JEPonline Journal of Exercise Physiology online
- 格式:pdf
- 大小:59.81 KB
- 文档页数:8

延边大学医学学报 2022年3月 第45卷 第1期与疾病报告2020概要[J].中国循环杂志,2021,36(6):521-545.[2] MEDINA-LEYTE DJ,ZEPEDA-GARCíA O,DOMíNG-UEZPéREZ M,et al..Endothelial dysfunction,inflam-mation and coronary artery disease:potential biomark-ers and promising therapeutical approaches[J].Int JMol Sci,2021,22(8):3850.[3] HUANG C,HUANG WH,WANG R,et al..Ulinastatininhibits the proliferation,invasion and phenotypicswitching of PDGF-BB-induced VSMCs via Akt/eNOS/NO/cGMP signaling pathway[J].Drug Des Devel Ther,2020,14:5505-5514.[4] 赵高峰.延边地区朝鲜族SMARCA4基因SNPrs1122608与冠心病相关性研究[D].延吉:延边大学,2020.[5] IM PK,MILLWOOD IY,KARTSONAKI C,et al..Al-cohol drinking and risks of total and site-specific canc-ers in China:A 10-year prospective study of 0.5millionadults[J].Int J Cancer,2021,149(3):522-534.[6] ROSCH PJ.Could the benefits of drinking alcohol out-weigh all its risks[J].Stress Health,2013.[7] WALLERATH T,POLEO D,LI H,et al..Red wine in-creases the expression of human endothelial nitric oxidesynthase:a mechanism that may contribute to its bene-ficial cardiovascular effects[J].JACC,2003,41(3):471-478.[8] ZHAO F,JI Z,CHI J,et al..Effects of Chinese yellowwine on nitric oxide synthase and intercellular adhesionmolecule-1expressions in rat vascular endothelial cells[J].Acta Cardiologica,2016,71(1):27-34.[9] 金春花,刘文博,关宏锏,等.一氧化氮合酶各种亚型的信号通路对心力衰竭的作用机制与治疗的研究进展[J].中国循环杂志,2017,32(8):830-832.[10]WAGNER MC,YELIGAR SM,LOU AB,et al..PPARγligands regulate NADPH oxidase,eNOS,and barrierfunction in the lung following chronic alcohol ingestion[J].Alcohol Clin Exp Res,2012,36(2):197-206.[11]TIRAPELLI CR,LEONE AF,YOGI A,et al..Ethanolconsumption increases blood pressure and alters the re-sponsiveness of the mesenteric vasculature in rats[J].J Pharm Pharmacol,2008,60(3):331-341.[12]CHEN SS,WANG ZY,ZHOU H,et al..Icariin re-duces high glucose-induced endothelial progenitor celldysfunction via inhibiting the p38/CREB pathway andactivating the Akt/eNOS/NO pathway[J].Exp TherMed,2019,18(6):4774-4780.■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■[基金项目] 国家自然科学基金项目(编号:81560179).[收稿日期] 2022-01-03[作者简介] 黄银珠(1995—),女,硕士,研究方向为口腔颌面外科.[通信作者] 金成日,Email:jcr1105@163.com.初级纤毛相关基因KIF3A通过Hedgehog信号通路影响口腔鳞状细胞癌增殖研究黄银珠,李京旭,金成日(延边大学附属医院口腔科,吉林延吉133000)[摘要] [目的]探讨初级纤毛相关基因KIF3A通过Hedgehog信号通路对口腔鳞状细胞癌增殖能力产生的影响.[方法]选择人舌鳞癌Tca8113细胞株作为研究对象,分为KIF3A-siRNA转染组和对照组.采用蛋白印迹实验和实时荧光定量实验检测各组KIF3A、Shh、Gli1蛋白及mRNA的相对表达水平;利用MTT细胞增殖实验观察Tca8113细胞增殖能力的变化.[结果]蛋白印迹实验结果显示,与对照组比较,KIF3A-siRNA转染组KIF3A、Shh、Gli1蛋白表达水平均明显下调(P<0.05);实时荧光定量实验结果显示,与对照组比较,KIF3A-siRNA转染组KIF3A、Shh、Gli1mRNA表达水平均明显下调(P<0.01);MTT细胞增殖实验结果显示,细胞培养24h时两组细胞增殖间差异无统计学意义(P>0.05),细胞培养48、72h时KIF3A-siRNA转染组增殖率较对照组明显降低(P<0.01).[结论]沉默初级纤毛相关基因KIF3A可能通过调控Hedgehog信号通路抑制口腔鳞癌的增殖能力.[关键词] 口腔鳞状细胞癌;初级纤毛;KIF3A;Hedgehog信号通路DOI:10.16068/j.1000-1824.2022.01.002[中图分类号] R 739.8 [文献标志码] A [文章编号] 1000-1824(2022)01-0007-04·7·■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■Journal of Medical Science Yanbian University Mar.2022 Vol.45 No.1Effects of primary cilium-associated gene KIF3Aon oral squamous cellcarcinoma proliferation by Hedgehog signaling pathwayHUANG Yinzhu,LI Jingxu,JIN Chengri(Department of Stomatology,Affiliated Hospital of Yanbian University,Yanji 133000,Jilin,China)ABSTRACT:OBJECTIVETo explore the effect of primary cilium-associated gene KIF3Aon oral squamouscell carcinoma proliferation through Hedgehog signaling pathway.METHODS Human tongue squamous cellcarcinoma Tca8113cell line was selected and divided into KIF3A-siRNA transfection group and controlgroup.Western blot and real-time qPCR were used to detect the relative expression levels of KIF3A,Shhand Gli1proteins and mRNA in each group.MTT cell proliferation assay was used to observe theproliferation of Tca8113cells.RESULTS Western blot results showed that compared with the control group,the protein expression levels of KIF3A,Shh and Gli1in the KIF3A-SiRNA transfection group weresignificantly down-regulated(P<0.05).Real-time qPCR results showed that compared with the controlgroup,the mRNA expression levels of KIF3A,Shh and Gli1in the KIF3A-SiRNA transfection group weresignificantly down-regulated(P<0.01).MTT cell proliferation assay showed that there was no significantdifference in cell proliferation between the two groups after 24hcell culture(P>0.05).The proliferationrate of the KIF3A-SiRNA transfection group was significantly lower than that of the control group after cellculture for 48and 72h(P<0.01).CONCLUSIONSilencing primary cilium-associated gene KIF3Amayinhibit the proliferation of oral squamous cell carcinoma by regulating Hedgehog signaling pathway.Keywords:oral squamous cell carcinoma;primary cilia;KIF3A;Hedgehog signaling pathway 口腔癌人数约占全世界罹患恶性肿瘤人数的3%,其中鳞状细胞癌比例超过90%.2005年,WHO将口腔鳞状细胞癌(OSCC)定义为一种具有不同分化程度和侵袭性的肿瘤,早期可出现广泛的淋巴结转移,好发于40~70岁的烟酒爱好者[1].口腔癌患者5年生存率约为50%[2],患者术后预后差和5年生存率低的主要原因为患者未及时就诊、淋巴结转移及复发[3].控制OSCC的侵袭转移能力对预后至关重要,认为可偿试从分子水平出发找出更好的治疗方法[4].初生纤毛属于一种细胞表面的细胞器,在脊椎动物发育和人类遗传疾病中发挥着重要作用.纤毛对发育信号的响应是必需的,而越来越多的证据表明初级纤毛是专属于Hedgehog信号传导的[5],而Hedgehog信号传导对肿瘤的发生发展起着重要作用.研究[6]结果显示,多种肿瘤组织中均存在Hedgehog信号的异常激活,如肺小细胞癌、胰腺癌、前列腺癌、乳腺癌、恶性胶质瘤、髓母细胞瘤及多发性骨髓瘤等.KIF3A属于初级纤毛内运输系统,具有维持和发挥初级纤毛正常功能的作用[7].本研究探讨了初级纤毛相关基因KIF3A通过Hedgehog信号通路对OSCC增殖能力产生的影响.1 材料与方法1.1 细胞 人舌鳞癌细胞株Tca8113购自美国菌种保藏中心(ATCC),置于延边大学附属医院中心实验室封存.1.2细胞培养与转染 取Tca8113细胞置于温度37℃、二氧化碳体积分数5%、饱和湿度95%的恒温孵育箱中,用含10%(质量分数)FBS、青链霉素的DMEM高糖型培养基传代培养.在转染前1d,将Tca8113细胞分为对照组和KIF3A-siRNA转染组后接种至6孔板中,待生长至80%时进行细胞转染.KIF3A-siRNA转染组转染KIF3A-siRNA,对照组未进行转染.转染操作按照LipofectamineTM2000Re-agen转染说明书进行.KIF3A-siRNA序列(5′-3′):UAA GGA AUG CUG AAG AAG ACG AGU C.1.3 蛋白印迹实验 转染24h后分别收集两组细胞,并加入RIPA与PMSF混合(100:1)冰上裂解细胞,提取细胞总蛋白,采用BCA试剂盒测定总蛋白水平.取各组标本行8%(质量分数)十二烷基硫酸钠聚丙烯酰胺凝胶(SDS-PAGE)电泳,转膜,室温下用5%(质量分数)脱脂奶粉封闭2h.TBST洗涤3·8·■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■延边大学医学学报 2022年3月 第45卷 第1期次,每次10min,加入兔抗人Kif3a一抗(1:1 000)、兔抗人Shh一抗(1:2 000)、兔抗人Gli1(1:1 000)及兔抗人GAPDH(1:1 000)后置于湿盒中,于4℃冰箱中孵育过夜;次日TBST洗涤3次,每次10min,分别加入山羊抗兔IgG二抗(1:10 000)后置于湿盒中室温孵育2h;TBST洗涤3次,每次10min,加入增强型化学发光剂ECL显影.以GAPDH为内参,采用Imagine J软件分析蛋白条带灰度值.1.4 实时荧光定量实验 转染24h后分别收集两组细胞,采用TRNzol法提取细胞中的总RNA,反转录为cDNA,按照RT-PCR试剂盒说明书以cDNA为模板进行扩增.测定循环阈值(Ct),采用2-△△Ct法进行统计.实验重复3次.引物序列,KIF3A-正向:5′-AGA GCG TCA ACG AGG TGT TT-3′;KIF3A-反向:5′-TAT TGA TCG GCA TCT TGG CCC-3′;Shh-正向:5′-CTC GCT GCT GGT ATG CTC G-3′;Shh-反向:5′-ATC GCT CGG AGT TTC TGGAGA-3′;Gli1-正向:5′-AGC GTG AGC CTG AATCTG TG-3′;Gli1-反向:5′-CAG CAT GTA CTGGGC TTT GAA-3′;GAPDH-正向:5′-GGA GCGAGA TCC CTC CAA AAT-3′;GAPDH-反向:5′-GGC TGT TGTCATACT TCTCATGG-3′.1.5 MTT细胞增殖实验 转染24h后分别收集两组细胞,以5 000个/孔接种于96孔板中,每组设置5个复孔.培养24、48、72h后弃去旧培养液,每孔加入100μL MTT(5 g/L),于37℃、5%(体积分数)二氧化碳、95%饱和湿度的恒温孵育箱中培养4 h,弃去上清液终止反应,每孔加入100μL DMSO,置于振荡器3 min溶解结晶后在酶标仪波长490 nm处测定各孔的光密度(OD)值,计算相对细胞增殖率.相对细胞增殖率(%)=(实验组OD值/对照组OD值)×100%.1.6 统计学分析 采用Graphpad Prism 7.0软件进行数据分析.计量数据以均数±标准偏差( x±s)表示,进行两个样本均数的t检验,以P<0.05为差异有统计学意义.2 结果2.1 沉默KIF3A基因后对Tca8113细胞Hedgehog信号通路中Shh、Gli1蛋白表达水平的影响 与对照组比较,KIF3A-siRNA转染组24h时KIF3A、Shh及Gli1蛋白表达水平均明显降低,差异均具有统计学意义(P<0.05),见Figure 1.compared with control,*P<0.05.Figure 1 The protein expression levels of KIF3A,Shhand Gli1decreased significantly aftertransfection of Tca8113cells2.2 沉默KIF3A基因后对Tca8113细胞中Hedgehog信号通路Shh、Gli1mRNA表达水平的影响 与对照组比较,KIF3A-siRNA转染24h组KIF3A、Shh及Gli1mRNA表达水平均显著降低,差异均具有统计学意义(P<0.01),见Figure 2.compared with control,**P<0.01.Figure 2 The mRNA expression levels of KIF3A,Shhand Gli1decreased significantly aftertransfection of Tca8113cells2.3 沉默KIF3A基因后对Tca8113细胞增殖能力的影响 MTT检测结果显示,细胞培养24h时KIF3A-siRNA转染组与对照组细胞增殖间差异无统计学意义(P>0.05);细胞培养48、72h时,KIF3A-siRNA转染组与对照组比较增殖显著受到抑制(P<0.01),见Figure 3.·9·■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■Journal of Medical Science Yanbian University Mar.2022 Vol.45 No.1compared with control,**P<0.01.Figure 3 The proliferation rate of Tca8113cells aftertransfection3 讨论初级纤毛为多数分化型细胞表面上的感觉附属物,具有化学感觉和机械感觉功能,在细胞周期控制中具有重要的作用.以往的研究认为初级纤毛在快速增殖的细胞上是找不到的,如癌症细胞,但KOWAL等[8]通过一种初级纤毛标志物Arl13b在HeLa和MG63细胞中发现了初级纤毛.