONSTRUCTION AND BUILDING MATERIALS工程技术3 1.834 CMC-COMPUT MATER CON CMC-Computers Materials & Continua工程技术30.972 NONLINEAR DYNAM NONLINEAR DYNAMICS工程技术3 1.247 IEEE T DEPEND SECURE IEEE Transactions on Dependable and Secure C工程技术3 1.140 COMPUT IND ENG COMPUTERS & INDUSTRIAL ENGINEERING工程技术3 1.589 MICROELECTRON ENG MICROELECTRONIC ENGINEERING工程技术3 1.557 ADV ENG INFORM ADVANCED ENGINEERING INFORMATICS工程技术3 1.489 EARTHQ ENG STRUCT D EARTHQUAKE ENGINEERING & STRUCTURA工程技术3 1.778 INT J PHOTOENERGY INTERNATIONAL JOURNAL OF PHOTOENER工程技术3 1.769 B EARTHQ ENG Bulletin of Earthquake Engineering工程技术3 1.559 INFORM SOFTWARE TECH INFORMATION AND SOFTWARE TECHNOLO工程技术3 1.250 COMPUT ELECTRON AGR COMPUTERS AND ELECTRONICS IN AGRICU工程技术3 1.846 SCI TECHNOL WELD JOI SCIENCE AND TECHNOLOGY OF WELDING 工程技术3 1.735 IND MANAGE DATA SYST INDUSTRIAL MANAGEMENT & DATA SYSTEM工程技术3 1.472 INFORM PROCESS MANAG INFORMATION PROCESSING & MANAGEME工程技术3 1.119 IEEE SENS J IEEE SENSORS JOURNAL工程技术3 1.520 IEEE T NUCL SCI IEEE TRANSACTIONS ON NUCLEAR SCIENC工程技术3 1.447 ARTIF INTELL MED ARTIFICIAL INTELLIGENCE IN MEDICINE工程技术3 1.345 COMP MATER SCI COMPUTATIONAL MATERIALS SCIENCE工程技术3 1.574 IEEE T COMMUN IEEE TRANSACTIONS ON COMMUNICATION工程技术3 1.677 J SOL-GEL SCI TECHN JOURNAL OF SOL-GEL SCIENCE AND TECH工程技术3 1.632 FLAVOUR FRAG J FLAVOUR AND FRAGRANCE JOURNAL工程技术3 1.424 IEEE T COMPUT IEEE TRANSACTIONS ON COMPUTERS工程技术3 1.103 STRUCT MULTIDISCIP O STRUCTURAL AND MULTIDISCIPLINARY OP工程技术3 1.488 INT J IMPACT ENG INTERNATIONAL JOURNAL OF IMPACT ENG工程技术3 1.701 EUR FOOD RES TECHNOL EUROPEAN FOOD RESEARCH AND TECHNO工程技术3 1.566 J MATER RES JOURNAL OF MATERIALS RESEARCH工程技术3 1.434 TRANSPORT SCI TRANSPORTATION SCIENCE工程技术3 1.507 MATER CHARACT MATERIALS CHARACTERIZATION工程技术3 1.572 ENERGIES Energies工程技术3 1.865 REV SCI INSTRUM REVIEW OF SCIENTIFIC INSTRUMENTS工程技术3 1.367 IEEE T HAPTICS IEEE Transactions on Haptics工程技术3 1.490 EUR J INFORM SYST EUROPEAN JOURNAL OF INFORMATION SY工程技术3 1.500 NEUROCOMPUTING NEUROCOMPUTING工程技术3 1.580 ENG APPL ARTIF INTEL ENGINEERING APPLICATIONS OF ARTIFICIA工程技术3 1.665 ELECTRON COMMER R A Electronic Commerce Research and Application工程技术3 1.472 INFORMATICA-LITHUAN INFORMATICA工程技术3 1.627 COMPUT AIDED DESIGN COMPUTER-AIDED DESIGN工程技术3 1.234 SYST CONTROL LETT SYSTEMS & CONTROL LETTERS工程技术3 1.222 J POLYM ENVIRON JOURNAL OF POLYMERS AND THE ENVIRO工程技术3 1.349 J POLYM SCI POL PHYS JOURNAL OF POLYMER SCIENCE PART B-P工程技术3 1.531 COMPUT MATH APPL COMPUTERS & MATHEMATICS WITH APPLI工程技术3 1.747 J INF SCI JOURNAL OF INFORMATION SCIENCE工程技术3 1.299 TRANSPORTATION TRANSPORTATION工程技术3 1.023 J SENS STUD JOURNAL OF SENSORY STUDIES工程技术3 1.597 IEEE T SERV COMPUT IEEE Transactions on Services Computing工程技术3 1.468 IEEE GEOSCI REMOTE S IEEE Geoscience and Remote Sensing Letters工程技术3 1.560 J NANOSCI NANOTECHNO JOURNAL OF NANOSCIENCE AND NANOTEC工程技术3 1.563 INT J WEB GRID SERV International Journal of Web and Grid Services工程技术3 1.919 ENG ANAL BOUND ELEM ENGINEERING ANALYSIS WITH BOUNDARY工程技术3 1.451 J SOUND VIB JOURNAL OF SOUND AND VIBRATION工程技术3 1.588。
SRIM –The stopping and range of ions in matter (2010)James F.Ziegler a,*,M.D.Ziegler b ,J.P.Biersack caUnited States Naval Academy,Physics Dept.,Annapolis,MD 21402,USA bUniversity of California at Los Angeles,Los Angeles,CA 90066,USA cBerlin,Germanya r t i c l e i n f o Article history:Available online 26February 2010Keywords:SRIMIon stopping Stopping power Stopping force Ion rangea b s t r a c tSRIM is a software package concerning the S topping and R ange of I ons in M atter.Since its introduction in 1985,major upgrades are made about every six years.Currently,more than 700scientific citations are made to SRIM every year.For SRIM-2010,the following major improvements have been made:(1)About 2800new experimental stopping powers were added to the database,increasing it to over 28,000stop-ping values.(2)Improved corrections were made for the stopping of ions in compounds.(3)New heavy ion stopping calculations have led to significant improvements on SRIM stopping accuracy.(4)A self-con-tained SRIM module has been included to allow SRIM stopping and range values to be controlled and read by other software applications.(5)Individual interatomic potentials have been included for all ion/atom collisions,and these potentials are now included in the SRIM package.A full catalog of stopping power plots can be downloaded at .Over 500plots show the accuracy of the stopping and ranges produced by SRIM along with 27,000experimental data points.References to the citations which reported the experimental data are included.Published by Elsevier B.V.1.IntroductionSRIM is a software package concerning the S topping and R ange of I ons in M atter.It has been continuously upgraded since its intro-duction in 1985[1].A recent textbook ‘‘SRIM –The Stopping and Range of Ions in Matter ”describes in detail the fundamental phys-ics of the software [2].Since this time,corrections have been made based on new experimental data [3].Major changes occur in SRIM about every six years.The last major changes were in 1995and 1998and 2003.In 1995a complete overhaul was made of the stop-ping of relativistic light ions with energies above 1MeV/u.In 1998,special attention was made to the Barkas Effect and the theoretical stopping of Li ions.In 2010,significant changes were made to cor-rect the stopping of ions in compounds.All the figures in this paper are also available on the SRIM website,in considerably more detail.2.SRIM-2010stopping accuracyShown in Table 1are the statistical improvements in SRIM’s stopping power accuracy when compared to experimental data and also compared to SRIM-1998.The right two columns show the percentage of data points within 5%and within 10%of the SRIM calculation.The experimental stopping powers for heavy ions contain far more scatter than for light ions,hence there are larger errors for heavy ions,Be–U.The accuracy of SRIM-2010for individual ions or targets can be reviewed by viewing plots which compare experimental values and the equivalent SRIM calculations.Fig.1shows a typical com-parison for a light ion,He,in Ag.Fig.2shows a similar plot for all heavy ions,Be(4)–U(92)in Ag.Here,the various ion stopping powers have been normalized to the stopping of Al ions in Al,(nor-malization means that for any ion,the relative error of its experi-mental value to that calculated by SRIM is plotted with a similar displacement from the stopping of Al ions in Ag).Note that the scatter of data points is much higher than for the case of He ions in Ag,which increases the perceived error of SRIM.Higher resolu-tion figures for each heavy ion and all elemental targets are avail-able at .3.Stopping of ions in compoundsBragg and Kleeman,in 1903,conducted stopping experiments with a radium source in organic gases such as methyl bromide and methyl iodide to find how alpha stopping depended on the atomic weight of the target.They also calculated the stopping contribution of hydrogen and carbon atoms in hydrocarbon target gases by assuming a linear addition based on the chemical composition of H and C atoms in the targets.The concept that the stopping power of a compound may be estimated by the linear combination of the stopping powers of its individual elements has come to be known as Bragg’s Rule [4].This rule is reasonably accurate,and the measured stopping of ions in compounds usually deviates less than 20%from that0168-583X/$-see front matter Published by Elsevier B.V.doi:10.1016/j.nimb.2010.02.091*Corresponding author.E-mail address:Ziegler@ (J.F.Ziegler).Nuclear Instruments and Methods in Physics Research B 268(2010)1818–1823Contents lists available at ScienceDirectNuclear Instruments and Methods in Physics Research Bjournal homepage:www.els ev i e r.c o m /l o c a t e /n i mbpredicted by Bragg’s rule.The accuracy of Bragg’s rule is limited be-cause the energy loss to the electrons in any material depends on the detailed orbital and excitation structure of the matter,and any differences between bonding in elemental materials and in compounds will cause Bragg’s rule to become inaccurate.Further,bonding changes may also alter the charge state of the transitionTable 1Accuracy of SRIM stopping calculations.Approx.data pts.SRIM-1998(%)SRIM-2010(%)SRIM-2010(within 5%)SRIM-2010(within 10%)H ions 9000 4.5 3.968%85%He ions 6800 4.6 3.570%87%Li ions 1700 6.4 4.668%81%Be–U Ions10,6008.1 5.655%78%Overall accuracy28,1006.14.364%85%Notes to Table 1:The above table compares all 28,000data points to SRIM calculations.If wacko points are omitted (those differing from SRIM by more than 25%)then most of the above heavy ion accuracy numbers would be reduced by about 25%.The overall accuracy of SRIM-2010then reduces to 3.9%instead of 4.3%.Approx.data points :Current total data points used in SRIM plots.SRIM-1998:Comparison of SRIM-1998stopping to experimental data.SRIM-1998was the last major change in SRIM stopping powers.SRIM-2010:Current stopping power calculation.SRIM-2010(within 5%):Percentage of experimental data within 5%of the SRIM values.SRIM-2010(within 10%):Percentage of experimental data within 10%of the SRIMvalues.Fig.1.The stopping of He ions in Ag targets.The plot shows experimental values of He ion stopping in Ag targets.It shows the actual stopping,in units of eV/(1015atoms/cm 2).At the right is a listing of the original data citations.As noted,there is a total of 439data points taken from 44papers,and they vary from SRIM calculations by an average of 3.9%.Also noted is the mean ionization potential used for Al (<I >=488eV)and the Fermi velocity ratio for Ag,V /V F =1.254.The <I >value is only used for high energy stopping (>1MeV/u),while the Fermi velocity is important for lower velocities.A higher resolution plot is available at .Fig.2.The stopping of heavy ions in Ag targets.The plot shows experimental stopping values of heavy ions (atomic numbers 4–92)in Ag targets.The plot is organized similar to that of Fig.1.There are 930experimental data points taken from 131citations,and the mean error of SRIM is 4.5%.Ag targets are easy to make and these targets tend to have small grains without texture and contain few contaminants.So the accuracy of SRIM is better than normal when compared to experimental heavy ion data due to the consistency of the targets.A higher resolution plot is available at .J.F.Ziegler et al./Nuclear Instruments and Methods in Physics Research B 268(2010)1818–18231819ion,thus changing the strength of its interaction with the target medium.Detailed experimental studies of Bragg’s rule started in the 1960’s,and wide discrepancies were found from simple additivity of stopping powers.A classic example is shown in Fig.3for targets containing H and C atoms,which show non-additivity of stopping in simple hydrocarbons[5].In thisfigure,the stopping of He ions in various hydrocarbons was measured for pairs of compounds,and the relative contribution of H and C was extracted for each pair (solving two equations with two unknowns).It was found that the relative stopping contributions of H and C differ by almost 2Âover the range of compounds.Similar work studied more com-plex hydrocarbons but instead of adding H and C bonds,they added extra hydrocarbon molecules.In this study,it was found that by adding identical molecules to hydrocarbon strings, stopping linearity returned[6].Adding new molecules to a target just scaled the stopping by the extra number of atoms.These results showed that atomic bonding had large effects on stopping powers in simple molecular targets,while extra agglomeration of molecules to the target compounds had a small stopping effect.Since these early experiments,theorists have shown that extensive calculations can predict the stopping of light ions (usually protons)in hydrocarbon compounds.Much of this work has been based on a seminal paper by Peter Sigmund that devel-oped methods to account for detailed internal motion within a medium[7].This theory allows for arbitrary electronic configura-tions in the target.Sabin and collaborators used this approach to calculate stopping powers for protons in hydrocarbons with good success[8].Sabin’s calculation follows what is sometimes called the‘‘Köln Core and Bond”(CAB)approach which is discussed in detail below.The Core and Bond(CAB)approach suggested that stopping powers in compounds can be predicted using the superposition of stopping by atomic‘‘cores”and then adding the stopping corre-sponding to the bonding electrons[9].The core stopping would simply follow Bragg’s rule for the atoms of the compound,where we linearly add the stopping from each of the atoms in the com-pounds.The chemical bonds of the compound would then contain the necessary stopping correction.They would be evaluated depending on the simple chemical nature of the compound.For example,for hydrocarbons,carbon in C–C,C@C and C…C struc-tures would have different bonding contributions(C@C indicates a double-bond structure and C…C is a triple bond).SRIM uses this CAB approach to generate corrections between Bragg’s rule and compounds containing the common elements in compounds:H,C,N,O,F,S and Cl.These light atoms have the larg-est bonding effect on stopping powers.Heavier atoms are assumed not to contribute anomalously to stopping because of their bonds (discussed later in Stopping of High Energy Heavy Ions).When you use SRIM,you have the option to use the Compound Dictionary which contains the chemical bonding information for about150 common compounds.The compounds with available corrections are shown with a Star symbol,q,next to the name.When these compounds are selected,SRIM shows the chemical bonding dia-gram and calculates the best stopping correction.The correction is a variation from unity(1.0=no correction).Some corrections are quite big:carbon atoms have almost a4Âchange in stopping power from single bonds to triple bonds.This large change indi-cates the importance of making some sort of correction for the stopping of ions in compounds.The CAB corrections that SRIM uses have been extracted from the stopping of H,He and Li ions in more than100compounds, from162experiments.The details of applying this correction are described in Ref.[10].SRIM correctly predicts the stopping of H and He ions in compounds with an accuracy of better than2%at the peak of their stopping power curve,$125keV/u.An example of a large correction for compound targets is the9% correction necessary for a target of water,H2O,see Fig.4.The stop-ping of He ions in gaseous H2and O2is shown with the lower two dotted lines.The stopping in gaseous water vapor is essentially the Bragg’s Rule sum since its bonding correction is only1%,see the upper dotted line.However,for solid water(ice),the sum of stop-ping in H2O is shown as the upper solid line.With the H2O phase correction,which reduces the stopping by9%at the peak,SRIM shows good agreement between predicted stopping and the data from the ten experimental reports[11].The limitations of the CAB approach should be mentioned.A.The most important limitation might be that of the targetband-gap.Experiments on insulating targets dominate the experimental results that we use.For compounds which are conducting,there might be an error with thecalculatedFig.3.Accuracy of Bragg’s Rule in hydrocarbon compounds.In thisfigure,the stopping of He ions(at500keV)in various hydrocarbons is shown for pairs of compounds,with the relative contributions of stopping in H and C extracted assuming Bragg’s Rule and solving using two unknowns[5].This classic paper shows with clarity the errors associated with Bragg’s Rule.The units of the ordinate and abscissa are reduced stopping units,e[18].It is found that the various determinations of stopping by H and C atoms differ by almost2Âover the range of compounds.The result is a clear indication of the importance of including bonding corrections in stopping powers.(Figure from Ref.[5]). 1820J.F.Ziegler et al./Nuclear Instruments and Methods in Physics Research B268(2010)1818–1823stopping correction being too small.Theoretically,band-gap materials are expected to have lower stopping powers than equivalent conductors because the small energy transfers to target electrons are not available in insulators.It is not clear what the magnitude of this effect is,but about 50papers have discussed the stopping of ions in metals and their oxides,e.g.targets of Fe,Fe 2O 3and Fe 3O 4.These exper-iments evaluated similar materials with and without band-gaps.No significant differences were found that could be attributed to the band-gap.Measurements have also been made of the stopping of H and He ions into ice (solid water)with various dopings of salt (NaCl).No change of energy loss was observed for up to 6orders of magnitude change in resistivity of the ice [11].B.The scaling of ion stopping from H to He to Li ions is assumed to be independent of target material.This assump-tion has been evaluated with 27targets which have been measured for two of the three ions (at the same ion velocity)and 6of these targets have been measured for all three ions (see listings in Ref.[11]).In all cases,the stopping scaled identically within 4%.That is,for H (125keV)and He (500keV)and Li (875keV)the scaling of stopping powers was 1:2.7:4.7for the 27targets (average error was <4%).(For those unfamiliar with stopping theory,the primary parameter for the scaling of stopping powers is the ion velocity,which reduces to scaling in units of keV/a.).C.The light elements of He and Ne are missing from the above list of target bonding atoms.No comparative experiments have been done on the stopping into elemental He in solid/gas phases.However studies of stopping into targets of Ne and Ar have been conducted in both gas and solid form.These papers show no significant difference between the stopping in gas and solid phases.It appears that the Van der Waals forces,which hold noble gases together in frozen form,are too weak to effect the energy loss of ions.Of particular note is the extensive work done in a PhD paper by Besenbacher.[12].D.The light target atoms of Li,Be and B are missing from the list of bonding atoms with corrections.This is a serious defect.The number of papers that have looked at com-pounds which contain significant amounts of these three elements is too limited to allow their evaluation.Target atoms of these three elements are considered by SRIM to have no bonding correction,which is clearly not true.But without experimental data,there is no reliable way to eval-uate the contribution of their bonds in compounds.E.Bragg’s Rule and Heavy Target Elements.We have concen-trated on the analysis of the stopping of ions in compounds made up of light elements.For compounds with heavier atoms,many experiments have shown that deviations from Bragg’s rule disappear.In Table 2are shown representative examples of ion stopping in various compounds containing heavy elements.None show measurable deviations from Bragg’s rule.These and other similar results were reviewed in the 1980s [13,14].4.Stopping of high energy heavy ionsThe stopping powers of high energy (E >1MeV/u)heavy ions (Z >3)have two separate components.First is the charge state of these ions,which is traditionally addressed by using the Brandt–Kitagawa approximation,and then the many high velocity effects are combined into modern Bethe–Bloch theory.The Brandt–Kitagawa (BK)theory [15]is easiest to understand relative to the Bohr theory of the average charge state of heavy ions [16].Bohr suggested the simple picture that the energetic heavy ion would lose any of its electrons whose classical velocity was slower than the ion’s velocity.This concept lasted for more than 30years,with remarkable success.The concept was then improved by the suggestion of BK that one should consider instead the loss of any electrons whose velocity was slower than the relative velocity of the ion to the target medium.This lowered the charge state of heavy ions since the relative velocity of the ion was always lower than its absolute velocity.BK then presented a simple method of calculating this relative velocity based on considering the target to be a perfect Fermi conductor.This significantly improved the calculation of stopping powers [1].Modern approaches to Bethe–Bloch stopping equation have been reviewed in detail in Ref.[17].In Bethe–Bloch,twolargeFig.4.Corrections for stopping in compounds:He ions in water.The effects on stopping of target phase are illustrated in the figure for the stopping of He ions in water (solid and gaseous).Data from 14citations are shown.The special bonding of H–O in water is approximately the same for H–H and O–O bonds,so the stopping in the gaseous H 2O is almost the same as found using Bragg’s Rule.However,a large 9%phase correction must be applied to calculate the stopping of H 2O in solid forms,ice and water (see text).A higher resolution plot is available at .J.F.Ziegler et al./Nuclear Instruments and Methods in Physics Research B 268(2010)1818–18231821components are not well described by pure theoretical consider-ations:(1)the mean ionization energy of the target,commonly symbolized using<I>,and(2)the shell corrections for the target, called C/Z2.The<I>value for a target corrects for the quantized en-ergy levels of the target electrons and also any band-gap and target phase correction.The C/Z2term corrects for the Bethe–Bloch assumption that the ion velocity is much larger than the target electron velocities.This term is usually calculated by detailed accounting of the particle’s interaction with each electronic orbit in various elements.Since both of these terms are only dependent on the target,they are assumed to be the same for heavy ions and lighter ions.An example of SRIM’s stopping accuracy for heavy ions is shown in Fig.5.It shows the ratio of experimental stopping values to SRIM calculation for heavy ions in Al targets.