未在OSCC中查看是否存在初级纤毛是本研究的不足之处.KIF3A属于初级纤毛内运输系统,在维持和发挥初级纤毛功能中起重要作用.HOANG-MINH等[9]的研究结果显示,沉默KIF3A基因表达后可降低胶质母细胞瘤中初级纤毛的数量,最终促进肿瘤的发生.玄云泽等[10]首次在OSCC中发现有初级纤毛,后来有学者亦在口腔白斑与OSCC组织中发现了初级纤毛,EGFR-Aurora A异常信号被激活后,口腔黏膜癌变过程中原发性纤毛逐渐减少[11].本研究结果显示,利用siRNA技术沉默KIF3A基因后mRNA和蛋白表达水平明显降低,提示初级纤毛的检出率降低.Hedgehog信号通路对胚胎发育及肿瘤的发生发展具有重要意义.研究[12-13]结果显示,Hedgehog信号通路的异常激活与皮肤、乳腺、肺及消化道等多种恶性肿瘤的发生、增殖密切相关.程志芬等[12]利用Hedgehog信号通路特异性抑制剂环巴胺处理Tca8113细胞株后,Smoothened的mRNA和蛋白表达明显减少,且细胞增殖受到抑制.KURODA等[14]的研究结果显示,给予Hedgehog信号通路抑制剂可抑制肿瘤前沿血管内皮细胞的增殖、迁移,进而抑制肿瘤的增殖和迁移.在小细胞肺癌中,沉默KIF3A后初级纤毛明显减少,且Shh信号通路介导的Gli1mRNA表达水平明显降低[15],此结果与本研究结果相符.研究[16]结果显示,Hedgehog信号通路转录因子Gli1的激活是恶性肿瘤的一个不良预后因素,利用环巴胺阻断Hedgehog信号通路可抑制Gli1激活,并增强头颈部鳞癌对放疗的敏感性,提示Hedgehog信号通路影响头颈部鳞癌的放疗预后.总之,本研究揭示了初级纤毛相关基因KIF3A在人舌鳞癌Tca8113细胞Hedgehog信号通路中的作用,初步解释了沉默KIF3A后可能通过抑制Hedgehog信号通路影响OSCC细胞的增殖能力,为OSCC患者的靶向治疗提供了理论依据.[参 考 文 献][1] DI PARDO BJ,BRONSON NW,DIGGS BS,et al..Theglobal burden of esophageal cancer:a disability-adjustedlife-year approach[J].World J Surg,2016,40(2):395-401.[2] BROCKLEHURST PR,BAKER SR,SPEIGHT PM.Oralcancer screening:what have we learnt and what is therestill to achieve[J].Future Oncol,2010,6(2):299-304.[3] BAVLE RM,VENUGOPAL R,KONDA P,et al..Mo-lecular classification of oral squamous cell carcinoma[J].JCDR,2016,10(9):Z18-Z21.[4] 陈新,徐文华,周健,等.口腔鳞状细胞癌现状[J].口腔医学,2017,37(5):462-465.[5] GOETZ SC,ANDERSON KV.The primary cilium:a signalling centre during vertebrate development[J].Nat Rev Gene,2010,11(5):331-344.[6] 那玉岩,刘万林.纤毛相关疾病:细胞学机制及转化应用进展[J].中国组织工程研究,2016,20(24):3642-3648.[7] NIEWIADOMSKI P,NIEDZIółKA SM,MARKIEWICZŁ,et al..Gli proteins:regulation in development andcancer[J].Cells,2019,8(2):147.[8] KOWAL TJ,FALK MM.Primary cilia found on HeLaand other cancer cells[J].Cell Biol Int,2015,39(11):1341-1347.[9] HOANG-MINH LB,DELEYROLLE LP,SIEBZEH-NRUBL D,et al..Disruption of KIF3Ain patient-de-rived glioblastoma cells:effects on ciliogenesis,hedge-hog sensitivity,and tumorigenesis[J].Oncotarget,2016,7(6):7029-7043.[10]玄云泽,李书进,金成日,等.初级纤毛相关基因KIF3A通过调控EMT过程在口腔鳞状细胞癌中的作用机制研究[J].重庆医学,2020,49(1):7-12.[11]YIN F,CHEN Q,SHI Y,et al..Activation of EGFR-Au-rora A induces loss of primary cilia in oral squamous cellcarcinoma[J].Oral Dis,2022,28(3):621-630.[12]程志芬,崔演,玄延花.Ptch和Smo在舌鳞状细胞癌Tca8113细胞中的表达及其意义[J].口腔医学研究,2015,31(5):433-436.[13]KONSTANTINOU D,BERTAUX-SKEIRIK N,ZAVROSY.Hedgehog signaling in the stomach[J].Curr OpinPharmacol,2016,31:76-82.[14]KURODA H,KURIO N,SHIMO T,et al..Oral squamouscell carcinoma-derived sonic hedgehog promotes angiogene-sis[J].Anticancer Res,2017,37(12):6731-6737.[15]COCHRANE CR,VAGHJIANI V,SZCZEPNY A,et al..Trp53and Rb1regulate autophagy and ligand-dependentHedgehog signaling[J].J Clin Invest,2020,130(8):4006-4018.[16]GAN GN,EAGLES J,KEYSAR SB,et al..Hedgehogsignaling drives radioresistance and stroma-driventumor repopulation in head and neck squamous cancers[J].Cancer Res,2014,74(23):7024-7036.■·01·■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■。



第47卷第6期2021年11月吉林大学学报(医学版)Journal of Jilin University(Medicine Edition)Vol.47No.6Nov.2021DOI:10.13481/j.1671‑587X.20210606棕榈酸对骨骼肌细胞脂质沉积的诱导作用何勇,洪莉,黄国涛,赵芷涵(武汉大学人民医院妇产科,湖北武汉430060)[摘要]目的目的:探讨棕榈酸(PA)对骨骼肌细胞脂质沉积的作用及其最佳作用浓度和时间。
方法:在C2C12细胞生长至70%~80%时分为对照组和不同浓度(50、75、100和125μmol·L-1)PA组。
不同浓度PA组细胞采用含不同浓度PA的生长培养基培养,对照组细胞不给予PA(0μmol·L-1PA)。
CCK-8法检测各组细胞增殖活性,定量检测细胞中甘油三酯(TG)水平,油红O染色观察细胞中脂滴形成情况,2%马血清诱导细胞分化并评价各组C2C12细胞分化状态。
结果结果:与对照组比较,50、75和100μmol·L-1PA组C2C12细胞增殖活性差异无统计学意义(P>0.05),125μmol·L-1PA组C2C12细胞增殖活性明显降低(P<0.01),50、75和100μmol·L-1PA组细胞中TG水平升高(P<0.01),100μmol·L-1PA组C2C12细胞中TG水平升高最明显(P<0.01);与培养24h比较,100μmol·L-1 PA培养48h后,C2C12细胞中TG水平升高(P<0.01)。
分化培养后,PA组和对照组C2C12细胞中均有较多肌管形成。
结论结论:100μmol·L-1PA作用48h可有效促进C2C12细胞脂质沉积。
[关键词]盆底功能障碍;肥胖;高脂;棕榈酸;脂质沉积[中图分类号]R711.4[文献标志码]AEffect of palmitic acid on induction of lipid deposition inskeletal muscle cellsHE Yong,HONG Li,HUANG Guotao,ZHAO Zhihan(Department of Obstetrics and Gynecology,People’s Hospital,Wuhan University,Wuhan430060,China)ABSTRACT Objective:To explore the effect of palmitic acid(PA)on the lipid deposition in the skeletal muscle cells,and to discuss the optimum action concentration and time.Methods:The C2C12cells were divided into control group and different concentrations(50,75,100,and125μmol·L-1)of PA groups when the C2C12cells grew to70%-80%.The C2C12cells in different concentrations of PA groups were given growth media containing different concentrations of PA,while the C2C12cells in control group were not treated with PA(0μmol·L-1PA).CCK-8method was used to detect the cell proliferation activities of cells in various groups,the triglyceride(TG)levels in the C2C12cells in various groups were detected,Oil Red O staining was used to observe the formation of lipid droplets in the C2C12cells in various groups,and2% horse serum was used to induce the cell differentiation and evaluate the differentiation of C2C12cells in various groups.Results:Compared with control group,there were no significant differences in the proliferation activities of the C2C12cells in50,75,and100μmol·L-1PA groups(P>0.05),the [文章编号]1671‑587X(2021)06‑1380‑06[收稿日期]2021‑02‑03[基金项目]国家自然科学基金面上项目(81971364,81771562)[作者简介]何勇(1994-),男,湖北省恩施州人,在读硕士研究生,主要从事女性盆底功能障碍性疾病和妇科肿瘤方面的研究。

DOI: 10.1126/science.1094786, 441 (2004);304Science et al.Mitchell S. Abrahamsen,Cryptosporidium parvum Complete Genome Sequence of the Apicomplexan, (this information is current as of October 7, 2009 ):The following resources related to this article are available online at/cgi/content/full/304/5669/441version of this article at:including high-resolution figures, can be found in the online Updated information and services,/cgi/content/full/1094786/DC1 can be found at:Supporting Online Material/cgi/content/full/304/5669/441#otherarticles , 9 of which can be accessed for free: cites 25 articles This article 239 article(s) on the ISI Web of Science. cited by This article has been /cgi/content/full/304/5669/441#otherarticles 53 articles hosted by HighWire Press; see: cited by This article has been/cgi/collection/genetics Genetics: subject collections This article appears in the following/about/permissions.dtl in whole or in part can be found at: this article permission to reproduce of this article or about obtaining reprints Information about obtaining registered trademark of AAAS.is a Science 2004 by the American Association for the Advancement of Science; all rights reserved. The title Copyright American Association for the Advancement of Science, 1200 New York Avenue NW, Washington, DC 20005. (print ISSN 0036-8075; online ISSN 1095-9203) is published weekly, except the last week in December, by the Science o n O c t o b e r 7, 2009w w w .s c i e n c e m a g .o r g D o w n l o a d e d f r o m3.R.Jackendoff,Foundations of Language:Brain,Gram-mar,Evolution(Oxford Univ.Press,Oxford,2003).4.Although for Frege(1),reference was established rela-tive to objects in the world,here we follow Jackendoff’s suggestion(3)that this is done relative to objects and the state of affairs as mentally represented.5.S.Zola-Morgan,L.R.Squire,in The Development andNeural Bases of Higher Cognitive Functions(New York Academy of Sciences,New York,1990),pp.434–456.6.N.Chomsky,Reflections on Language(Pantheon,New York,1975).7.J.Katz,Semantic Theory(Harper&Row,New York,1972).8.D.Sperber,D.Wilson,Relevance(Harvard Univ.Press,Cambridge,MA,1986).9.K.I.Forster,in Sentence Processing,W.E.Cooper,C.T.Walker,Eds.