(Al targets seem to be the most reliable target to make,since the data scatter about an aver-age value is the least of that for any solid).The data shown are from 135papers,and represents720data points for ion energies over 1MeV/u.There are several heavy ion data points at about 100MeV/u which show about5%higher experimental values than SRIM values.This is of the order of the estimated nuclear reaction losses,and is always a problem with very high energy ions (>10MeV/u).5.Anomalous heavy Ion stopping valuesSRIM uses several different stopping theories to evaluate the accuracy of experimental stopping powers.Specifically,calcula-tions are made for all ions in individual targets(which eliminates common difficulties with target dependent quantities such as shell corrections and mean ionization potentials,discussed above).Cal-culations are also made of one heavy ion in all solids,which elim-inates some of the difficulties with ion dependent quantities such as the degree of ion stripping.Also,calculations are made from fun-damental theories like the Brandt–Kitagawa theory and LSS theory [18].If the experimental values are within reasonable agreement with this set of theoretical calculations,then the experimental val-ues are weighted with the theoretical values to obtainfinal values. However,at times,significant errors occur in experimental stop-ping values and they deviate so far from theoretical values that they are totally ignored.Table2Bragg’s rule accuracy in heavy compounds.Compound Deviation from Bragg’s rule(%)Compound Deviation from Bragg’s rule(%)Compound Deviation from Bragg’s rule(%)Al2O3<1HfSi2<2Si3N4<2Au–Ag alloys<1NbC<2Ta2O5<1Au–Cu alloys<2NbN<2TiO2<1BaCl2<2Nb2O5<1W2N3<2BaF2<2RhSi<2WO3<2Fe2O3<1SiC<2ZnO<1Fe3O4<1Note:For compounds which contain elements with atomic numbers greater than12,it is possible to combine the CAB approach with Bragg’s rule.The CAB approach can beused for the small atomic number cores and bonds,and these can be combined with the normal stopping contribution of the other components of thecompound.Fig.5.Stopping of high energy heavy ions in aluminum.Thefigure shows the ratio of experimental stopping to SRIM calculation for high energy(>1MeV/u)ions in aluminum.The data shown is from135papers,and represents720data points over1MeV/u.The mean error is2.7%.There are several heavy ion data points at about 100MeV/u which show about5%higher experimental values than SRIM values.This is of the order of estimated nuclear reaction losses,and is always a problem with very high energy ions(>10MeV/u).A higher resolution plot is available at .1822J.F.Ziegler et al./Nuclear Instruments and Methods in Physics Research B268(2010)1818–1823Shown in Fig.6is the stopping of Mg ions in all solids.Note the large number of experimental data points below 100keV/u,which diverge from the SRIM stopping by up to 200%.For Mg ions,SRIM has an average accuracy of about 9%,the worst for any ion.Almost lost by the large number of data points which disagree with SRIM are those from seven citations which showed values almost identi-cal to SRIM.All of the deviant experimental stopping values were deter-mined by a technique called ‘‘Inverted Doppler Shift Attenuation ”,IDSA [19].This technique relies on the knowledge of the life-time of an excited nuclear state and is fraught with potential errors.The technique requires a nuclear reaction to occur in the target,result-ing in an emitted gamma ray.The gamma ray energy may be shifted due to motion of the recoiling particle.A particular source of error occurs if the differential of the particle energy loss with ion velocity changes much while the particle is slowing down.Note that in the energy range of 10–100keV/u,the energy loss is chang-ing rapidly with ion velocity,and this is where the maximum devi-ation occurs between IDSA stopping values and SRIM.As also shown,SRIM agrees well with 7papers which measured stopping using other methods.The advantage of the IDSA technique is that it can be used to determine stopping in difficult targets such as liquids and also to evaluate bonding effects in compounds.However,it is often used without full consideration of its sensitivity to non-linear effects.6.SRIM sub-routine moduleA ‘‘module”has been made so that the stopping and ranges of SRIM may be run as a batch sub-program for other applications [20].This allows the user to use SRIM as a sub-routine of another application that needs stopping powers and ranges.The user cre-ates a control file and executes the file ‘‘SRModule.exe”which will generate an output table similar to those normally made by SRIM.The user can generate the standard file (with stopping and ranges)or can generate a file which contains stopping powers for a specific list of energies.AcknowledgementsThe author is particularly indebted to the many users of SRIM who helped debug the first twenty five years of SRIM,leading to SRIM-2010.Without your significant help and enthusiasm,SRIM would not be the robust and versatile program that it is.References[1]J.F.Ziegler,J.Biersack,U.Littmark,‘‘The Stopping and Range of Ions in Matter”,Pergamon Press,1985.[2]J.F.Ziegler,J.P.Biersack,M.D.Ziegler,SRIM –The Stopping and Range of Ions inMatter”,Ion Implantation Press,2008./content/1524197.[3]See .More than 500plots are included showing more than28,000experimental data points as compared to SRIM calculations.[4]W.H.Bragg,R.Kleeman,Phil.Mag.10(1905)318.[5]A.S.Lodhi,D.Powers,Phys.Rev.A10(1974)2131.[6]D.Powers,Acc.Chem.Res.13(1980)433.[7]P.Sigmund,Phys.Rev.A26(1982)2497.[8]J.R.Sabin,J.Oddershede,Nucl.Instrum.Methods B27(1987)280.[9]G.Both,R.Krotz,K.Lohman,W.Neuwirth,Phys.Rev.A28(1983)3212.[10]The most recent core and bond values used in SRIM are shown at: n SRIM n CompoundsCABTheory.htm .The modeling technique used to extract these values was originally described in:J.F.Ziegler,J.M.Manoyan,Nucl.Instrum.Methods,B35(1988)215.[11]The data plotted in Fig.4are from papers listed at: n SRIM n Compounds.htm .This website also describes in detail how corrections are made for target phase changes (solid or gas phases)and for target compound binding.Also,citations are listed for compounds containing heavy atoms,and also the effects of variations of the target band-gap on stopping powers.[12]F.Besenbacher,J.Bottiger,O.Graversen,J.Hanse,H.Sorensen,Nucl.Instrum.Methods 188(1981)657–667.[13]D.I.Thwaites,Nucl.Instrum.Methods B12(1985)84.[14]D.I.Thwaites,Nucl.Instrum.Methods B27(1987)293.[15]W.Brandt,M.Kitagawa,Phys.Rev.25B (1982)5631.[16]N.Bohr,Mat.-Fys.Medd.K.Dan.Selse 18(1948)1.[17]J.F.Ziegler,Applied physics reviews,J.Appl.Phys.85(1999)1249–1272.[18]J.Lindhard,M.Scharff,H.E.Schiott,Kgl.Danske Vid.Sels.Mat.-Fys.Medd.33(1963)1.[19]P.Petkova, A.Dewaldb,P.von Brentano,‘‘A new procedure for lifetimedetermination using the Doppler-shift attenuation method”,Nucl.Instrum.Methods A-560(2006)564–570.[20]Details of using the Stopping and Range module are included in the SRIM-2010package.See the SRIM directory,.../SR Module/HELP SRModule.rtf.Fig.6.The stopping of Mg ions in all solids.The plot shows experimental stopping values for Mg ions in all solids.This plot shows a considerable number of data points which differ from SRIM calculations,especially for low energy ions (<100keV/u).The variation arises from the use of ‘‘Inverted Doppler Shift Attenuation”,IDSA,as a method to measure stopping powers.This technique is quite complex and relies on the knowledge of the life-time of an excited nuclear state (see text)and is fraught with potential errors.As shown,SRIM calculations are in serious disagreement with the lower energy Mg values of which were determined by IDSA,however it agrees with 7papers which measured stopping at the same energies,using other methods.A higher resolution plot is available at .J.F.Ziegler et al./Nuclear Instruments and Methods in Physics Research B 268(2010)1818–18231823。
第 21 卷 第 12 期2023 年 12 月Vol.21,No.12Dec.,2023太赫兹科学与电子信息学报Journal of Terahertz Science and Electronic Information Technology新型太赫兹光子晶体OAM光纤设计杨婧翾1,李巍2,成利敏1(1.廊坊师范学院电子信息工程学院,河北廊坊065000;2.北华航天工业学院电子与控制工程学院,河北廊坊065000)摘要:太赫兹通信兼具微波通信和光波通信的优势,是解决通信容量紧缺难题的最有效技术手段之一。
针对太赫兹波段吸收损耗严重及抗外在扰动差,难以支持长距传输问题,设计了一种基于环形光子晶体光纤(PCF)结构的新型太赫兹光纤。
以现有常见材料作为光纤基底材质,通过创新光纤结构中空气孔排布方式,抵消材料高吸收损耗,以支持高性能轨道角动量(OAM)模式传输。
选择最优参数,实现6个OAM模式群的高模式质量、低限制损耗和宽带宽的稳定传输。
在0.2~0.9 THz宽波段内,实现模式纯度超过88.9%,限制损耗小于10-7 dB/m。
通过软件仿真实验设计,解决了太赫兹与OAM技术相结合的关键问题,为模分复用(MDM)技术在太赫兹通信系统的应用奠定了理论研究基础。
关键词:轨道角动量;太赫兹通信;光子晶体光纤;模分复用中图分类号:TN914 文献标志码:A doi:10.11805/TKYDA2023089Design of new terahertz photonic crystal fiber forOrbital Angular Momentum modes transmissionYANG Jingxuan1,LI Wei2,CHENG Limin1(1.School of Electrical Information Engineering,Langfang Normal University,Langfang Hebei 065000,China;2.School of Electronic and Control Engineering,North China Institute of Aerospace Engineering,Langfang Hebei 065000,China)AbstractAbstract::Terahertz communication has the advantages of both microwave communication and optical communication, which is one of the most effective technical means to solve the problem ofcommunication capacity shortage. In order to solve the problems of serious absorption loss and poorexternal disturbance resistance in terahertz band, a new terahertz fiber based on circular PhotonicCrystal Fiber(PCF) structure is designed to support high performance Orbital Angular Momentum(OAM)modes transmission. The existing common materials are used as the fiber base material, and the highabsorption loss of materials is offset by the innovation of hollow porosity arrangement in the fiberstructure. The optimal parameters are selected to realize the stable transmission of six OAM mode groupswith high mode quality, low confinement loss and wide bandwidth. The mode purity is above 88.9% andthe confinement loss is below 10-7 dB/m in the 0.2~0.9 THz band. Through simulation, the key problemof combining terahertz and OAM technology is solved, which lays a theoretical foundation for theapplication of Mode Division Multiplexing(MDM) technology in terahertz communication system.KeywordsKeywords::Orbital Angular Momentum;terahertz communication;Photonic Crystal Fiber;Mode Division Multiplexing随着信息互联网技术创新的快速发展,人工智能、高清视频、直播等新的应用方式受到了社会各界的广泛关注[1-3]。
阈值阈值3.4585225序号123456789101112131415161718192021222324252627282930 31 32NanoNanotechnology Biology andPROPHOTOVOLTAICSPROCEEDIEEEJOURNALPROGRESGROWTH ANDCHARACTERIZATION OFMATERIALSACM COMPUPolymCHEMISTRYACPROGRESSCIENCETREBIOTECHNOLOGYADVANCEMATERIALSNANOANNUALBIOMEDICAL ENGINEERINGLAB OINTERMATERIALS REVIEWSBIOTECADVANCESCURRENBIOTECHNOLOGYMaterBIOMAnnual Revand Biomolecular EngineeringSADVANCEANNUALMATERIALS RESEARCHNATURENature NNATURE BIEnergy EducTechnologyPROGRESSSCIENCEMATERIAENGINEERING R-REPORTSNanPROGRESAND COMBUSTION刊111111111111111111111111377分区111111111549-9634NANOMED-NANOTECHNOL1062-7995PROG PHOTOVOLTAICS0018-9219P IEEE1369-7021MATER TODAY0142-9612BIOMATERIALS0021-9517J CATAL1473-0197LAB CHIP0950-6608INT MATER REV0960-8974PROG CRYST GROWTH CH0360-0300ACM COMPUT SURV1558-3724POLYM REV1616-301X ADV FUNCT MATER0734-9750BIOTECHNOL ADV0897-4756CHEM MATER1947-5438ANNU REV CHEM BIOMOL1613-6810SMALL0958-1669CURR OPIN BIOTECH1531-7331ANNU REV MATER RES1936-0851ACS NANO0079-6816PROG SURF SCI0167-7799TRENDS BIOTECHNOLISSN刊名简称1476-1122NAT MATER1748-3387NAT NANOTECHNOL1523-9829ANNU REV BIOMED ENG1区2区期刊数期刊数3区阈值1.931 1.1121087-0156NAT BIOTECHNOL1301-8361ENERGY EDUC SCI TECH0079-6425PROG MATER SCI0935-9648ADV MATER0927-796X MAT SCI ENG R1748-0132NANO TODAY0360-1285PROG ENERG COMBUST1530-6984NANO LETT33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 5051 52 5354 55 56 57 58 59 6061 62CRITICALFOOD SCIENCE ANDCURRENSOLID STATE & MATERIALSIEEE TRANINDUSTRIAL ELECTRONICSMACROMOLCOMMUNICATIONSSOLAMATERIALS AND SOLARJOURNASOURCESINTERNATIOF PLASTICITYBiotechnolMIS QSIAM JouSciencesBIORTECHNOLOGYADVANCEMECHANICSELECTROCOMMUNICATIONSMOLECULAFOOD RESEARCHJOURNALCHEMISTRYMETABOLICSoActa BNaIEEE CoSurveys and TutorialsIEEE SIGNAMAGAZINECAIEEE TRANPATTERN ANALYSIS ANDMACHINE INTELLIGENCEMACRORENESUSTAINABLE ENERGYCRITICALBIOTECHNOLOGYNPG ABIOSEBIOELECTRONICSPROGRESELECTRONICSMRS1111111111111111111111111111110378-7753J POWER SOURCES1040-8398CRIT REV FOOD SCI1388-2481ELECTROCHEM COMMUN1359-0286CURR OPIN SOLID ST M0278-0046IEEE T IND ELECTRON1022-1336MACROMOL RAPID COMM0927-0248SOL ENERG MAT SOL C1613-4125MOL NUTR FOOD RES1744-683X SOFT MATTER1742-7061ACTA BIOMATER0960-8524BIORESOURCE TECHNOL0065-2156ADV APPL MECH1754-6834BIOTECHNOL BIOFUELS0276-7783MIS QUART1053-5888IEEE SIGNAL PROC MAG0008-6223CARBON2040-3364NANOSCALE1936-4954SIAM J IMAGING SCI0162-8828IEEE T PATTERN ANAL0024-9297MACROMOLECULES0749-6419INT J PLASTICITY1553-877X IEEE COMMUN SURV TUT0883-7694MRS BULL0959-9428J MATER CHEM1364-0321RENEW SUST ENERG REV0738-8551CRIT REV BIOTECHNOL1096-7176METAB ENG1884-4049NPG ASIA MATER0956-5663BIOSENS BIOELECTRON0079-6727PROG QUANT ELECTRON63 64 65 6667 68 69 70 71 7273 74 7576 77 7879 8081 82 83 8485 86878889 90 91 92 93 94SENSACTUATORS B-CHEMICALAPPLIED SREVIEWSFOOD CPlaJournal of NMicrofluidicsENIEEE JOSELECTED TOPICS INQUANTUM ELECTRONICSIEEE TRANFUZZY SYSTEMSBIOTECHBIOENGINEERINGIEEE TRANPOWER ELECTRONICSBIOMASSELECTROAnnual RScience and TechnologyNANOTEACM TRANGRAPHICSJOURNALSCIENCETRENDS IN& TECHNOLOGYIEEE JOSELECTED AREAS INORGANICACTA MINTERNATIOF COMPUTER VISIONIEEE TRANEVOLUTIONARYAPPLIEACS ApplInterfacesINTERNATIOF HYDROGEN ENERGYJOURNAL OMATERIALSMEDICAL IMHUMANINTERACTIONMicrobialInternationalSystems222211112222111211111111111111111741-2560J NEURAL ENG1613-4982MICROFLUID NANOFLUID0308-8146FOOD CHEM0360-5442ENERGY0016-2361FUEL1557-1955PLASMONICS0376-7388J MEMBRANE SCI0925-4005SENSOR ACTUAT B-CHEM1077-260X IEEE J SEL TOP QUANT1063-6706IEEE T FUZZY SYST0961-9534BIOMASS BIOENERG0570-4928APPL SPECTROSC REV1941-1413ANNU REV FOOD SCI T0957-4484NANOTECHNOLOGY0730-0301ACM T GRAPHIC0013-4686ELECTROCHIM ACTA0306-2619APPL ENERG1944-8244ACS APPL MATER INTER0006-3592BIOTECHNOL BIOENG0885-8993IEEE T POWER ELECTR0733-8716IEEE J SEL AREA COMM1566-1199ORG ELECTRON1361-8415MED IMAGE ANAL0737-0024HUM-COMPUT INTERACT0360-3199INT J HYDROGEN ENERG1359-6454ACTA MATER1475-2859MICROB CELL FACT0129-0657INT J NEURAL SYST0924-2244TRENDS FOOD SCI TECH0304-3894J HAZARD MATER0920-5691INT J COMPUT VISION1089-778X IEEE T EVOLUT COMPUT9596 979899100 101 102 103 104 105106 107108 109 110 111 112 113 114 115116 117 118119 120 121 122 123 124DENTALCOMPOSAND TECHNOLOGYJOUBIOTECHNOLOGYIEEE CIntelligence MagazineJournal ofBehavior of BiomedicalCOMBUSTIINTERNATIOF ROBOTICS RESEARCHENVIRMODELLING & SOFTWARESCIETECHNOLOGY OFFOOD HYDJournal of Cand ManagementIEEE TRANIMAGE PROCESSINGIEEE COMMAGAZINEFuJOUNANOPARTICLE RESEARCHMARINE BIOJournal of SINFORMATCORROSMICROMICROANALYSISINTERNATIOF NONLINEAR SCIENCESAND NUMERICALSIMULATIONFOOD MIBIOMMICRODEVICESAPPLIED MAND BIOTECHNOLOGYIEEE JOURSTATE CIRCUITSINTERNATIOF FOOD MICROBIOLOGYInternatioNanomedicineCHEMICALJOURNALCurrenBiomechanicMechanobiology2222222222222222222222222222221392-3730J CIV ENG MANAG1057-7149IEEE T IMAGE PROCESS0266-3538COMPOS SCI TECHNOL0168-1656J BIOTECHNOL1388-0764J NANOPART RES1436-2228MAR BIOTECHNOL0163-6804IEEE COMMUN MAG1556-603X IEEE COMPUT INTELL M1548-7660J STAT SOFTW0020-0255INFORM SCIENCES0109-5641DENT MATER1615-6846FUEL CELLS1431-9276MICROSC MICROANAL1751-6161J MECH BEHAV BIOMED0278-3649INT J ROBOT RES1364-8152ENVIRON MODELL SOFTW1178-2013INT J NANOMED0010-2180COMBUST FLAME1468-6996SCI TECHNOL ADV MAT0268-005X FOOD HYDROCOLLOID0010-938X CORROS SCI1385-8947CHEM ENG J1570-1646CURR PROTEOMICS1617-7959BIOMECH MODEL MECHAN0018-9200IEEE J SOLID-ST CIRC0168-1605INT J FOOD MICROBIOL0740-0020FOOD MICROBIOL1387-2176BIOMED MICRODEVICES0175-7598APPL MICROBIOL BIOT1565-1339INT J NONLIN SCI NUM125126127128129130131132133134135136137138139140141142143144145146147148149150151152153Science of ANanoscale IEEE ELECLETTERSIEEE TRANINFORMATION THEORY FUEL PTECHNOLOGY PROGRESSSCIENCES BiofIEEE TRANSOFTWARE ENGINEERINGFOODINTERNATIONAL COMPUTEAND INFRASTRUCTUREENGINEERING JOURNAL OMATERIALS RESEARCHInnovativeEmerging Technologies Journal of Nand Rehabilitation SCRIPTA IBM JORESEARCH AND JOURNAL JOURNALAND COMPATIBLE JOURNALLEARNING RESEARCHVLDB JOUSUPERCRITICAL FLUIDS IEEE TRANNEURAL NETWORKS IEEE TRANSYSTEMS MAN AND CYBERNETICS PART B-CYBERNETICS InternatioElectrochemical Science PROCEEDCOMBUSTION INSTITUTE DYES AN IEEE IndusMagazineSEPARPURIFICATION CEL GOLD 222222222222222222222222222220098-5589IEEE T SOFTWARE ENG1947-2935SCI ADV MATER0963-9969FOOD RES INT 1466-8564INNOV FOOD SCI EMERG 1743-0003J NEUROENG REHABIL 0376-0421PROG AEROSP SCI 1758-5082BIOFABRICATION 0741-3106IEEE ELECTR DEVICE L 0018-9448IEEE T INFORM THEORY 0004-5411J ACM0883-9115J BIOACT COMPAT POL 1359-6462SCRIPTA MATER 0378-3820FUEL PROCESS TECHNOL 1532-4435J MACH LEARN RES 1066-8888VLDB J1931-7573NANOSCALE RES LETT 0018-8646IBM J RES DEV0017-1557GOLD BULL 1093-9687COMPUT-AIDED CIV INF 0896-8446J SUPERCRIT FLUID 1045-9227IEEE T NEURAL NETWOR1540-7489P COMBUST INST 1549-3296J BIOMED MATER RES A 1932-4529IEEE IND ELECTRON M 1383-5866SEP PURIF TECHNOL0969-0239CELLULOSE 0143-7208DYES PIGMENTS 1083-4419IEEE T SYST MAN CY B 1452-3981INT J ELECTROCHEM SC154155 156157158 159 160161 162 163164 165166167168 169 170171 172173174 175 176 177178 179 180181 182 183 184COMPOSAPPLIED SCIENCE ANDMANUFACTURINGIEEE TRANENERGY CONVERSIONISPRS JPHOTOGRAMMETRY ANDREMOTE SENSINGIEEE PoMagazineBIOCENGINEERING JOURNALPATTERNIEEEEARTHQUJournal ofIEEE TRANSIGNAL PROCESSINGREACTIVEPOLYMERSENERGFOOD ANTOXICOLOGYIEEE TraRoboticsAUTIEEECOMPUTINGANNUALINFORMATION SCIENCEAND TECHNOLOGYIEEE INSYSTEMSJournal ofARTIFICIALADVABIOCHEMICALENGINEERING /BIOTECHNOLOGYINTEGRATEAIDED ENGINEERINGIEEE TRANGEOSCIENCE AND REMOTESENSINGIEEE TRANINTELLIGENTTRANSPORTATIONSYSTEMSRENEWAIEEE-ASMEON MECHATRONICSBIOTECOMPREHEIN FOOD SCIENCE ANDFOOD SAFETYBMC BIOTFOOD22222222222222222222222222222220887-0624ENERG FUEL1359-835X COMPOS PART A-APPL S0924-2716ISPRS J PHOTOGRAMM1540-7977IEEE POWER ENERGY M0272-1732IEEE MICRO0885-8969IEEE T ENERGY CONVER1570-8268J WEB SEMANT1053-587X IEEE T SIGNAL PROCES1381-5148REACT FUNCT POLYM8755-2930EARTHQ SPECTRA1069-2509INTEGR COMPUT-AID E0196-2892IEEE T GEOSCI REMOTE1369-703X BIOCHEM ENG J0031-3203PATTERN RECOGN1552-3098IEEE T ROBOT0005-1098AUTOMATICA1083-4435IEEE-ASME T MECH0736-6205BIOTECHNIQUES1524-9050IEEE T INTELL TRANSP1089-7801IEEE INTERNET COMPUT1942-0862MABS-AUSTIN1541-4337COMPR REV FOOD SCI F0278-6915FOOD CHEM TOXICOL0960-1481RENEW ENERG0956-7135FOOD CONTROL0066-4200ANNU REV INFORM SCI1556-4959J FIELD ROBOT0004-3702ARTIF INTELL1541-1672IEEE INTELL SYST0724-6145ADV BIOCHEM ENG BIOT1472-6750BMC BIOTECHNOL185 186187 188189 190 191 192 193 194 195 196 197 198 199 200 201 202 203 204205 206 207 208 209 210 211 212JOURNEUROPEAN CERAMICIEEE PhoINTERNAJOURNALPLANT FOONUTRITIONIEEE CONTMAGAZINECOMPUINTELLIGENCEIEEE TRANPOWER SYSTEMSLEBENWISSENSCHAFT UND-TECHNOLOGIE-FOODSCIENCE ANDJOURNAL OMICROBIOLOGY &BIOTECHNOLOGYCHEMICALSCIENCEEXPERT SAPPLICATIONSIEEE TRANELECTRON DEVICESJOURNAL OTECHNOLOGYJOURNASCIENCETRANSRESEARCH PART B-JOURNALTHERMODYNAMICSAPPLIED SOINFORMANAGEMENTJournal ofMateriaEngineering C-Materials forARCCOMPUTATIONALANNALS OENGINEERINGCEMENT ARESEARCHIEEE TRANMOBILE COMPUTINGCOMPUTEREnterprisSystemsJOURNAL OTECHNOLOGYJOURNELECTROCHEMICAL22222222222222222222222222221367-5435J IND MICROBIOL BIOT0009-2509CHEM ENG SCI0885-8950IEEE T POWER SYST0023-6438LWT-FOOD SCI TECHNOL0018-9383IEEE T ELECTRON DEV0955-2219J EUR CERAM SOC0958-6946INT DAIRY J0921-9668PLANT FOOD HUM NUTR1748-0221J INSTRUM1943-0655IEEE PHOTONICS J1066-033X IEEE CONTR SYST MAG0824-7935COMPUT INTELL-US0957-4174EXPERT SYST APPL0928-4931MAT SCI ENG C-MATER1134-3060ARCH COMPUT METHOD E0090-6964ANN BIOMED ENG1568-4946APPL SOFT COMPUT0378-7206INFORM MANAGE-AMSTER0733-5210J CEREAL SCI0191-2615TRANSPORT RES B-METH0360-1315COMPUT EDUC1751-7575ENTERP INF SYST-UK0008-8846CEMENT CONCRETE RES0021-9614J CHEM THERMODYN0268-3962J INF TECHNOL0013-4651J ELECTROCHEM SOC0733-8724J LIGHTWAVE TECHNOL1536-1233IEEE T MOBILE COMPUT213214 215 216217 218219 220 221 222 223 224 225 226 227 228 229 230 231 232 233 234 235 236 237238239 240INTERNATIOF COAL GEOLOGYJOURNAMERICAN SOCIETY FORINFORMATION SCIENCEAND TECHNOLOGYMATERIALAND PHYSICSULTRAMFUTURECOMPUTER SYSTEMSJOURNALINFORMATION SYSTEMSJOURNAL OCOMPOUNDSPhotonics anFundamentals andJOURNAL OMATERIALS RESEARCHPART B-APPLIEDBIOMATERIALSCOMPUTEAPPLIED MECHANICS ANDENGINEERINGJOURNALSCIENCE-MATERIALS INMEDICINEBiomedFOOD QPREFERENCEIEEE WCOMMUNICATIONSIEEE JourTopics in Signal ProcessingSOLAIEEE TRANWIRELESSCOMMUNICACMBioinIEEE TRANAUTOMATIC CONTROLJOURNENGINEERINGAUSTRALIAGRAPE AND WINEMEATIEEE PCOMPUTINGUSER MOUSER-ADAPTEDEVOLCOMPUTATIONIEEE TraAutonomous MentalBIOTECPROGRESS22222222222222222222222222221532-2882J AM SOC INF SCI TEC0925-8388J ALLOY COMPD1569-4410PHOTONIC NANOSTRUCT0167-739X FUTURE GENER COMP SY1552-4973J BIOMED MATER RES B0045-7825COMPUT METHOD APPL M0166-5162INT J COAL GEOL0963-8687J STRATEGIC INF SYST0018-9286IEEE T AUTOMAT CONTR0254-0584MATER CHEM PHYS0957-4530J MATER SCI-MATER M1748-6041BIOMED MATER1932-4553IEEE J-STSP0304-3991ULTRAMICROSCOPY1536-1276IEEE T WIREL COMMUN0001-0782COMMUN ACM1934-8630BIOINTERPHASES0038-092X SOL ENERGY0924-1868USER MODEL USER-ADAP1063-6560EVOL COMPUT0950-3293FOOD QUAL PREFER1536-1284IEEE WIREL COMMUN1322-7130AUST J GRAPE WINE R0309-1740MEAT SCI1536-1268IEEE PERVAS COMPUT1943-0604IEEE T AUTON MENT DE8756-7938BIOTECHNOL PROGR0260-8774J FOOD ENG241 242243 244 245 246 247 248 249250 251252 253 254255 256257 258259 260261262 263 264 265 266267 268269COMPUTERENGINEERINGJOUBIOMATERIALSENERGY COMANAGEMENTIEEE TraIndustrial InformaticsIEEE TRANBIOMEDICAL ENGINEERINGBUILDENVIRONMENTIEEE JOQUANTUM ELECTRONICSIEEECOMPOSITIEEE-ACM TON NETWORKINGMATERIAJOURNAL OINTERIEEE RAUTOMATION MAGAZINECHEMOMINTELLIGENT LABORATORYSYSTEMSINTERNATIOF HEAT AND MASSFLUID PHAIEEE TRANSYSTEMS MAN ANDCYBERNETICS PART A-SYSTEMS AND HUMANSAICHENEURAL CInternatioSemantic Web andIEEE TRANVISUALIZATION ANDCOMPUTER GRAPHICSDESAKNOWLINFORMATION SYSTEMSARTIFJOURNAMERICAN CERAMICJOUMICROMECHANICS ANDJOURNCOMPOSITION ANDDECISIOSYSTEMS222222222222222222222222222220933-2790J CRYPTOL0966-9795INTERMETALLICS0098-1354COMPUT CHEM ENG0196-8904ENERG CONVERS MANAGE1551-3203IEEE T IND INFORM0017-9310INT J HEAT MASS TRAN0378-3812FLUID PHASE EQUILIBR1070-9932IEEE ROBOT AUTOM MAG0018-9294IEEE T BIO-MED ENG1083-4427IEEE T SYST MAN CY A0001-1541AICHE J0885-3282J BIOMATER APPL0169-7439CHEMOMETR INTELL LAB1552-6283INT J SEMANT WEB INF0360-1323BUILD ENVIRON0890-8044IEEE NETWORK0263-8223COMPOS STRUCT0002-7820J AM CERAM SOC0018-9197IEEE J QUANTUM ELECT1063-6692IEEE ACM T NETWORK0167-577X MATER LETT0899-7667NEURAL COMPUT0960-1317J MICROMECH MICROENG0889-1575J FOOD COMPOS ANAL0167-9236DECIS SUPPORT SYST0219-1377KNOWL INF SYST1064-5462ARTIF LIFE1077-2626IEEE T VIS COMPUT GR0011-9164DESALINATION270271272 273 274 275 276 277 278 279 280 281 282 283284 285 286287288 289290 291 292ACM TransaNetworksHYDROMENERGY AMATERIALSENGINEERING A-STRUCTURAL