(Erlbaum,Hillsdale,NJ,1989),pp.27–85.10.H.H.Clark,Using Language(Cambridge Univ.Press,Cambridge,1996).11.Often word meanings can only be fully determined byinvokingworld knowledg e.For instance,the meaningof “flat”in a“flat road”implies the absence of holes.However,in the expression“aflat tire,”it indicates the presence of a hole.The meaningof“finish”in the phrase “Billfinished the book”implies that Bill completed readingthe book.However,the phrase“the g oatfin-ished the book”can only be interpreted as the goat eatingor destroyingthe book.The examples illustrate that word meaningis often underdetermined and nec-essarily intertwined with general world knowledge.In such cases,it is hard to see how the integration of lexical meaning and general world knowledge could be strictly separated(3,31).12.W.Marslen-Wilson,C.M.Brown,L.K.Tyler,Lang.Cognit.Process.3,1(1988).13.ERPs for30subjects were averaged time-locked to theonset of the critical words,with40items per condition.Sentences were presented word by word on the centerof a computer screen,with a stimulus onset asynchronyof600ms.While subjects were readingthe sentences,their EEG was recorded and amplified with a high-cut-off frequency of70Hz,a time constant of8s,and asamplingfrequency of200Hz.14.Materials and methods are available as supportingmaterial on Science Online.15.M.Kutas,S.A.Hillyard,Science207,203(1980).16.C.Brown,P.Hagoort,J.Cognit.Neurosci.5,34(1993).17.C.M.Brown,P.Hagoort,in Architectures and Mech-anisms for Language Processing,M.W.Crocker,M.Pickering,C.Clifton Jr.,Eds.(Cambridge Univ.Press,Cambridge,1999),pp.213–237.18.F.Varela et al.,Nature Rev.Neurosci.2,229(2001).19.We obtained TFRs of the single-trial EEG data by con-volvingcomplex Morlet wavelets with the EEG data andcomputingthe squared norm for the result of theconvolution.We used wavelets with a7-cycle width,with frequencies ranging from1to70Hz,in1-Hz steps.Power values thus obtained were expressed as a per-centage change relative to the power in a baselineinterval,which was taken from150to0ms before theonset of the critical word.This was done in order tonormalize for individual differences in EEG power anddifferences in baseline power between different fre-quency bands.Two relevant time-frequency compo-nents were identified:(i)a theta component,rangingfrom4to7Hz and from300to800ms after wordonset,and(ii)a gamma component,ranging from35to45Hz and from400to600ms after word onset.20.C.Tallon-Baudry,O.Bertrand,Trends Cognit.Sci.3,151(1999).tner et al.,Nature397,434(1999).22.M.Bastiaansen,P.Hagoort,Cortex39(2003).23.O.Jensen,C.D.Tesche,Eur.J.Neurosci.15,1395(2002).24.Whole brain T2*-weighted echo planar imaging bloodoxygen level–dependent(EPI-BOLD)fMRI data wereacquired with a Siemens Sonata1.5-T magnetic reso-nance scanner with interleaved slice ordering,a volumerepetition time of2.48s,an echo time of40ms,a90°flip angle,31horizontal slices,a64ϫ64slice matrix,and isotropic voxel size of3.5ϫ3.5ϫ3.5mm.For thestructural magnetic resonance image,we used a high-resolution(isotropic voxels of1mm3)T1-weightedmagnetization-prepared rapid gradient-echo pulse se-quence.The fMRI data were preprocessed and analyzedby statistical parametric mappingwith SPM99software(http://www.fi/spm99).25.S.E.Petersen et al.,Nature331,585(1988).26.B.T.Gold,R.L.Buckner,Neuron35,803(2002).27.E.Halgren et al.,J.Psychophysiol.88,1(1994).28.E.Halgren et al.,Neuroimage17,1101(2002).29.M.K.Tanenhaus et al.,Science268,1632(1995).30.J.J.A.van Berkum et al.,J.Cognit.Neurosci.11,657(1999).31.P.A.M.Seuren,Discourse Semantics(Basil Blackwell,Oxford,1985).32.We thank P.Indefrey,P.Fries,P.A.M.Seuren,and M.van Turennout for helpful discussions.Supported bythe Netherlands Organization for Scientific Research,grant no.400-56-384(P.H.).Supporting Online Material/cgi/content/full/1095455/DC1Materials and MethodsFig.S1References and Notes8January2004;accepted9March2004Published online18March2004;10.1126/science.1095455Include this information when citingthis paper.Complete Genome Sequence ofthe Apicomplexan,Cryptosporidium parvumMitchell S.Abrahamsen,1,2*†Thomas J.Templeton,3†Shinichiro Enomoto,1Juan E.Abrahante,1Guan Zhu,4 Cheryl ncto,1Mingqi Deng,1Chang Liu,1‡Giovanni Widmer,5Saul Tzipori,5GregoryA.Buck,6Ping Xu,6 Alan T.Bankier,7Paul H.Dear,7Bernard A.Konfortov,7 Helen F.Spriggs,7Lakshminarayan Iyer,8Vivek Anantharaman,8L.Aravind,8Vivek Kapur2,9The apicomplexan Cryptosporidium parvum is an intestinal parasite that affects healthy humans and animals,and causes an unrelenting infection in immuno-compromised individuals such as AIDS patients.We report the complete ge-nome sequence of C.parvum,type II isolate.Genome analysis identifies ex-tremely streamlined metabolic pathways and a reliance on the host for nu-trients.In contrast to Plasmodium and Toxoplasma,the parasite lacks an api-coplast and its genome,and possesses a degenerate mitochondrion that has lost its genome.Several novel classes of cell-surface and secreted proteins with a potential role in host interactions and pathogenesis were also detected.Elu-cidation of the core metabolism,including enzymes with high similarities to bacterial and plant counterparts,opens new avenues for drug development.Cryptosporidium parvum is a globally impor-tant intracellular pathogen of humans and animals.The duration of infection and patho-genesis of cryptosporidiosis depends on host immune status,ranging from a severe but self-limiting diarrhea in immunocompetent individuals to a life-threatening,prolonged infection in immunocompromised patients.Asubstantial degree of morbidity and mortalityis associated with infections in AIDS pa-tients.Despite intensive efforts over the past20years,there is currently no effective ther-apy for treating or preventing C.parvuminfection in humans.Cryptosporidium belongs to the phylumApicomplexa,whose members share a com-mon apical secretory apparatus mediating lo-comotion and tissue or cellular invasion.Many apicomplexans are of medical or vet-erinary importance,including Plasmodium,Babesia,Toxoplasma,Neosprora,Sarcocys-tis,Cyclospora,and Eimeria.The life cycle ofC.parvum is similar to that of other cyst-forming apicomplexans(e.g.,Eimeria and Tox-oplasma),resulting in the formation of oocysts1Department of Veterinary and Biomedical Science,College of Veterinary Medicine,2Biomedical Genom-ics Center,University of Minnesota,St.Paul,MN55108,USA.3Department of Microbiology and Immu-nology,Weill Medical College and Program in Immu-nology,Weill Graduate School of Medical Sciences ofCornell University,New York,NY10021,USA.4De-partment of Veterinary Pathobiology,College of Vet-erinary Medicine,Texas A&M University,College Sta-tion,TX77843,USA.5Division of Infectious Diseases,Tufts University School of Veterinary Medicine,NorthGrafton,MA01536,USA.6Center for the Study ofBiological Complexity and Department of Microbiol-ogy and Immunology,Virginia Commonwealth Uni-versity,Richmond,VA23198,USA.7MRC Laboratoryof Molecular Biology,Hills Road,Cambridge CB22QH,UK.8National Center for Biotechnology Infor-mation,National Library of Medicine,National Insti-tutes of Health,Bethesda,MD20894,USA.9Depart-ment of Microbiology,University of Minnesota,Min-neapolis,MN55455,USA.*To whom correspondence should be addressed.E-mail:abe@†These authors contributed equally to this work.‡Present address:Bioinformatics Division,Genetic Re-search,GlaxoSmithKline Pharmaceuticals,5MooreDrive,Research Triangle Park,NC27009,USA.R E P O R T S SCIENCE VOL30416APRIL2004441o n O c t o b e r 7 , 2 0 0 9 w w w . s c i e n c e m a g . o r g D o w n l o a d e d f r o mthat are shed in the feces of infected hosts.C.parvum oocysts are highly resistant to environ-mental stresses,including chlorine treatment of community water supplies;hence,the parasite is an important water-and food-borne pathogen (1).The obligate intracellular nature of the par-asite ’s life cycle and the inability to culture the parasite continuously in vitro greatly impair researchers ’ability to obtain purified samples of the different developmental stages.The par-asite cannot be genetically manipulated,and transformation methodologies are currently un-available.To begin to address these limitations,we have obtained the complete C.parvum ge-nome sequence and its predicted protein com-plement.