MATERIALSPROPERTIES MICROSTFOOD ADCONTAMINANTSJOURNAL OEDUCATIONIEEE PTECHNOLOGY LETTERSIEEE ENGMEDICINE AND BIOLOGYMAGAZINEINDUENGINEERING CHEMISTRYEUROPEJOURNAL EINTERNATIOF MACHINE TOOLS &MANUFACTUREJOUBIOMATERIALS SCIENCE-NEURALJOURNALTECHNOLOGY ANDBIOTECHNOLOGYBaltic JourBridge EngineeringINTERNATIOF ELECTRICAL POWER &ENERGY SYSTEMSIEEE TRANCIRCUITS AND SYSTEMSFOR VIDEO TECHNOLOGYJOURNAL OELECTROCHEMISTRYJOUMANAGEMENTJOUMICROELECTROMECHANICAL SYSTEMSIEEE TRANSYSTEMS MAN ANDCYBERNETICS PART C-APPLICATIONS AND REJournal of thInformation SystemsMATERIALBULLETIN222222222222222222222221041-1135IEEE PHOTONIC TECH L0739-5175IEEE ENG MED BIOL0304-386X HYDROMETALLURGY0378-7788ENERG BUILDINGS0890-6955INT J MACH TOOL MANU0920-5063J BIOMAT SCI-POLYM E0888-5885IND ENG CHEM RES0921-5093MAT SCI ENG A-STRUCT0893-6080NEURAL NETWORKS0268-2575J CHEM TECHNOL BIOT1550-4859ACM T SENSOR NETWORK1292-8941EUR PHYS J E0025-5408MATER RES BULL1944-0049FOOD ADDIT CONTAM A1822-427X BALT J ROAD BRIDGE E0142-0615INT J ELEC POWER1432-8488J SOLID STATE ELECTR1069-4730J ENG EDUC1057-7157J MICROELECTROMECH S1094-6977IEEE T SYST MAN CY C1536-9323J ASSOC INF SYST0742-1222J MANAGE INFORM SYST1051-8215IEEE T CIRC SYST VID293294295 296297 298 299300 301 302303 304 305 306 307 308 309 310 311312 313314 315 316317 318 319 320 321BIOPROBIOSYSTEMSSTRUCTUMONITORING-ANINTERNATIONAL JOURNALTRANSRESEARCH PART A-POLICYMECHANIAND SIGNAL PROCESSINGMACROMATERIALS ANDCEMENTCOMPOSITESCOASTALIET NanoDATA MKNOWLEDGE DISCOVERYJournal of DiBIODEGIEEE TRANANTENNAS ANDMECHANICSSMART MSTRUCTURESSTRUCTUINTERNATIOF APPROXIMATEPOWDERJOURNAL OAND ENGINEERINGIEEE TRANNANOTECHNOLOGYELECTROCSOLID STATE LETTERSAPPLIEENGINEERINGSURFACETECHNOLOGYIEEE TRANKNOWLEDGE AND DATAENGINEERINGINTERNATIOF ENERGY RESEARCHCOMPULINGUISTICSINTERNATIOF ADHESION ANDINTERNATIFOR NUMERICAL METHODSIN ENGINEERINGFunctionalIEEE TRANMICROWAVE THEORY ANDTECHNIQUES333333333332222222222222222220965-8564TRANSPORT RES A-POL1438-7492MACROMOL MATER ENG0888-3270MECH SYST SIGNAL PR0888-613X INT J APPROX REASON0032-5910POWDER TECHNOL1615-7591BIOPROC BIOSYST ENG1475-9217STRUCT HEALTH MONIT0378-3839COAST ENG1751-8741IET NANOBIOTECHNOL1099-0062ELECTROCHEM SOLID ST1359-4311APPL THERM ENG1226-086X J IND ENG CHEM1384-5810DATA MIN KNOWL DISC0257-8972SURF COAT TECH1041-4347IEEE T KNOWL DATA EN0958-9465CEMENT CONCRETE COMP1536-125X IEEE T NANOTECHNOL0891-2017COMPUT LINGUIST1551-319X J DISP TECHNOL0018-926X IEEE T ANTENN PROPAG0167-6636MECH MATER1793-6047FUNCT MATER LETT0923-9820BIODEGRADATION0964-1726SMART MATER STRUCT0167-4730STRUCT SAF0363-907X INT J ENERG RES0018-9480IEEE T MICROW THEORY0143-7496INT J ADHES ADHES0029-5981INT J NUMER METH ENG322323 324325 326327 328 329 330 331332 333 334335 336337 338 339 340 341 342 343 344 345 346 347 348349 350 351CHEMIDEPOSITIONSENSACTUATORS A-PHYSICALMATERIAIEEE-ACMComputational Biology andBioinformaticsFoodPLASMA CPLASMA PROCESSINGOPTICALARTIFICAPPLIESCIENCEPROGRESCOATINGSIEEE TraInformation Forensics andJOURNALAND ENGINEERING DATAIEEE TraBiomedical Circuits andACM TRANCOMPUTER SYSTEMSENERGSYNTHEACM TransaCOMACM TRANMATHEMATICALADSORPTIOTHE INTERNATIONALADSORPTION SOCIETYDIAMONDMATERIALSTRANSRESEARCH PART E-LOGISTICS ANDTRANSPORTATION REVIEWTHIN SCOMPUTEIMAGE UNDERSTANDINGIEEE TRANMULTIMEDIAJOURNALCONTROLINTERNATIOF THERMAL SCIENCESJournal of FJOURNPROTECTIONCOMPOPERATIONS RESEARCH3333333333333333333333333333330925-3467OPT MATER0948-1907CHEM VAPOR DEPOS0261-3069MATER DESIGN0734-2071ACM T COMPUT SYST0924-4247SENSOR ACTUAT A-PHYS1545-5963IEEE ACM T COMPUT BI1876-4517FOOD SECUR0272-4324PLASMA CHEM PLASMA P0195-6574ENERG J0018-9162COMPUTER0098-3500ACM T MATH SOFTWARE0021-9568J CHEM ENG DATA1932-4545IEEE T BIOMED CIRC S0169-4332APPL SURF SCI0300-9440PROG ORG COAT1366-5545TRANSPORT RES E-LOG0040-6090THIN SOLID FILMS0929-5607ADSORPTION1556-6013IEEE T INF FOREN SEC1077-3142COMPUT VIS IMAGE UND1520-9210IEEE T MULTIMEDIA0160-564X ARTIF ORGANS0925-9635DIAM RELAT MATER0305-0548COMPUT OPER RES0379-6779SYNTHETIC MET0959-1524J PROCESS CONTR1290-0729INT J THERM SCI1559-1131ACM T WEB1756-4646J FUNCT FOODS0362-028X J FOOD PROTECT352 353354 355 356 357 358359 360361362 363 364 365366 367368 369370 371 372373 374375376377378379380381SIAM JOCOMPUTINGMACHINMEDICAL EPHYSICSGPS SINTERNATIOF DAMAGE MECHANICSBioinspiratioJOURNAMERICAN OIL CHEMISTSCOMPOSENGINEERINGFoodWJOURNALMATERIALSEMPIRICAENGINEERINGAPPLIEAAPGAPPLIED BAND BIOTECHNOLOGYFOOD ANBULLETININTERNATIOF HEAT AND FLUID FLOWSEIET RenGenerationPOLYMADVANCEDKNOWLESYSTEMSIEEE MICWIRELESS COMPONENTSLETTERSInformPOLYMIEEE TRANBROADCASTINGCHEMICALAND PROCESSINGINTERNATIOF CIRCUIT THEORY ANDAPPLICATIONSTRANSRESEARCH PART C-EMERGING TECHNOLOGIESJOURNFRANKLIN INSTITUTE-ENGINEERING ANDAPPLIED MATHEMATICSJOURNALSCIENCE3333333333333333333333333333331056-7895INT J DAMAGE MECH1748-3182BIOINSPIR BIOMIM1350-4533MED ENG PHYS0003-021X J AM OIL CHEM SOC1080-5370GPS SOLUT0142-727X INT J HEAT FLUID FL0097-5397SIAM J COMPUT1359-8368COMPOS PART B-ENG1557-1858FOOD BIOPHYS1382-3256EMPIR SOFTW ENG0885-6125MACH LEARN0149-1423AAPG BULL0273-2289APPL BIOCHEM BIOTECH0379-5721FOOD NUTR BULL1559-128X APPL OPTICS1566-2535INFORM FUSION0142-9418POLYM TEST0043-1648WEAR0022-3115J NUCL MATER1752-1416IET RENEW POWER GEN1042-7147POLYM ADVAN TECHNOL0098-9886INT J CIRC THEOR APP0968-090X TRANSPORT RES C-EMER0018-9316IEEE T BROADCAST0950-7051KNOWL-BASED SYST0016-0032J FRANKLIN I0022-2461J MATER SCI1424-8220SENSORS-BASEL0255-2701CHEM ENG PROCESS1531-1309IEEE MICROW WIREL CO382 383384 385 386387 388389390 391392 393 394395 396397 398399 400 401 402 403 404 405 406407 408IEEE TRANCIRCUITS AND SYSTEMS I-FUNDAMENTAL THEORYAND APPLICATJOUMICROENCAPSULATIONDigesNanomaterials andMODESIMULATION IN MATERIALSSCIENCE ANDENGINEERINGJOURNALMANAGEMENTAd HoEUROPEALIPID SCIENCE ANDMacromolEngineeringCIRPMANUFACTURINGTECHNOLOGYJOURNALSPRAY TECHNOLOGYIEEE TRANDEVICE AND MATERIALSRELIABILITYACM TRANSOFTWARE ENGINEERINGAND METHODOLOGYJOURNSCIENCEAPPLIEDMATERIALS SCIENCE &COMPSTRUCTURESMEXPERIMEINTERNATIOF HUMAN-COMPUTERCurrentJOUMICROSCOPY-OXFORDIEEE TRANINFORMATIONTECHNOLOGY INBIOMEDICINEMETIEEE SIEEE TRANCONTROL SYSTEMSTECHNOLOGYANNUALCONTROLBIOTECHNOMACRORESEARCH3333333333333333333333333330265-2048J MICROENCAPSUL1530-4388IEEE T DEVICE MAT RE1049-331X ACM T SOFTW ENG METH0007-8506CIRP ANN-MANUF TECHN0022-1147J FOOD SCI0947-8396APPL PHYS A-MATER1549-8328IEEE T CIRCUITS-I1059-9630J THERM SPRAY TECHN1996-1944MATERIALS1842-3582DIG J NANOMATER BIOS1063-8016J DATABASE MANAGE1570-8705AD HOC NETW0740-7459IEEE SOFTWARE0965-0393MODEL SIMUL MATER SC1438-7697EUR J LIPID SCI TECH1862-832X MACROMOL REACT ENG0045-7949COMPUT STRUCT1063-6536IEEE T CONTR SYST T1367-5788ANNU REV CONTROL0141-5492BIOTECHNOL LETT1089-7771IEEE T INF TECHNOL B0026-1394METROLOGIA1071-5819INT J HUM-COMPUT ST1573-4137CURR NANOSCI0022-2720J MICROSC-OXFORD1598-5032MACROMOL RES0723-4864EXP FLUIDS409410 411412 413 414 415 416 417 418 419 420 421 422 423 424 425 426 427 428 429 430431432433 434 435 436TRIBOLOJOURNALINTELLIGENCE RESEARCHTRIINTERNATIONALMJournal of thof Chemical EngineersDATA & KENGINEERINGDRYING TCOMPUTFORUMMACROTHEORY AND SIMULATIONSCONTROLPRACTICENanoscaleThermophysical EngineeringMETALLUMATERIALSTRANSACTIONS A-PHYSICAL METALLURGYAND MATERIALAUTONOMAND MULTI-AGENTWINDINTERNATIOF REFRACTORY METALS& HARD MATERIALSIEEE TraComputational Intelligence andBIOLOGICAIEEE TransSpeech and LanguageJOURNAELECTROCHEMISTRYIEEE CGRAPHICS ANDJOURNAL OREASONINGCERAMICSLETTERMICROBIOLOGYIEEE TRANVEHICULAR TECHNOLOGYAPPLIED SIEEE TRANULTRASONICSFERROELECTRICS ANDFREQUENCY CONTROLINTERNATIOF ROBUST ANDNONLINEAR CONTROLINTERNATIOF FATIGUE33333333333333333333333333330737-3937DRY TECHNOL0167-7055COMPUT GRAPH FORUM1076-9757J ARTIF INTELL RES0301-679X TRIBOL INT1556-7265NANOSC MICROSC THERM1073-5623METALL MATER TRANS A1022-1344MACROMOL THEOR SIMUL0968-4328MICRON1387-2532AUTON AGENT MULTI-AG1095-4244WIND ENERGY1023-8883TRIBOL LETT0967-0661CONTROL ENG PRACT0018-9545IEEE T VEH TECHNOL1876-1070J TAIWAN INST CHEM E0263-4368INT J REFRACT MET H1943-068X IEEE T COMP INTEL AI0021-891X J APPL ELECTROCHEM0169-023X DATA KNOWL ENG0168-7433J AUTOM REASONING0272-8842CERAM INT0266-8254LETT APPL MICROBIOL0272-1716IEEE COMPUT GRAPH1049-8923INT J ROBUST NONLIN0142-1123INT J FATIGUE0340-1200BIOL CYBERN1558-7916IEEE T AUDIO SPEECH0003-7028APPL SPECTROSC0885-3010IEEE T ULTRASON FERR。
PHYSICAL REVIEW B84,104112(2011)High-pressure vibrational and optical study of Bi2Te3R.Vilaplana,1,*O.Gomis,1F.J.Manj´o n,2A.Segura,3E.P´e rez-Gonz´a lez,4P.Rodr´ıguez-Hern´a ndez,4A.Mu˜n oz,4 J.Gonz´a lez,5,6V.Mar´ın-Borr´a s,3V.Mu˜n oz-Sanjos´e,3C.Drasar,7and V.Kucek71Centro de Tecnolog´ıas F´ısicas,MALTA Consolider Team,Universitat Polit`e cnica de Val`e ncia,46022Valencia,Spain 2Instituto de Dise˜n o para la Fabricaci´o n y Producci´o n Automatizada,MALTA Consolider Team,Universitat Polit`e cnica de Val`e ncia,46022Valencia,Spain3Instituto de Ciencia de Materiales de la Universidad de Valencia—MALTA Consolider Team—Departamento de F´ısica Aplicada,Universitatde Val`e ncia,46100Burjassot,Valencia,Spain4MALTA Consolider Team—Departamento de F´ısica Fundamental II and Instituto Universitario de Materiales y Nanotecnolog´ıa,Universidad de La Laguna,La Laguna,Tenerife,Spain5DCITIMAC,MALTA Consolider Team,Universidad de Cantabria,Avda.de Los Castros s/n,39005Santander,Spain6Centro de Estudios de Semiconductores,Universidad de los Andes,M´e rida5201,Venezuela7Faculty of Chemical Technology,University of Pardubice,Studentsk´a95,53210-Pardubice,Czech Republic(Received8June2011;revised manuscript received25July2011;published9September2011)We report an experimental and theoretical lattice dynamics study of bismuth telluride(Bi2Te3)up to23GPa together with an experimental and theoretical study of the optical absorption and reflection up to10GPa.The indirect bandgap of the low-pressure rhombohedral(R-3m)phase(α-Bi2Te3)was observed to decrease withpressure at a rate of−6meV/GPa.In regard to lattice dynamics,Raman-active modes ofα-Bi2Te3were observedup to7.4GPa.The pressure dependence of their frequency and width provides evidence of the presence of anelectronic-topological transition around4.0GPa.Above7.4GPa a phase transition is detected to the C2/mstructure.On further increasing pressure two additional phase transitions,attributed to the C2/c and disorderedbcc(Im-3m)phases,have been observed near15.5and21.6GPa in good agreement with the structures recentlyobserved by means of x-ray diffraction at high pressures in Bi2Te3.After release of pressure the sample reverts backto the original rhombohedral phase after considerable hysteresis.Raman-and IR-mode symmetries,frequencies,and pressure coefficients in the different phases are reported and discussed.DOI:10.1103/PhysRevB.84.104112PACS number(s):61.50.Ks,62.50.−p,78.20.Ci,78.30.