(This whole-genome shotgun project has been deposited at DDBJ/EMBL/GenBank under the project accession AAEE00000000.The version described in this paper is the first version,AAEE01000000.)The random shotgun approach was used to obtain the complete DNA sequence (2)of the Iowa “type II ”isolate of C.parvum .This isolate readily transmits disease among numerous mammals,including humans.The resulting ge-nome sequence has roughly 13ϫgenome cov-erage containing five gaps and 9.1Mb of totalDNA sequence within eight chromosomes.The C.parvum genome is thus quite compact rela-tive to the 23-Mb,14-chromosome genome of Plasmodium falciparum (3);this size difference is predominantly the result of shorter intergenic regions,fewer introns,and a smaller number of genes (Table 1).Comparison of the assembled sequence of chromosome VI to that of the recently published sequence of chromosome VI (4)revealed that our assembly contains an ad-ditional 160kb of sequence and a single gap versus two,with the common sequences dis-playing a 99.993%sequence identity (2).The relative paucity of introns greatly simplified gene predictions and facilitated an-notation (2)of predicted open reading frames (ORFs).These analyses provided an estimate of 3807protein-encoding genes for the C.parvum genome,far fewer than the estimated 5300genes predicted for the Plasmodium genome (3).This difference is primarily due to the absence of an apicoplast and mitochondrial genome,as well as the pres-ence of fewer genes encoding metabolic functions and variant surface proteins,such as the P.falciparum var and rifin molecules (Table 2).An analysis of the encoded pro-tein sequences with the program SEG (5)shows that these protein-encoding genes are not enriched in low-complexity se-quences (34%)to the extent observed in the proteins from Plasmodium (70%).Our sequence analysis indicates that Cryptosporidium ,unlike Plasmodium and Toxoplasma ,lacks both mitochondrion and apicoplast genomes.The overall complete-ness of the genome sequence,together with the fact that similar DNA extraction proce-dures used to isolate total genomic DNA from C.parvum efficiently yielded mito-chondrion and apicoplast genomes from Ei-meria sp.and Toxoplasma (6,7),indicates that the absence of organellar genomes was unlikely to have been the result of method-ological error.These conclusions are con-sistent with the absence of nuclear genes for the DNA replication and translation machinery characteristic of mitochondria and apicoplasts,and with the lack of mito-chondrial or apicoplast targeting signals for tRNA synthetases.A number of putative mitochondrial pro-teins were identified,including components of a mitochondrial protein import apparatus,chaperones,uncoupling proteins,and solute translocators (table S1).However,the ge-nome does not encode any Krebs cycle en-zymes,nor the components constituting the mitochondrial complexes I to IV;this finding indicates that the parasite does not rely on complete oxidation and respiratory chains for synthesizing adenosine triphosphate (ATP).Similar to Plasmodium ,no orthologs for the ␥,␦,or εsubunits or the c subunit of the F 0proton channel were detected (whereas all subunits were found for a V-type ATPase).Cryptosporidium ,like Eimeria (8)and Plas-modium ,possesses a pyridine nucleotide tran-shydrogenase integral membrane protein that may couple reduced nicotinamide adenine dinucleotide (NADH)and reduced nico-tinamide adenine dinucleotide phosphate (NADPH)redox to proton translocation across the inner mitochondrial membrane.Unlike Plasmodium ,the parasite has two copies of the pyridine nucleotide transhydrogenase gene.Also present is a likely mitochondrial membrane –associated,cyanide-resistant alter-native oxidase (AOX )that catalyzes the reduction of molecular oxygen by ubiquinol to produce H 2O,but not superoxide or H 2O 2.Several genes were identified as involved in biogenesis of iron-sulfur [Fe-S]complexes with potential mitochondrial targeting signals (e.g.,nifS,nifU,frataxin,and ferredoxin),supporting the presence of a limited electron flux in the mitochondrial remnant (table S2).Our sequence analysis confirms the absence of a plastid genome (7)and,additionally,the loss of plastid-associated metabolic pathways including the type II fatty acid synthases (FASs)and isoprenoid synthetic enzymes thatTable 1.General features of the C.parvum genome and comparison with other single-celled eukaryotes.Values are derived from respective genome project summaries (3,26–28).ND,not determined.FeatureC.parvum P.falciparum S.pombe S.cerevisiae E.cuniculiSize (Mbp)9.122.912.512.5 2.5(G ϩC)content (%)3019.43638.347No.of genes 38075268492957701997Mean gene length (bp)excluding introns 1795228314261424ND Gene density (bp per gene)23824338252820881256Percent coding75.352.657.570.590Genes with introns (%)553.9435ND Intergenic regions (G ϩC)content %23.913.632.435.145Mean length (bp)5661694952515129RNAsNo.of tRNA genes 454317429944No.of 5S rRNA genes 6330100–2003No.of 5.8S ,18S ,and 28S rRNA units 57200–400100–20022Table parison between predicted C.parvum and P.falciparum proteins.FeatureC.parvum P.falciparum *Common †Total predicted proteins380752681883Mitochondrial targeted/encoded 17(0.45%)246(4.7%)15Apicoplast targeted/encoded 0581(11.0%)0var/rif/stevor ‡0236(4.5%)0Annotated as protease §50(1.3%)31(0.59%)27Annotated as transporter 69(1.8%)34(0.65%)34Assigned EC function ¶167(4.4%)389(7.4%)113Hypothetical proteins925(24.3%)3208(60.9%)126*Values indicated for P.falciparum are as reported (3)with the exception of those for proteins annotated as protease or transporter.†TBLASTN hits (e Ͻ–5)between C.parvum and P.falciparum .‡As reported in (3).§Pre-dicted proteins annotated as “protease or peptidase”for C.parvum (CryptoGenome database,)and P.falciparum (PlasmoDB database,).Predicted proteins annotated as “trans-porter,permease of P-type ATPase”for C.parvum (CryptoGenome)and P.falciparum (PlasmoDB).¶Bidirectional BLAST hit (e Ͻ–15)to orthologs with assigned Enzyme Commission (EC)numbers.Does not include EC assignment numbers for protein kinases or protein phosphatases (due to inconsistent annotation across genomes),or DNA polymerases or RNA polymerases,as a result of issues related to subunit inclusion.(For consistency,46proteins were excluded from the reported P.falciparum values.)R E P O R T S16APRIL 2004VOL 304SCIENCE 442 o n O c t o b e r 7, 2009w w w .s c i e n c e m a g .o r g D o w n l o a d e d f r o mare otherwise localized to the plastid in other apicomplexans.C.parvum fatty acid biosynthe-sis appears to be cytoplasmic,conducted by a large(8252amino acids)modular type I FAS (9)and possibly by another large enzyme that is related to the multidomain bacterial polyketide synthase(10).Comprehensive screening of the C.parvum genome sequence also did not detect orthologs of Plasmodium nuclear-encoded genes that contain apicoplast-targeting and transit sequences(11).C.parvum metabolism is greatly stream-lined relative to that of Plasmodium,and in certain ways it is reminiscent of that of another obligate eukaryotic parasite,the microsporidian Encephalitozoon.The degeneration of the mi-tochondrion and associated metabolic capabili-ties suggests that the parasite largely relies on glycolysis for energy production.The parasite is capable of uptake and catabolism of mono-sugars(e.g.,glucose and fructose)as well as synthesis,storage,and catabolism of polysac-charides such as trehalose and amylopectin. Like many anaerobic organisms,it economizes ATP through the use of pyrophosphate-dependent phosphofructokinases.The conver-sion of pyruvate to acetyl–coenzyme A(CoA) is catalyzed by an atypical pyruvate-NADPH oxidoreductase(Cp PNO)that contains an N-terminal pyruvate–ferredoxin oxidoreductase (PFO)domain fused with a C-terminal NADPH–cytochrome P450reductase domain (CPR).Such a PFO-CPR fusion has previously been observed only in the euglenozoan protist Euglena gracilis(12).Acetyl-CoA can be con-verted to malonyl-CoA,an important precursor for fatty acid and polyketide biosynthesis.Gly-colysis leads to several possible organic end products,including lactate,acetate,and ethanol. The production of acetate from acetyl-CoA may be economically beneficial to the parasite via coupling with ATP production.Ethanol is potentially produced via two in-dependent pathways:(i)from the combination of pyruvate decarboxylase and alcohol dehy-drogenase,or(ii)from acetyl-CoA by means of a bifunctional dehydrogenase(adhE)with ac-etaldehyde and alcohol dehydrogenase activi-ties;adhE first converts acetyl-CoA to acetal-dehyde and then reduces the latter to ethanol. AdhE predominantly occurs in bacteria but has recently been identified in several protozoans, including vertebrate gut parasites such as Enta-moeba and Giardia(13,14).Adjacent to the adhE gene resides a second gene encoding only the AdhE C-terminal Fe-dependent alcohol de-hydrogenase domain.This gene product may form a multisubunit complex with AdhE,or it may function as an alternative alcohol dehydro-genase that is specific to certain growth condi-tions.C.parvum has a glycerol3-phosphate dehydrogenase similar to those of plants,fungi, and the kinetoplastid Trypanosoma,but(unlike trypanosomes)the parasite lacks an ortholog of glycerol kinase and thus this pathway does not yield glycerol production.In addition to themodular fatty acid synthase(Cp FAS1)andpolyketide synthase homolog(Cp PKS1), C.parvum possesses several fatty acyl–CoA syn-thases and a fatty acyl elongase that may partici-pate in fatty acid metabolism.Further,enzymesfor the metabolism of complex lipids(e.g.,glyc-erolipid and inositol phosphate)were identified inthe genome.Fatty acids are apparently not anenergy source,because enzymes of the fatty acidoxidative pathway are absent,with the exceptionof a3-hydroxyacyl-CoA dehydrogenase.C.parvum purine metabolism is greatlysimplified,retaining only an adenosine ki-nase and enzymes catalyzing conversionsof adenosine5Ј-monophosphate(AMP)toinosine,xanthosine,and guanosine5Ј-monophosphates(IMP,XMP,and GMP).Among these enzymes,IMP dehydrogenase(IMPDH)is phylogenetically related toε-proteobacterial IMPDH and is strikinglydifferent from its counterparts in both thehost and other apicomplexans(15).In con-trast to other apicomplexans such as Toxo-plasma gondii and P.falciparum,no geneencoding hypoxanthine-xanthineguaninephosphoribosyltransferase(HXGPRT)is de-tected,in contrast to a previous report on theactivity of this enzyme in C.parvum sporo-zoites(16).The absence of HXGPRT sug-gests that the parasite may rely solely on asingle enzyme system including IMPDH toproduce GMP from AMP.In contrast to otherapicomplexans,the parasite appears to relyon adenosine for purine salvage,a modelsupported by the identification of an adeno-sine transporter.Unlike other apicomplexansand many parasitic protists that can synthe-size pyrimidines de novo,C.parvum relies onpyrimidine salvage and retains the ability forinterconversions among uridine and cytidine5Ј-monophosphates(UMP and CMP),theirdeoxy forms(dUMP and dCMP),and dAMP,as well as their corresponding di-and triphos-phonucleotides.The parasite has also largelyshed the ability to synthesize amino acids denovo,although it retains the ability to convertselect amino acids,and instead appears torely on amino acid uptake from the host bymeans of a set of at least11amino acidtransporters(table S2).Most of the Cryptosporidium core pro-cesses involved in DNA replication,repair,transcription,and translation conform to thebasic eukaryotic blueprint(2).The transcrip-tional apparatus resembles Plasmodium interms of basal transcription machinery.How-ever,a striking numerical difference is seenin the complements of two RNA bindingdomains,Sm and RRM,between P.falcipa-rum(17and71domains,respectively)and C.parvum(9and51domains).This reductionresults in part from the loss of conservedproteins belonging to the spliceosomal ma-chinery,including all genes encoding Smdomain proteins belonging to the U6spliceo-somal particle,which suggests that this par-ticle activity is degenerate or entirely lost.This reduction in spliceosomal machinery isconsistent with the reduced number of pre-dicted introns in Cryptosporidium(5%)rela-tive to Plasmodium(Ͼ50%).In addition,keycomponents of the small RNA–mediatedposttranscriptional gene silencing system aremissing,such as the RNA-dependent RNApolymerase,Argonaute,and Dicer orthologs;hence,RNA interference–related technolo-gies are unlikely to be of much value intargeted disruption of genes in C.parvum.Cryptosporidium invasion of columnarbrush border epithelial cells has been de-scribed as“intracellular,but extracytoplas-mic,”as the parasite resides on the surface ofthe intestinal epithelium but lies underneaththe host cell membrane.This niche may al-low the parasite to evade immune surveil-lance but take advantage of solute transportacross the host microvillus membrane or theextensively convoluted parasitophorous vac-uole.Indeed,Cryptosporidium has numerousgenes(table S2)encoding families of putativesugar transporters(up to9genes)and aminoacid transporters(11genes).This is in starkcontrast to Plasmodium,which has fewersugar transporters and only one putative ami-no acid transporter(GenBank identificationnumber23612372).As a first step toward identification ofmulti–drug-resistant pumps,the genome se-quence was analyzed for all occurrences ofgenes encoding multitransmembrane proteins.Notable are a set of four paralogous proteinsthat belong to the sbmA family(table S2)thatare involved in the transport of peptide antibi-otics in bacteria.A putative ortholog of thePlasmodium chloroquine resistance–linkedgene Pf CRT(17)was also identified,althoughthe parasite does not possess a food vacuole likethe one seen in Plasmodium.Unlike Plasmodium,C.parvum does notpossess extensive subtelomeric clusters of anti-genically variant proteins(exemplified by thelarge families of var and rif/stevor genes)thatare involved in immune evasion.In contrast,more than20genes were identified that encodemucin-like proteins(18,19)having hallmarksof extensive Thr or Ser stretches suggestive ofglycosylation and signal peptide sequences sug-gesting secretion(table S2).One notable exam-ple is an11,700–amino acid protein with anuninterrupted stretch of308Thr residues(cgd3_720).Although large families of secretedproteins analogous to the Plasmodium multi-gene families were not found,several smallermultigene clusters were observed that encodepredicted secreted proteins,with no detectablesimilarity to proteins from other organisms(Fig.1,A and B).Within this group,at leastfour distinct families appear to have emergedthrough gene expansions specific to the Cryp-R E P O R T S SCIENCE VOL30416APRIL2004443o n O c t o b e r 7 , 2 0 0 9 w w w . s c i e n c e m a g . o r g D o w n l o a d e d f r o mtosporidium clade.These families —SKSR,MEDLE,WYLE,FGLN,and GGC —were named after well-conserved sequence motifs (table S2).Reverse transcription polymerase chain reaction (RT-PCR)expression analysis (20)of one cluster,a locus of seven adjacent CpLSP genes (Fig.1B),shows coexpression during the course of in vitro development (Fig.1C).An additional eight genes were identified that encode proteins having a periodic cysteine structure similar to the Cryptosporidium oocyst wall protein;these eight genes are similarly expressed during the onset of oocyst formation and likely participate in the formation of the coccidian rigid oocyst wall in both Cryptospo-ridium and Toxoplasma (21).Whereas the extracellular proteins described above are of apparent apicomplexan or lineage-specific in-vention,Cryptosporidium possesses many genesencodingsecretedproteinshavinglineage-specific