−jI.INTRODUCTIONBismuth telluride(Bi2Te3)is a layered chalcogenide with a tremendous impact for thermoelectric applications.1The thermoelectric properties of Bi2Te3and their alloys have been extensively studied due to their promising operation in the temperature range of300–400K.In fact Bi2Te3is the material with the best thermoelectric performance at ambient temperature.2,3Recently,it has been shown that Bi2Te3can be exfoliated like graphene and that a single layer exhibits high electrical conductivity and low thermal conductivity so that a new nanostructure route can be envisaged to improve dramatically the thermoelectrical properties of this compound by means of either charge-carrier confinement or acoustic-phonon confinement.4,5Bi2Te3is a narrow bandgap semiconductor with tetradymite crystal structure[R-3m,space group(S.G.)166,Z=3].6 This rhombohedral-layered structure is formed by layers, which containfive hexagonal close-packed atomic sublayers (Te-Bi-Te-Bi-Te)and is named a quintuple linked by van der Waals forces.The same layered structure is common to other narrow bandgap semiconductor chalcogenides,like Bi2Se3and Sb2Te3,and has been found in As2Te3at high pressures.7 Bi2Te3,as well as Bi2Se3and Sb2Te3,has been recently predicted to behave as a topological insulator8;i.e.,a new class of materials that behave as insulators in the bulk but conduct electrical current in the surface.The topological insulators are characterized by the presence of a strong spin-orbit(SO) coupling that leads to the opening of a narrow bandgap and causes certain topological invariants in the bulk to differ from their values in vacuum.The sudden change of invariants at the interface results in metallic,time-reversal invariant-surface states whose properties are useful for applications in spintronics and quantum computation.9,10Therefore,in recent years a number of papers have been devoted to the search of the 3D-topological insulators among Sb2Te3,Bi2Te3,and Bi2Se3, and different works observed the features of the topological nature of the band structure in the three compounds.11–13 High-pressure studies are very useful to understand mate-rials properties and design new materials because the increase in pressure allows us to reduce the interatomic distances and tofinely tune the materials properties.It has been verified that the thermoelectric properties of semiconductor chalcogenides improve with increasing pressure,and that the study of the properties of these materials could help in the design of better thermoelectric materials by substituting external pressure by chemical pressure.14–18Therefore,the electrical and thermoelectric properties of Sb2Te3,Bi2Te3,and Bi2Se3, as well as their electronic-band structure,have been studied at high pressures.19–27In fact a decrease of the bandgap energy with increasing pressure was found in Bi2Te3.19,20 Furthermore,recent high-pressure studies in these compounds have shown a pressure-induced superconductivity28,29that has further stimulated high-pressure studies.30However,the pressure dependence of many properties of these layered chalcogenides is still not known.In particular the determi-nation of the crystalline structures of these materials at high pressures has been a long puzzle15,23,31,32and the space groups of the high-pressure phases of Bi2Te3have been elucidatedR.VILAPLANA et al.PHYSICAL REVIEW B84,104112(2011)only recently by powder x-ray diffraction measurements at synchrotron-radiation sources33,34specially with the use of particle-swarm optimization algorithms for crystal-structure prediction.34Recent high-pressure powder x-ray diffraction measure-ments have evidenced a pressure-induced electronic topolog-ical transition(ETT)in Bi2Te3around3.2GPa as a changein compressibility.29,31,32,35,36An ETT or Lifshitz transitionoccurs when an extreme of the electronic-band structure,whichis associated to a Van Hove singularity in the density of states,crosses the Fermi-energy level.37This crossing,which canbe driven by pressure,temperature,doping,etc.,results in achange in the topology of the Fermi surface that changes theelectronic density of states near the Fermi energy.An ETTis a2.5transition in the Ehrenfest description of the phasetransitions so no discontinuity of the volume(first derivativeof the Gibbs free energy)but a change in the compressibility(second derivative of the Gibbs free energy)is expected inthe vicinity of the ETT.Anomalies in the phonon spectrumare also expected for materials undergoing an ETT38,39andhave been observed in a number of materials40,41as well as inSb1.5Bi0.5Te3.31The lattice dynamics of Bi2Te3have been studied ex-perimentally at room pressure42–44and a recent study sug-gests that Raman spectroscopy can be used to monitorthe number of single quintuple layers in nanostructuredBi2Te3,as in graphene.45Theoretical studies of the lat-tice dynamics of Bi2Te3at room pressure have also beenperformed;46–49however,Raman measurements at high pres-sures have only been reported up to0.5GPa,50and to ourknowledge there is no theoretical study of the lattice dynamicsproperties of Bi2Te3under high pressure.As a part of oursystematic study of the structural stability and the vibrationalproperties of the semiconductor chalcogenide family,wereport in this work room-temperature Raman-scattering mea-surements in Bi2Te3up to23GPa together with total-energyand lattice-dynamical ab initio calculations at different pres-sures.We discuss the recent observation of a pressure-inducedETT in the rhombohedral phase ofα-Bi2Te3and study whetherthe Raman-scattering signal of the Bi2Te3at pressures above7.4GPa match with the proposed high-pressure phases recentlyreported for this compound33,34and which have also beenfound in Sb2Te3at high pressures.51II.EXPERIMENTAL DETAILSWe have used single crystals of p-type Bi2Te3that weregrown using a modified Bridgman technique.A polycrystallineingot was synthesized by the reaction of stoichiometricquantities of the constituting elements(5N).Afterward,thepolycrystalline material was annealed and submitted to thegrowth process in a vertical Bridgman furnace.Preliminaryroom-temperature measurements on single crystalline samples(15mm×4mm×0.3mm)yield in-plane electrical resistivityρ⊥c=1.7·10−5 m and Hall coefficient R H(B c)= g0.52cm3C−1.Following the calculation presented in Ref.52,the latter gives hole concentration of7.2·1018cm−3andminority electron concentration of2.1·1017cm−3.A smallflake of the single crystal(100μm×100μm×5μm)was inserted in a membrane-type diamond anvil cell(DAC)with a4:1methanol-ethanol mixture as pressure-transmitting medium,which ensures hydrostatic con-ditions up to10GPa and quasihydrostatic conditions between 10and23GPa.53,54Pressure was determined by the ruby luminescence method.55Unpolarized room-temperature Raman-scattering measure-ments at high pressures were performed in backscattering geometry using two setups:(i)A Horiba Jobin Yvon LabRAM HR microspectrometer equipped with a TE-cooled multichan-nel CCD detector and a spectral resolution below2cm−1. HeNe laser(6328˚A line)was used for excitation.(ii)A Horiba Jobin Yvon T64000triple-axis spectrometer with resolution of1cm−1.In this case an Ar+laser(6470˚A line)was used for excitation.In order not to burn the sample,power levels below2mW were used inside the DAC.This power is higher than that used in Raman measurements at room pressure due to superior cooling of the sample in direct contact with the pressure-transmitting media and the diamonds.Optical transmission and reflection measurements under pressure were performed by putting the DAC in a home-built Fourier Transform infrared(FTIR)setup operating in the mid-IR region(400–4000cm−1).The pressure-transmitting medium was KBr.The setup consists of a commercial TEO-400FTIR interferometer by ScienceTech S.L.,which includes a Globar thermal-infrared source and a Michelson interferome-ter,and a liquid-nitrogen cooled Mercury-Cadmium-Telluride (MCT)detector with wavelength cutoff at25μm(400cm−1) from IR Associates Inc.A gold-coated parabolic mirror focuses the collimated IR beam onto a calibrated iris of1 to3mm diameter.A gold-coated X15Cassegrain microscope objective focuses the IR beam inside the DAC to a size of70–200μm.A second Cassegrain microscope objective collects the transmitted IR beam and sends it to the detector after being focused by another parabolic mirror.In the reflection configuration,aflat gold mirror is placed at45◦before the focusing Cassegrain objective,blocking half of the IR beam. The half-beam let into the DAC is reflected by the sample, then by theflat gold mirror,andfinally focused on the MCT detector by another parabolic mirror.III.AB INITIO CALCULATIONSTwo recent works have reported the structures of the high-pressure phases of Bi2Te3up to52GPa.33,34The rhombohedral(R-3m)structure(α-Bi2Te3)is suggested to transform to the C2/m(β-Bi2Te3,S.G.12,Z=4)and the C2/c (γ-Bi2Te3,S.G.15,Z=4)structures above8.2and13.4GPa, respectively.34Furthermore,a fourth phase(δ-Bi2Te3)has been found above14.5GPa and assigned to a disordered bcc structure(Im-3m,S.G.229,Z=1).33,34In order to explore the relative stability of these phases in Bi2Te3we have performed ab initio total-energy calculations within the density functional theory(DFT)56using the plane-wave method and the pseudopotential theory with the Vienna ab initio simulation package(V ASP)57We have used the projector-augmented wave scheme(PAW)58implemented in this package.Ba-sis set,including plane waves up to an energy cutoff of 320eV,were used in order to achieve highly converged results and accurate descriptions of the electronic properties. We have used the generalized gradient approximation(GGA)HIGH-PRESSURE VIBRATIONAL AND OPTICAL STUDY...PHYSICAL REVIEW B84,104112(2011)for the description of the exchange-correlation energy with the PBEsol59exchange-correlation prescription.Dense special k-points sampling for the Brillouin zone(BZ)integration were performed in order to obtain very well-converged energies and forces.At each selected volume,the structures were fully relaxed to their equilibrium configuration through the calculation of the forces on atoms and the stress tensor.In the relaxed equilibrium configuration,the forces on the atoms are less than0.002eV/˚A and the deviation of the stress tensor from a diagonal hydrostatic form is less than1kbar(0.1GPa). Since the calculation of the disordered bcc phase was not possible to do,we have attempted to perform calculations for the bcc-like monoclinic C2/m structure proposed in Ref.34. The application of DFT-based total-energy calculations to the study of semiconductors properties under high pressure has been reviewed in Ref.60,showing that the phase stability, electronic and dynamical properties of compounds under pressure are well describe by DFT.Furthermore,since the calculation of the disordered bcc phase is not possible to do with the V ASP code,we have attempted to perform calculations for the bcc-like mono-clinic C2/m structure proposed in Ref.34.Also,because the thermodynamic-phase transition between two structures occurs when the Gibbs free energy(G)is the same for both phases,we have obtained the Gibbs free energy of the different phases using a quasiharmonic Debye model61that allows obtaining G at room temperature from calculations performed for T=0K in order to discuss the relative stability of the different phases proposed in the present work.In order to fully confirm whether the experimentally mea-sured Raman scattering of the high-pressure phases of Bi2Te3 agree with theoretical estimates for these phases,we have also performed lattice-dynamics calculations of the phonon modes in the R-3m,C2/m,and C2/c phases at the zone center ( point)of the BZ.Our theoretical results enable us to assign the Raman modes observed for the different phases of Bi2Te3. Furthermore,the calculations also provide information about the symmetry of the modes and polarization vectors,which is not readily accessible in the present experiment.Highly converged results on forces are required for the calculation of the dynamical matrix.We use the direct-force constant approach(or supercell method).62Highly converged results on forces are required for the calculation of the dynamical matrix. The construction of the dynamical matrix at the point of the BZ is particularly simple and involves separate calculations of the forces in which afixed displacement from the equilibrium configuration of the atoms within the primitive unit cell is considered.Symmetry aids by reducing the number of such independent displacements,reducing the computational effort in the study of the analyzed structures considered in this work.Diagonalization of the dynamical matrix provides both the frequencies of the normal modes and their polarization vectors.It allows to us to identify the irreducible representation and the character of the phonon’s modes at the point.In this work we provide and discuss the calculated frequencies and pressure coefficients of the Raman-active modes for the three calculated phases of Bi2Te3.The theoretical results obtained for infrared-active modes for the three calculated phases of Bi2Te3are given as supplementary material of this article.63Finally,we want to mention that we have also checked the effect of the SO coupling in the structural stability and the phonon frequencies of the different phases.We have found that the effect of the SO coupling is very small and did not affect our present results(small differences of1–3cm−1in the phonon frequencies at the point)but increased substantially the computer time so that the cost of the computation was very high for the more complex monoclinic high-pressure phases, as already discussed in Ref.34.Therefore,all the theoretical values corresponding to lattice-dynamics calculations in the present paper do not include the SO coupling.In order to test our calculations,we show in Table I the calculated lattice parameters in the different phases of Bi2Te3at different pressures.For the sake of comparison we show in Table I other theoretical calculations and experimental results available.As far as the R-3m phase is concerned,our calculated lattice parameters are in relatively good agreement with experimental values from Refs.6and36.Our calculations with GGA-PBEsol give values which are intermediate between those calculated with GGA-PBE and local density approximation (LDA),as it is generally known.Additionally,we give the calculated lattice parameters of Bi2Te3in the monoclinic C2/m and C2/c structures at7.7and15.5GPa,respectively,for comparison with experimental data.Note that in Table I the a and b lattice parameters of the C2/m and C2/c structures at7.7and15.5GPa are very similar to those reported by Zhu et al.;34however,the c lattice parameter andβangle for monoclinic C2/m and C2/c structures differ from those obtained by Zhu et al.34The reason is the results of our ab initio calculations are given in the standard setting for the monoclinic structures,in contrast with Ref.34,for a better comparison to future experiments since many experimentalists use the standard setting.IV.RESULTS AND DISCUSSIONA.Optical absorption ofα-Bi2Te3under pressureIt is known thatα-Bi2Te3has an indirect forbidden bandgap, E gap,between130and170meV.19,64–66Figure1shows the optical transmittance of ourα-Bi2Te3sample in the mid-IR region at room pressure outside the DAC.The spectrum near the fundamental absorption edge is dominated by large interferences.The large amplitude of the interference fringe pattern in the transparent region is a result of the high value of the refractive index,that is larger than9.42,65,66The sample transmittance and the interference-fringe amplitude decreases at low-photon energy due to the onset of free-carrier absorption and to high energies due to the fundamental absorption edge caused by band-to-band absorption.The absorption coefficient can be accurately determined from the transmittance spectrum only in a small photon energy range between the end of the interference pattern and the photon energy at which the transmitted intensity merges into noise.In this interval the absorption coefficient exhibits an exponential dependence on the photon energy.This prevents a detailed analysis of the absorption edge shape.Consequently,the optical bandgap has been determined byfitting a calculated transmittance to the experimental one.We calculate the transmittance by assumingR.VILAPLANA et al.PHYSICAL REVIEW B 84,104112(2011)TABLE I.Calculated (th.)and experimental (exp.)lattice parameters,bulk modulus (B 0),and its derivative (B 0 )of Bi 2Te 3in the R -3m structure at ambient pressure and calculated lattice parameters of Bi 2Te 3in the C 2/m and C 2/c structures at 8.4and 15.5GPa,respectively.a(˚A)b(˚A)c(˚A)β(∞)B 0(GPa)B 0 Ref.α-Bi 2Te 3(0GPa)th.(GGA-PBEsol)4.38029.98241.924.89This work th.(GGA-PBESol)a 4.37530.16741.614.68This workth.(GGA-PBE)4.4531.6349th.(GGA-PBE)a 4.4731.1249th.(LDA)a 4.3630.3847exp.4.38530.4976exp.4.38330.38032.5b 10.1b 3640.9c3.2c β-Bi 2Te 3(8.4GPa)th.(GGA-PBESol)14.8834.0669.12189.7341.254.06This workth.(GGA-PBE)d 14.8654.05617.468148.3934exp.d14.6454.09617.251148.4834γ-Bi 2Te 3(15.5GPa)th.(GGA-PBESol)9.8956.9627.70970.3045.283.57This workth.(GGA-PBE)e 9.9567.14610.415134.8634exp.e10.2336.95510.503136.034a Calculations including the SO coupling.bAt room pressure.cAbove 3.2GPa.dAround 12-12.6GPa.eAround 14-14.4GPa.an absorption coefficient with two termsα(E )=A E 2+Be−E gap −E (1)where the first one corresponds to the free-carrier contribution and the second one corresponds to the Urbach tail of the fundamental absorption edge.Equation (1)was used to fit the calculated transmittance spectra to the experimental ones.The dotted line in Fig.1was calculated with Equation (1)by using only A and E gap as fitting parameters,where E gap =159meV at roompressure.FIG.1.(Color online)Experimental transmittance of a 7-μm-thick α-Bi 2Te 3sample at room pressure outside the DAC (solid line).Dotted line indicates the fit of the experimental spectrum.Figure 2shows the Bi 2Te 3transmittance spectrum for several pressures up to 5.5GPa.Above that pressure the signal-to-noise ratio is too low to determine the optical bandgap energy.Figure 3shows the pressure dependence of the optical bandgap of Bi 2Te 3,as determined from the previously described procedure.The pressure coefficient turns out to be −6.4±0.6meV /GPa.This pressure coefficient of the optical bandgap is close to the value we obtained for the pressure dependence of the indirect bandgap from ab initio calculations (−10meV /GPa).From this result it appears that,even if the sample becomes opaque at 5.5GPa,Bi 2Te 3still has a finite bandgap of some 120meV.FIG.2.(Color online)Experimental transmittance of α-Bi 2Te 3at different pressures up to 5.5.GPa.A shift of the absorption edge to low energies is observed with increasing pressure.HIGH-PRESSURE VIBRATIONAL AND OPTICAL STUDY...PHYSICAL REVIEW B84,104112(2011)FIG.3.(Color online)Pressure dependence of the optical bandgap ofα-Bi2Te3according to reflectance(red squares)and to transmittance(black circles)measurements.Sample opacity above5.5GPa seems to be then a result of the free-carrier absorption tail shifting to higher energies as the carrier concentration increases.Consequently,the sample opacity is likely caused by the overlap of the free-carrier absorption tail with the fundamental-absorption tail rather than a real closure of the bandgap.We have to note that our pressure coefficient of the optical bandgap is somewhat smaller in module than the pressure coefficient previously reported for the indirect bandgap:−22meV/GPa19;−12meV/GPa below3GPa;and−60meV/GPa above3GPa.20We have to consider that the estimation of these pressure coefficients in Refs.19and20were indirectly obtained from the pressure dependence of the electrical conductivity and those estimations suffer considerable errors since they assume that the change in resistivity is only due to the change of the indirect bandgap energy,which is not a well-founded assumption in extrinsic degenerate semiconductors.In order to confirm our results on optical absorption we have performed high-pressure reflectance measurements in a3-μm-thick sample whose results are shown in Fig.4.The reflectance spectrum also exhibits a large interference fringe pattern in the transparency region,with amplitude decreasing to lowand high photon energies.The reflectance spectrum at6GPaFIG.4.(Color online)Experimental reflectance ofα-Bi2Te3at different pressures.shows that the sample exhibits a clear onset of the fundamental absorption edge at around120meV and also that the free-carrier absorption edge,even if it has shifted to higher energies, has not overlapped the fundamental absorption.Therefore our reflectance measurements allow us to confirm the results obtained from absorption measurements.Furthermore,the bandgap pressure coefficient,as determined from the shift of the photon energy at which interferences disappear,agrees with the one determined from the transmission spectra.At 7GPa,a clear change in the reflectance occurs,with a large increase of the reflectance by80%in the low-energy range.A large reflectance minimum(not shown here)appears at some 4000cm−1(500meV),suggesting a phase transition to a metallic phase.The metallic nature of the high-pressure phases is in good agreement with previously reported resistivity measurements.17,21,28–30If the reflectance minimum is taken as an estimation of the plasma frequency of the high-pressure phase above7GPa,the carrier concentration would be larger than1021cm−3(assuming the same dielectric constant as in the rhombohedral phase).If the dielectric constant inβphase is much smaller,the carrier concentration should be close to1022cm−3,which is more consistent with the observed superconducting behavior.28–30The shift of the free-carrier absorption tail follows the in-crease of the free-carrier plasma frequency.Then the pressure dependence of the plasma frequency can be estimated from the shift of the photon energy at which the free-carrier absorption tail quenches the interference fringe pattern.Reflectance measurements outside the cell show that the plasma frequency at ambient pressure is below50meV,consistently with the hole concentration that is of the order of7·1018cm−3,as measured by Hall effect.At4.3GPa interference fringes are observed down to some60meV(560cm−1).This upper limit to the plasma frequency would correspond to hole concentration of lower than1019cm−3,typical of a degenerate semiconductor.This increase in the hole concentration should result in a Burstein-Moss positive contribution to the optical bandgap, which explains the discrepancy between the experimental and theoretical value of the bandgap pressure coefficient.The