multidomain architectures composed of animal-and bacterial-like extracellular adhe-sive domains (fig.S1).Lineage-specific expansions were ob-served for several proteases (table S2),in-cluding an aspartyl protease (six genes),a subtilisin-like protease,a cryptopain-like cys-teine protease (five genes),and a Plas-modium falcilysin-like (insulin degrading enzyme –like)protease (19genes).Nine of the Cryptosporidium falcilysin genes lack the Zn-chelating “HXXEH ”active site motif and are likely to be catalytically inactive copies that may have been reused for specific protein-protein interactions on the cell sur-face.In contrast to the Plasmodium falcilysin,the Cryptosporidium genes possess signal peptide sequences and are likely trafficked to a secretory pathway.The expansion of this family suggests either that the proteins have distinct cleavage specificities or that their diversity may be related to evasion of a host immune response.Completion of the C.parvum genome se-quence has highlighted the lack of conven-tional drug targets currently pursued for the control and treatment of other parasitic protists.On the basis of molecular and bio-chemical studies and drug screening of other apicomplexans,several putative Cryptospo-ridium metabolic pathways or enzymes have been erroneously proposed to be potential drug targets (22),including the apicoplast and its associated metabolic pathways,the shikimate pathway,the mannitol cycle,the electron transport chain,and HXGPRT.Nonetheless,complete genome sequence analysis identifies a number of classic and novel molecular candidates for drug explora-tion,including numerous plant-like and bacterial-like enzymes (tables S3and S4).Although the C.parvum genome lacks HXGPRT,a potent drug target in other api-complexans,it has only the single pathway dependent on IMPDH to convert AMP to GMP.The bacterial-type IMPDH may be a promising target because it differs substan-tially from that of eukaryotic enzymes (15).Because of the lack of de novo biosynthetic capacity for purines,pyrimidines,and amino acids,C.parvum relies solely on scavenge from the host via a series of transporters,which may be exploited for chemotherapy.C.parvum possesses a bacterial-type thymidine kinase,and the role of this enzyme in pyrim-idine metabolism and its drug target candida-cy should be pursued.The presence of an alternative oxidase,likely targeted to the remnant mitochondrion,gives promise to the study of salicylhydroxamic acid (SHAM),as-cofuranone,and their analogs as inhibitors of energy metabolism in the parasite (23).Cryptosporidium possesses at least 15“plant-like ”enzymes that are either absent in or highly divergent from those typically found in mammals (table S3).Within the glycolytic pathway,the plant-like PPi-PFK has been shown to be a potential target in other parasites including T.gondii ,and PEPCL and PGI ap-pear to be plant-type enzymes in C.parvum .Another example is a trehalose-6-phosphate synthase/phosphatase catalyzing trehalose bio-synthesis from glucose-6-phosphate and uridine diphosphate –glucose.Trehalose may serve as a sugar storage source or may function as an antidesiccant,antioxidant,or protein stability agent in oocysts,playing a role similar to that of mannitol in Eimeria oocysts (24).Orthologs of putative Eimeria mannitol synthesis enzymes were not found.However,two oxidoreductases (table S2)were identified in C.parvum ,one of which belongs to the same families as the plant mannose dehydrogenases (25)and the other to the plant cinnamyl alcohol dehydrogenases.In principle,these enzymes could synthesize protective polyol compounds,and the former enzyme could use host-derived mannose to syn-thesize mannitol.References and Notes1.D.G.Korich et al .,Appl.Environ.Microbiol.56,1423(1990).2.See supportingdata on Science Online.3.M.J.Gardner et al .,Nature 419,498(2002).4.A.T.Bankier et al .,Genome Res.13,1787(2003).5.J.C.Wootton,Comput.Chem.18,269(1994).Fig.1.(A )Schematic showing the chromosomal locations of clusters of potentially secreted proteins.Numbers of adjacent genes are indicated in paren-theses.Arrows indicate direc-tion of clusters containinguni-directional genes (encoded on the same strand);squares indi-cate clusters containingg enes encoded on both strands.Non-paralogous genes are indicated by solid gray squares or direc-tional triangles;SKSR (green triangles),FGLN (red trian-gles),and MEDLE (blue trian-gles)indicate three C.parvum –specific families of paralogous genes predominantly located at telomeres.Insl (yellow tri-angles)indicates an insulinase/falcilysin-like paralogous gene family.Cp LSP (white square)indicates the location of a clus-ter of adjacent large secreted proteins (table S2)that are cotranscriptionally regulated.Identified anchored telomeric repeat sequences are indicated by circles.(B )Schematic show-inga select locus containinga cluster of coexpressed large secreted proteins (Cp LSP).Genes and intergenic regions (regions between identified genes)are drawn to scale at the nucleotide level.The length of the intergenic re-gions is indicated above or be-low the locus.(C )Relative ex-pression levels of CpLSP (red lines)and,as a control,C.parvum Hedgehog-type HINT domain gene (blue line)duringin vitro development,as determined by semiquantitative RT-PCR usingg ene-specific primers correspondingto the seven adjacent g enes within the CpLSP locus as shown in (B).Expression levels from three independent time-course experiments are represented as the ratio of the expression of each gene to that of C.parvum 18S rRNA present in each of the infected samples (20).R E P O R T S16APRIL 2004VOL 304SCIENCE 444 o n O c t o b e r 7, 2009w w w .s c i e n c e m a g .o r g D o w n l o a d e d f r o m。

• 244 •中国运动医学杂志2022年3月第41卷第3期Chin J Sports Med,Mar. 2022,V〇1.41,N〇.3[41] 孙民康,孙小玲,李良,等.从“渐进合作”到“多边融 合”:新中国学生体育健康促进政策的历史回眸与现实审 思[J].南京体育学院学报,2021, 20(5): 51-59.[42] 柳鸣毅,王梅,徐杰,等.“健康中国2030”背景下中国 青少年体育公共政策研究[•!].体育科学,2018, 38(2): 91-97.[43] 陈华东,廖晓阳,刘长明,等.《健康中国行动(2019~ 2030年)》之重大专项行动核心要点解读与启示:全科医生 视角[J].中国卫生事业管理,2020, 37(12): 958-960.[44] 陈华荣.实施全民健身国家战略的政策法规体系研究[J]. 体育科学,2017,37(4): 74-86.[45] 刘惠琴,李盈,郑莉,等.中国抗癌协会系列期刊集群化 办刊实践与思考[J].科技与出版,2015, 247(7): 16-19.[46] Hyuna S, Jcaques F, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J]. CA Cancer J Clin, 2021, 71(3): 209-249.[47] Zheng RS, ChunFeng Q,Siwei Z, et al. Liver cancerincidence and mortality in China: temporal trends and projections to 2030[J]. Chin J Cancer Res, 2018,30 (6): 571-579.[48] Yang S,Shuai L, Na L, et al. Burden, trends, and risk factors of esophageal cancer in China from 1990 to 2017: an up to date overview and comparison with those in Japan and South Korea[J]. J Hematol Oncol,2021,