bandgap around5GPa is in fact smaller than the measured optical gap.Given the band structure of Bi2Te3,67with six equivalent minima in the valence band,the density of states is very large and the hole concentration per minimum would be only of some1.5×1018cm−3,which would lead to a Burstein-Moss shift of some50meV for a hole effective mass of0.09m0.68Then even taking into account the Burstein-Moss shift,Bi2Te3at5GPa would still be a low-gap semiconductor. In fact this estimation of the Burstein-Moss shift is based on the ambient-pressure electronic structure.At pressures above the ETT transition the density of states in the valence-band maximum is expected to be much larger as the ellipsoids merge into a thoroidal ring,as proposed by Istkevitch et al.69 Consequently,the Burstein-Moss shift above the ETT should be much lower than50meV.Finally,we must note that our analysis of the optical absorption edge in Bi2Te3have not allowed us to detect any change in the pressure dependence of the indirect bandgap around3GPa to confirm the presence of an ETT as observed in other works.20,29,31,32,35,36The very small change in the pressure coefficient of the indirect bandgap seems not toR.VILAPLANA et al.PHYSICAL REVIEW B 84,104112(2011)FIG.5.Experimental Raman spectra of α-Bi 2Te 3at pressures between room pressure and 7.4GPa.be affected by the ETT since there is no change in volume but in volume compressibility,and the change is very subtle to be measured in our transmission or reflection spectra in comparison with the drastic effects observed in transport measurements or even in the parameters of the Raman modes (as will be discussed in the next section).B.Raman scattering of α-Bi 2Te 3under pressureThe rhombohedral structure of α-Bi 2Te 3is a centrosym-metric structure that has a primitive cell with one Te atom located in a 3a Wyckoff position and the remaining Bi(2)and Te(2)atoms occupying 6c Wyckoff sites.Therefore,group theory allows 10zone-center modes,which decompose in the irreducible representations as follows 70:10=2A 1g +3A 2u +2E g +3E u .(2)The two acoustic branches come from one A 2u and a doubly degenerated E u mode,while the rest correspond to optic modes.Gerade (g)modes are Raman active while ungerade (u)modes are infrared (IR)active.Therefore,there are four Raman-active modes (2A 1g +2E g )and four IR-active modes (2A 2u +2E u ).The E g modes correspond to atomic vibrations in the plane of the layers,while the A 1g modes correspond to vibrations along the c axis perpendicular to the layers.42–44,50Figure 5shows the experimental Raman spectra of α-Bi 2Te 3at different pressures up to 7.4GPa.We have observed and followed under pressure three out of the four Raman-active modes.The E g mode calculated to be close to 40cm −1has not been observed in our experiments as it was also not seen in previous Raman-scattering measurements at room and high pressures.42,50,71–73Figure 6(a)shows the experimental-pressure dependence of the frequencies of the three first-orderRaman modes measured in α-Bi 2Te 3,and Table II summarizes our experimental and theoretical first-order Raman-mode frequencies and pressure coefficients in the rhombohedral phase.Our experimental frequencies at room pressure are in good agreement with those already measured in Ref.42and Ref.50and with those recently measured in Refs.45and 71–73.On the other hand our theoretical frequencies at room pressure are also in good agreement with those reported in Ref.49without SO coupling (see Table II )and are slightly larger than those calculated including SO coupling (see Ref.49).In Fig.6(a)it can be observed that all the measured Raman modes exhibit a hardening with increasing pressure.The experimental values of the pressure coefficients of the Raman-mode frequencies are in a general good agreement with our theoretical calculations and with the values reported in Ref.50up to 0.5GPa;however,a decrease of the pressure coefficient of two modes around 4.0GPa should be noted [see dashed lines in Fig.6(b)].We have attributed the less positive pressure coefficient of these two Raman modes to the pressure-induced ETT observed in Sb 2Te 3and Bi 2Te 3.20,29,31,32,35,36In fact in a previous study in Sb 2Te 3under pressure we have found a change in the pressure coefficient of the frequency of all modes measured.51In order to support our hypothesis we also plot as Fig.6(b)the pressure dependence of the full width at half maximum (FWHM)of the three measured Raman modes.Curiously,it is observed that the FWHM changes its slope around 4GPa thus confirming an anomaly related to the ETT.Therefore,both our results of the pressure dependence of the frequency and linewidth give support to the observation of the ETT around 4.0GPa in α-Bi 2Te 3similarly to the case of α-Sb 2Te 3.51As previously commented,anomalies in the phonon spec-trum are also expected for materials undergoing a ETT and have been observed in Sb 1.5Bi 0.5Te 3.15In the latter work the high-frequency A 1g mode was not altered near the ETT in good agreement with our measurements;however,we have noted a change both in the lower A 1g and the higher-frequency E g modes.Since A 1g modes are polarized in the direction perpendicular to the layers while the E g modes are polarized along the layers,our observation of a less positive pressure coefficient at 4.0GPa of both modes in α-Bi 2Te 3suggests that the ETT in Bi 2Te 3is related to a change of the structural compressibility of both the direction perpendicular to the layers and the direction along the layers.This seems not to be in agreement with Polian et al.’s observations,which suggest that the ETT in Bi 2Te 3only affects the plane of the layers.36Consequently,more work is needed to understand the mechanism of the ETT in this material.To conclude this section regarding the rhombohedral structure of α-Bi 2Te 3,we want to make a comment on the pressure coefficients of the Raman modes of this structure in comparison to those recently measured in α-Sb 2Te 3.51It is known that in chalcogenide laminar materials,the two lowest frequency E and A modes are usually related to shear vibrations between adjacent layers along the a -b plane and to vibrations of one layer against the others along the c axis,respectively.It has been commented that the E mode displays the smallest pressure coefficient due to the weak bending force constant between the interlayer distances (in our case,Te-Te distances)while。
Spin-chargeseparationinthesingle-hole-dopedMottantiferromagnetZ.Y.Weng,1V.N.Muthukumar,2D.N.Sheng,1,3andC.S.Ting11TexasCenterforSuperconductivity,UniversityofHouston,Houston,Texas77204-5506
2JosephHenryLaboratoriesofPhysics,PrincetonUniversity,Princeton,NewJersey08544
3DepartmentofPhysicsandAstronomy,CaliforniaStateUniversity,Northridge,California91330
͑Received23August2000;published16January2001͒
ThemotionofasingleholeinaMottantiferromagnetisinvestigatedbasedonthet-Jmodel.Anexactexpressionoftheenergyspectrumisobtained,inwhichtheirreparablephasestringeffect͓Phys.Rev.Lett.77,5102͑1996͔͒isexplicitlypresent.Byidentifyingthephasestringeffectwithspinbackflow,wepointoutthatspin-chargeseparationmustexistinsuchasystem:thedopedholehastodecayintoaneutralspinonandaspinlessholon,togetherwiththephasestring.Weshowthatwhilethespinonremainscoherent,theholonmotionisdeterredbythephasestring,resultinginitslocalizationinspace.Wecalculatetheelectronspectralfunctionwhichexplainsthelineshapeofthespectralfunctionaswellasthe‘‘quasiparticle’’spectrumobservedinangle-resolvedphotoemissionexperiments.Otheranalyticandnumericalapproachesarediscussedbasedonthepresentframework.
DOI:10.1103/PhysRevB.63.075102PACSnumber͑s͒:71.27.ϩa,74.20.Mn,74.72.Ϫh
I.INTRODUCTIONSincethediscoveryofhigh-Tcsuperconductivity,therehasbeenmuchefforttoelucidatethepropertiesofdopedMottinsulators.Inthiscontext,thespecificcaseofoneholeinaMottinsulatingantiferromagnet͑half-filledband͒hasbeenthesubjectofextensivetheoreticalandexperimentalinvestigation.Thesestudiesaddressthebasicquestionofwhetherthemotionofaholeintheantiferromagnetcanpos-siblybedescribedwithinaquasiparticleapproach.Photo-emissionspectroscopyprovidesvaluableinformationthatcanhelpresolvethisissue.Experimentalresultsfromangle-resolvedphotoemissionspectroscopy͑ARPES͒arenowavailableforSr2CuO2Cl2aswellasforCa2CuO2Cl2.
1–5
BoththesematerialsareMottinsulatorsandareparentcom-poundsofthehigh-Tccuprates.TheresultsfromARPEScan
besummarizedasfollows:͑i͒thespectralfeaturesobservedarenotsharpatall.Quitetothecontrary,anintrinsicbroadfeatureextendingtoenergiesoftheorderof1.5eVbeforemergingintothemainvalencebandisseen.Thisistobecontrastedwithsharpspectralfeaturesthatoneexpectsinaquasiparticlescenario;͑ii͒theobserveddispersionisisotro-picaroundk0ϭ(/2,/2);͑iii͒themeasurementsofRon-
ningetal.3revealthepresenceofaso-calledremnantFermisurfaceofthemomentumstructureintheenergy-integratedspectralfunction.ThebroadspectralfeaturesseeninARPESstronglysug-gestthebreakdownofthequasiparticlepicture.Basedonsuchconsiderations,Laughlin6conjecturedthefailureof
quasiparticletheoryinthesematerials,andfurtherproposedthattheobservedisotropicdispersionhasitsoriginsinanunderlyingspinonspectrum.Thispictureenvisagesspin-chargeseparationwiththescaleoftheobserveddispersiondeterminedbythesuperexchangeJ.Anentirelydifferentpictureispresentedbytheself-consistentBornapproximation͑SCBA͒approach.7–10
Thoughthisschemeisbasedonthet-Jmodel,itdepictsaspin-polaronpictureforthesingleholecasewherethedopedholebehavesnotverydifferentfromaquasiparticleinthe
Landau-Fermiliquidtheory.TheSCBAresultshavealsobeensupportedbyexactnumericalcalculationsonfinitelattices11upto32sites12aswellasvariationalcalculations
onlargerlattices.13Here,theconsistencyamongdifferent
numericalmethodsmostlyconcernsthespectrum⑀k,usuallydefinedastheminimumenergyforagivenmomentumk.Thespectrumisanisotropicaroundthe‘‘Fermipoints’’k0
ϭ(Ϯ/2,Ϯ/2)inallthesecalculations.Itiscalleda‘‘qua-
siparticle’’spectrumsinceasharppeakinthespectralfunc-tionusuallyappearsat⑀kinboththeSCBAaswellasresults
fromexactdiagonalization.NotethattheseresultsarenotconsistentwithARPESwhichexhibitsanisotropicdisper-sionaroundk0,andratherbroadspectralfeatures.Thus,to
accountfortheformer,theinclusionofsecond(tЈ)andthird(tЉ)nearest-neighborhoppingstothet-Jmodelhavebeenproposedintheliterature.2,14–16However,thisapproachis
meaningfulonlywhenthequasiparticledescriptionholds,i.e.,onlyifasharpquasiparticlepeakexistsat⑀kinthet-J
model.AsillustratedinFig.1,ifinthethermodynamiclimit,thespectralfunctionforagivenmomentumkdoesnotshowasharpquasiparticlepeak,then,notwithstandinghowaccu-rately⑀kisdeterminedbyvarioustheoreticalmethods,ithas
noexperimentalimplication.For,inthiscase,thehigherenergy⑀k
Јatthebroad‘‘peak’’inFig.1,whichisobservable
experimentally,mayhavenothingtodowiththeanisotropic⑀k.
Onmoregeneralgrounds,thebreakdownoftheLandau-FermiquasiparticlepicturehasbeendiscussedbyAndersonforadopedMottinsulatorinthepresenceofanupperHub-bardband.17HearguesthatthequasiparticleweightZvan-ishesduetounrenormalizableFermi-surfacephaseshiftsin-ducedwhenaparticleisinjectedintotheMottinsulator.Indeed,basedonarigorousformulationofthesingle-electronGreen’sfunctionintheone-holecaseusingthet-Jmodel,ithasbeendemonstrated18,19thatthequasiparticle