期刊全称JCR期刊简称ISSN所属小类ACM COMPUTING SURVEYS ACM COMPUT SURV0360-0300C OMPUTER SC ACM Transactions on Intelligent Systems and Te ACM T INTEL SYST TEC2157-6904C OMPUTER SC ACM Transactions on Intelligent Systems and Te ACM T INTEL SYST TEC2157-6904C OMPUTER SC ACS Applied Materials & Interfaces ACS APPL MATER INTER1944-8244M ATERIALS S ACS Applied Materials & Interfaces ACS APPL MATER INTER1944-8244N ANOSCIENC ACS Macro Letters ACS MACRO LETT2161-1653P OLYMER SCI ACS Nano ACS NANO1936-0851M ATERIALS S ACS Nano ACS NANO1936-0851C HEMISTRY, MACS Nano ACS NANO1936-0851N ANOSCIENCACS Nano ACS NANO1936-0851C HEMISTRY, P Acta Biomaterialia ACTA BIOMATER1742-7061M ATERIALS S Acta Biomaterialia ACTA BIOMATER1742-7061E NGINEERING ADVANCES IN APPLIED MECHANICS ADV APPL MECH0065-2156E NGINEERING ADVANCES IN APPLIED MECHANICS ADV APPL MECH0065-2156M ECHANICS Advanced Energy Materials ADV ENERGY MATER1614-6832M ATERIALS S Advanced Energy Materials ADV ENERGY MATER1614-6832E NERGY & FU Advanced Energy Materials ADV ENERGY MATER1614-6832P HYSICS, CON Advanced Energy Materials ADV ENERGY MATER1614-6832P HYSICS, APP Advanced Energy Materials ADV ENERGY MATER1614-6832C HEMISTRY, P ADVANCED FUNCTIONAL MATERIALS ADV FUNCT MATER1616-301X M ATERIALS S ADVANCED FUNCTIONAL MATERIALS ADV FUNCT MATER1616-301X C HEMISTRY, M ADVANCED FUNCTIONAL MATERIALS ADV FUNCT MATER1616-301X N ANOSCIENC ADVANCED FUNCTIONAL MATERIALS ADV FUNCT MATER1616-301X P HYSICS, CON ADVANCED FUNCTIONAL MATERIALS ADV FUNCT MATER1616-301X P HYSICS, APP ADVANCED FUNCTIONAL MATERIALS ADV FUNCT MATER1616-301X C HEMISTRY, P Advanced Healthcare Materials ADV HEALTHC MATER2192-2640M ATERIALS SADVANCED MATERIALS ADV MATER0935-9648M ATERIALS S ADVANCED MATERIALS ADV MATER0935-9648C HEMISTRY, M ADVANCED MATERIALS ADV MATER0935-9648N ANOSCIENC ADVANCED MATERIALS ADV MATER0935-9648P HYSICS, CON ADVANCED MATERIALS ADV MATER0935-9648P HYSICS, APP ADVANCED MATERIALS ADV MATER0935-9648C HEMISTRY, PANNU REV BIOMED ENG1523-9829E NGINEERING ANNUAL REVIEW OF BIOMEDICAL ENGINEAnnual Review of Chemical and Biomolecular En ANNU REV CHEM BIOMOL1947-5438E NGINEERING Annual Review of Chemical and Biomolecular En ANNU REV CHEM BIOMOL1947-5438C HEMISTRY, A Annual Review of Food Science and Technology A NNU REV FOOD SCI T1941-1413F OOD SCIENCANNU REV MATER RES1531-7331M ATERIALS S ANNUAL REVIEW OF MATERIALS RESEARCAPPLIED ENERGY APPL ENERG0306-2619E NGINEERING APPLIED ENERGY APPL ENERG0306-2619E NERGY & FU BIOMATERIALS BIOMATERIALS0142-9612M ATERIALS S BIOMATERIALS BIOMATERIALS0142-9612E NGINEERING BIORESOURCE TECHNOLOGY BIORESOURCE TECHNOL0960-8524E NERGY & FU BIORESOURCE TECHNOLOGY BIORESOURCE TECHNOL0960-8524A GRICULTURA BIORESOURCE TECHNOLOGY BIORESOURCE TECHNOL0960-8524B IOTECHNOLO BIOSENSORS & BIOELECTRONICS BIOSENS BIOELECTRON0956-5663E LECTROCHE BIOSENSORS & BIOELECTRONICS BIOSENS BIOELECTRON0956-5663C HEMISTRY, A BIOSENSORS & BIOELECTRONICS BIOSENS BIOELECTRON0956-5663N ANOSCIENC BIOSENSORS & BIOELECTRONICS BIOSENS BIOELECTRON0956-5663B IOTECHNOLO BIOSENSORS & BIOELECTRONICS BIOSENS BIOELECTRON0956-5663B IOPHYSICS BIOTECHNOLOGY ADVANCES BIOTECHNOL ADV0734-9750B IOTECHNOLO Biotechnology for Biofuels BIOTECHNOL BIOFUELS1754-6834E NERGY & FU Biotechnology for Biofuels BIOTECHNOL BIOFUELS1754-6834B IOTECHNOLO CARBON CARBON0008-6223M ATERIALS S CARBON CARBON0008-6223C HEMISTRY, P CHEMISTRY OF MATERIALS CHEM MATER0897-4756M ATERIALS S CHEMISTRY OF MATERIALS CHEM MATER0897-4756C HEMISTRY, PCOMPR REV FOOD SCI F1541-4337F OOD SCIENC COMPREHENSIVE REVIEWS IN FOOD SCIENCOMPUT-AIDED CIV INF1093-9687E NGINEERING COMPUTER-AIDED CIVIL AND INFRASTRUCCOMPUTER-AIDED CIVIL AND INFRASTRUCCOMPUT-AIDED CIV INF1093-9687C OMPUTER SCCOMPUT-AIDED CIV INF1093-9687C ONSTRUCTIO COMPUTER-AIDED CIVIL AND INFRASTRUCCOMPUT-AIDED CIV INF1093-9687T RANSPORTA COMPUTER-AIDED CIVIL AND INFRASTRUCCRITICAL REVIEWS IN BIOTECHNOLOGY CRIT REV BIOTECHNOL0738-8551B IOTECHNOLOCRIT REV FOOD SCI1040-8398F OOD SCIENC CRITICAL REVIEWS IN FOOD SCIENCE ANDCRIT REV FOOD SCI1040-8398N UTRITION & CRITICAL REVIEWS IN FOOD SCIENCE ANDCURRENT OPINION IN BIOTECHNOLOGY CURR OPIN BIOTECH0958-1669B IOCHEMICAL CURRENT OPINION IN BIOTECHNOLOGY CURR OPIN BIOTECH0958-1669B IOTECHNOLOCURR OPIN SOLID ST M1359-0286M ATERIALS S CURRENT OPINION IN SOLID STATE & MATCURR OPIN SOLID ST M1359-0286P HYSICS, CON CURRENT OPINION IN SOLID STATE & MATCURRENT OPINION IN SOLID STATE & MATCURR OPIN SOLID ST M1359-0286P HYSICS, APP ELECTROCHEMISTRY COMMUNICATIONS E LECTROCHEM COMMUN1388-2481E LECTROCHEIEEE Communications Surveys and Tutorials IEEE COMMUN SURV TUT1553-877X T ELECOMMUN IEEE Communications Surveys and Tutorials IEEE COMMUN SURV TUT1553-877X C OMPUTER SC IEEE Industrial Electronics Magazine IEEE IND ELECTRON M1932-4529E NGINEERING IEEE SIGNAL PROCESSING MAGAZINE IEEE SIGNAL PROC MAG1053-5888E NGINEERINGIEEE TRANSACTIONS ON EVOLUTIONARY IEEE T EVOLUT COMPUT1089-778X C OMPUTER SCIEEE TRANSACTIONS ON EVOLUTIONARY IEEE T EVOLUT COMPUT1089-778X C OMPUTER SC IEEE TRANSACTIONS ON FUZZY SYSTEMS IEEE T FUZZY SYST1063-6706E NGINEERING IEEE TRANSACTIONS ON FUZZY SYSTEMS IEEE T FUZZY SYST1063-6706C OMPUTER SC IEEE TRANSACTIONS ON INDUSTRIAL ELE IEEE T IND ELECTRON0278-0046E NGINEERING IEEE TRANSACTIONS ON INDUSTRIAL ELE IEEE T IND ELECTRON0278-0046I NSTRUMENT IEEE TRANSACTIONS ON INDUSTRIAL ELE IEEE T IND ELECTRON0278-0046A UTOMATIONIEEE Transactions on Industrial Informatics IEEE T IND INFORM1551-3203E NGINEERING IEEE Transactions on Industrial Informatics IEEE T IND INFORM1551-3203C OMPUTER SC IEEE Transactions on Industrial Informatics IEEE T IND INFORM1551-3203A UTOMATION IEEE Transactions on Neural Networks and LearnIEEE T NEUR NET LEAR2162-237X E NGINEERINGIEEE T NEUR NET LEAR2162-237X C OMPUTER SC IEEE Transactions on Neural Networks and LearnIEEE Transactions on Neural Networks and LearnIEEE T NEUR NET LEAR2162-237X C OMPUTER SCIEEE T NEUR NET LEAR2162-237X C OMPUTER SC IEEE Transactions on Neural Networks and LearnIEEE TRANSACTIONS ON PATTERN ANALY IEEE T PATTERN ANAL0162-8828E NGINEERINGIEEE TRANSACTIONS ON PATTERN ANALY IEEE T PATTERN ANAL0162-8828C OMPUTER SC IEEE TRANSACTIONS ON POWER ELECTRO IEEE T POWER ELECTR0885-8993E NGINEERING IEEE Transactions on Smart Grid IEEE T SMART GRID1949-3053E NGINEERINGIEEE T THZ SCI TECHN2156-342X E NGINEERING IEEE Transactions on Terahertz Science and TechIEEE T THZ SCI TECHN2156-342X O PTICSIEEE Transactions on Terahertz Science and TechIEEE T THZ SCI TECHN2156-342X P HYSICS, APP IEEE Transactions on Terahertz Science and TechIEEE WIRELESS COMMUNICATIONS IEEE WIREL COMMUN1536-1284T ELECOMMUN IEEE WIRELESS COMMUNICATIONS IEEE WIREL COMMUN1536-1284E NGINEERING IEEE WIRELESS COMMUNICATIONS IEEE WIREL COMMUN1536-1284C OMPUTER SCIEEE WIRELESS COMMUNICATIONS IEEE WIREL COMMUN1536-1284C OMPUTER SCInternational Journal of Neural Systems INT J NEURAL SYST0129-0657C OMPUTER SC INTERNATIONAL JOURNAL OF PLASTICITYINT J PLASTICITY0749-6419M ATERIALS SINT J PLASTICITY0749-6419E NGINEERING INTERNATIONAL JOURNAL OF PLASTICITYINTERNATIONAL JOURNAL OF PLASTICITYINT J PLASTICITY0749-6419M ECHANICS INTERNATIONAL MATERIALS REVIEWS INT MATER REV0950-6608M ATERIALS S JOURNAL OF CATALYSIS J CATAL0021-9517E NGINEERINGJOURNAL OF CATALYSIS J CATAL0021-9517C HEMISTRY, P JOURNAL OF HAZARDOUS MATERIALS J HAZARD MATER0304-3894E NGINEERING JOURNAL OF HAZARDOUS MATERIALS J HAZARD MATER0304-3894E NGINEERING JOURNAL OF HAZARDOUS MATERIALS J HAZARD MATER0304-3894E NVIRONMEN Journal of Materials Chemistry A J MATER CHEM A2050-7488M ATERIALS S Journal of Materials Chemistry A J MATER CHEM A2050-7488E NERGY & FU Journal of Materials Chemistry A J MATER CHEM A2050-7488C HEMISTRY, P Journal of Materials Chemistry B J MATER CHEM B2050-750X M ATERIALS S Journal of Materials Chemistry C J MATER CHEM C2050-7526M ATERIALS S Journal of Materials Chemistry C J MATER CHEM C2050-7526P HYSICS, APP JOURNAL OF MEMBRANE SCIENCE J MEMBRANE SCI0376-7388P OLYMER SCI JOURNAL OF MEMBRANE SCIENCE J MEMBRANE SCI0376-7388E NGINEERING JOURNAL OF NANOBIOTECHNOLOGY J NANOBIOTECHNOL1477-3155N ANOSCIENC JOURNAL OF NANOBIOTECHNOLOGY J NANOBIOTECHNOL1477-3155B IOTECHNOLO JOURNAL OF POWER SOURCES J POWER SOURCES0378-7753E LECTROCHE JOURNAL OF POWER SOURCES J POWER SOURCES0378-7753E NERGY & FU Journal of Statistical Software J STAT SOFTW1548-7660C OMPUTER SC Journal of Statistical Software J STAT SOFTW1548-7660S TATISTICS & LAB ON A CHIP LAB CHIP1473-0197C HEMISTRY, M LAB ON A CHIP LAB CHIP1473-0197N ANOSCIENC LAB ON A CHIP LAB CHIP1473-0197B IOCHEMICAL mAbs MABS-AUSTIN1942-0862M EDICINE, RE MACROMOLECULAR RAPID COMMUNICAT MACROMOL RAPID COMM1022-1336P OLYMER SCIMACROMOLECULES MACROMOLECULES0024-9297P OLYMER SCIMATERIALS SCIENCE & ENGINEERING R-R MAT SCI ENG R0927-796X M ATERIALS S MATERIALS SCIENCE & ENGINEERING R-R MAT SCI ENG R0927-796X P HYSICS, APP Materials Today MATER TODAY1369-7021M ATERIALS S MEDICAL IMAGE ANALYSIS MED IMAGE ANAL1361-8415E NGINEERING MEDICAL IMAGE ANALYSIS MED IMAGE ANAL1361-8415R ADIOLOGY, MEDICAL IMAGE ANALYSIS MED IMAGE ANAL1361-8415C OMPUTER SC MEDICAL IMAGE ANALYSIS MED IMAGE ANAL1361-8415C OMPUTER SCMETABOLIC ENGINEERING METAB ENG1096-7176B IOTECHNOLOMIS QUARTERLY MIS QUART0276-7783C OMPUTER SCMOL NUTR FOOD RES1613-4125F OOD SCIENC MOLECULAR NUTRITION & FOOD RESEARCMRS BULLETIN MRS BULL0883-7694M ATERIALS S MRS BULLETIN MRS BULL0883-7694P HYSICS, APP Nano Energy NANO ENERGY2211-2855M ATERIALS S Nano Energy NANO ENERGY2211-2855N ANOSCIENC Nano Energy NANO ENERGY2211-2855P HYSICS, APP Nano Energy NANO ENERGY2211-2855C HEMISTRY, P NANO LETTERS NANO LETT1530-6984M ATERIALS S NANO LETTERS NANO LETT1530-6984C HEMISTRY, M NANO LETTERS NANO LETT1530-6984N ANOSCIENC NANO LETTERS NANO LETT1530-6984P HYSICS, CON NANO LETTERS NANO LETT1530-6984P HYSICS, APP NANO LETTERS NANO LETT1530-6984C HEMISTRY, P Nano Research NANO RES1998-0124M ATERIALS S Nano Research NANO RES1998-0124N ANOSCIENC Nano Research NANO RES1998-0124P HYSICS, APP Nano Research NANO RES1998-0124C HEMISTRY, P Nano Today NANO TODAY1748-0132M ATERIALS S Nano Today NANO TODAY1748-0132C HEMISTRY, M Nano Today NANO TODAY1748-0132N ANOSCIENC Nanomedicine-Nanotechnology Biology and Med NANOMED-NANOTECHNOL1549-9634N ANOSCIENC Nanomedicine-Nanotechnology Biology and Med NANOMED-NANOTECHNOL1549-9634M EDICINE, RE Nanoscale NANOSCALE2040-3364M ATERIALS S Nanoscale NANOSCALE2040-3364C HEMISTRY, M Nanoscale NANOSCALE2040-3364N ANOSCIENC Nanoscale NANOSCALE2040-3364P HYSICS, APP NATURE BIOTECHNOLOGY NAT BIOTECHNOL1087-0156B IOTECHNOLO NATURE MATERIALS NAT MATER1476-1122M ATERIALS S NATURE MATERIALS NAT MATER1476-1122P HYSICS, CON NATURE MATERIALS NAT MATER1476-1122P HYSICS, APP NATURE MATERIALS NAT MATER1476-1122C HEMISTRY, PNature Nanotechnology NAT NANOTECHNOL1748-3387M ATERIALS SNature Nanotechnology NAT NANOTECHNOL1748-3387N ANOSCIENC NPG Asia Materials NPG ASIA MATER1884-4049M ATERIALS S PROCEEDINGS OF THE IEEE P IEEE0018-9219E NGINEERING Polymer Reviews POLYM REV1558-3724P OLYMER SCIPROGRESS IN ENERGY AND COMBUSTION PROG ENERG COMBUST0360-1285E NGINEERING PROGRESS IN ENERGY AND COMBUSTION PROG ENERG COMBUST0360-1285E NGINEERINGPROGRESS IN ENERGY AND COMBUSTION PROG ENERG COMBUST0360-1285E NERGY & FU PROGRESS IN ENERGY AND COMBUSTION PROG ENERG COMBUST0360-1285T HERMODYNA PROGRESS IN MATERIALS SCIENCE PROG MATER SCI0079-6425M ATERIALS S PROGRESS IN PHOTOVOLTAICS PROG PHOTOVOLTAICS1062-7995M ATERIALS S PROGRESS IN PHOTOVOLTAICS PROG PHOTOVOLTAICS1062-7995E NERGY & FUPROGRESS IN PHOTOVOLTAICS PROG PHOTOVOLTAICS1062-7995P HYSICS, APP PROGRESS IN QUANTUM ELECTRONICS PROG QUANT ELECTRON0079-6727E NGINEERING PROGRESS IN SURFACE SCIENCE PROG SURF SCI0079-6816P HYSICS, CON PROGRESS IN SURFACE SCIENCE PROG SURF SCI0079-6816C HEMISTRY, P RENEWABLE & SUSTAINABLE ENERGY RE RENEW SUST ENERG REV1364-0321E NERGY & FU SMALL SMALL1613-6810M ATERIALS S SMALL SMALL1613-6810C HEMISTRY, M SMALL SMALL1613-6810N ANOSCIENC SMALL SMALL1613-6810P HYSICS, CON SMALL SMALL1613-6810P HYSICS, APP SMALL SMALL1613-6810C HEMISTRY, P Soft Matter SOFT MATTER1744-683X M ATERIALS S Soft Matter SOFT MATTER1744-683X P OLYMER SCI Soft Matter SOFT MATTER1744-683X P HYSICS, MUL Soft Matter SOFT MATTER1744-683X C HEMISTRY, PSOL ENERG MAT SOL C0927-0248M ATERIALS S SOLAR ENERGY MATERIALS AND SOLAR CSOL ENERG MAT SOL C0927-0248E NERGY & FU SOLAR ENERGY MATERIALS AND SOLAR CSOL ENERG MAT SOL C0927-0248P HYSICS, APP SOLAR ENERGY MATERIALS AND SOLAR CTRENDS IN BIOTECHNOLOGY TRENDS BIOTECHNOL0167-7799B IOTECHNOLO TRENDS IN FOOD SCIENCE & TECHNOLOG T RENDS FOOD SCI TECH0924-2244F OOD SCIENCACM TRANSACTIONS ON GRAPHICS ACM T GRAPHIC0730-0301C OMPUTER SCACM TRANSACTIONS ON MATHEMATICAL ACM T MATH SOFTWARE0098-3500C OMPUTER SC ACM TRANSACTIONS ON MATHEMATICAL ACM T MATH SOFTWARE0098-3500M ATHEMATIC ACTA MATERIALIA ACTA MATER1359-6454M ATERIALS S ACTA MATERIALIA ACTA MATER1359-6454M ETALLURGY ADVANCES IN BIOCHEMICAL ENGINEERIN ADV BIOCHEM ENG BIOT0724-6145B IOTECHNOLO AICHE JOURNAL AICHE J0001-1541E NGINEERING ANNALS OF BIOMEDICAL ENGINEERINGANN BIOMED ENG0090-6964E NGINEERING ANNUAL REVIEW OF INFORMATION SCIEN ANNU REV INFORM SCI0066-4200C OMPUTER SCAPPL MICROBIOL BIOT0175-7598B IOTECHNOLO APPLIED MICROBIOLOGY AND BIOTECHNOAPPLIED SOFT COMPUTING APPL SOFT COMPUT1568-4946C OMPUTER SC APPLIED SOFT COMPUTING APPL SOFT COMPUT1568-4946C OMPUTER SC APPLIED SPECTROSCOPY REVIEWS APPL SPECTROSC REV0570-4928S PECTROSCOP APPLIED SPECTROSCOPY REVIEWS APPL SPECTROSC REV0570-4928I NSTRUMENT APPLIED SURFACE SCIENCE APPL SURF SCI0169-4332M ATERIALS S APPLIED SURFACE SCIENCE APPL SURF SCI0169-4332P HYSICS, CON APPLIED SURFACE SCIENCE APPL SURF SCI0169-4332P HYSICS, APP APPLIED SURFACE SCIENCE APPL SURF SCI0169-4332C HEMISTRY, P APPLIED THERMAL ENGINEERING APPL THERM ENG1359-4311E NGINEERINGAPPLIED THERMAL ENGINEERING APPL THERM ENG1359-4311M ECHANICS APPLIED THERMAL ENGINEERING APPL THERM ENG1359-4311E NERGY & FU APPLIED THERMAL ENGINEERING APPL THERM ENG1359-4311T HERMODYNAARCH COMPUT METHOD E1134-3060E NGINEERING ARCHIVES OF COMPUTATIONAL METHODSARCH COMPUT METHOD E1134-3060C OMPUTER SC ARCHIVES OF COMPUTATIONAL METHODSARCH COMPUT METHOD E1134-3060M ATHEMATIC ARCHIVES OF COMPUTATIONAL METHODSARTIFICIAL INTELLIGENCE ARTIF INTELL0004-3702C OMPUTER SC AUSTRALIAN JOURNAL OF GRAPE AND WI AUST J GRAPE WINE R1322-7130F OOD SCIENC AUSTRALIAN JOURNAL OF GRAPE AND WI AUST J GRAPE WINE R1322-7130H ORTICULTUR AUTOMATICA AUTOMATICA0005-1098E NGINEERINGAUTOMATICA AUTOMATICA0005-1098A UTOMATION BIOCHEMICAL ENGINEERING JOURNAL BIOCHEM ENG J1369-703X E NGINEERING BIOCHEMICAL ENGINEERING JOURNAL BIOCHEM ENG J1369-703X B IOTECHNOLO BIODEGRADATION BIODEGRADATION0923-9820B IOTECHNOLO Biofabrication BIOFABRICATION1758-5082M ATERIALS S Biofabrication BIOFABRICATION1758-5082E NGINEERING Bioinspiration & Biomimetics BIOINSPIR BIOMIM1748-3182M ATERIALS S Bioinspiration & Biomimetics BIOINSPIR BIOMIM1748-3182E NGINEERING Bioinspiration & Biomimetics BIOINSPIR BIOMIM1748-3182R OBOTICS Biointerphases BIOINTERPHASES1934-8630M ATERIALS S Biointerphases BIOINTERPHASES1934-8630B IOPHYSICSBIOMASS & BIOENERGY BIOMASS BIOENERG0961-9534E NERGY & FU BIOMASS & BIOENERGY BIOMASS BIOENERG0961-9534A GRICULTURA BIOMASS & BIOENERGY BIOMASS BIOENERG0961-9534B IOTECHNOLO Biomechanics and Modeling in Mechanobiology B IOMECH MODEL MECHAN1617-7959E NGINEERINGBiomechanics and Modeling in Mechanobiology B IOMECH MODEL MECHAN1617-7959B IOPHYSICS Biomedical Materials BIOMED MATER1748-6041M ATERIALS S Biomedical Materials BIOMED MATER1748-6041E NGINEERING BIOMEDICAL MICRODEVICES BIOMED MICRODEVICES1387-2176E NGINEERING BIOMEDICAL MICRODEVICES BIOMED MICRODEVICES1387-2176N ANOSCIENC BIOTECHNIQUES BIOTECHNIQUES0736-6205B IOCHEMICAL BIOTECHNIQUES BIOTECHNIQUES0736-6205B IOCHEMISTR BIOTECHNOLOGY AND BIOENGINEERING BIOTECHNOL BIOENG0006-3592B IOTECHNOLO BIOTECHNOLOGY PROGRESS BIOTECHNOL PROGR8756-7938B IOTECHNOLO BIOTECHNOLOGY PROGRESS BIOTECHNOL PROGR8756-7938F OOD SCIENC BMC BIOTECHNOLOGY BMC BIOTECHNOL1472-6750B IOTECHNOLO BUILDING AND ENVIRONMENT BUILD ENVIRON0360-1323E NGINEERING BUILDING AND ENVIRONMENT BUILD ENVIRON0360-1323E NGINEERING BUILDING AND ENVIRONMENT BUILD ENVIRON0360-1323C ONSTRUCTIO CELLULOSE CELLULOSE0969-0239M ATERIALS SCELLULOSE CELLULOSE0969-0239M ATERIALS SCELLULOSE CELLULOSE0969-0239P OLYMER SCICEMENT & CONCRETE COMPOSITES CEMENT CONCRETE COMP0958-9465M ATERIALS S CEMENT & CONCRETE COMPOSITES CEMENT CONCRETE COMP0958-9465C ONSTRUCTIOCEMENT AND CONCRETE RESEARCH CEMENT CONCRETE RES0008-8846M ATERIALS SCEMENT AND CONCRETE RESEARCH CEMENT CONCRETE RES0008-8846C ONSTRUCTIO CHEMICAL ENGINEERING JOURNAL CHEM ENG J1385-8947E NGINEERING CHEMICAL ENGINEERING JOURNAL CHEM ENG J1385-8947E NGINEERING CHEMICAL ENGINEERING RESEARCH & DE CHEM ENG RES DES0263-8762E NGINEERING CHEMICAL ENGINEERING SCIENCE CHEM ENG SCI0009-2509E NGINEERING CHEMOMETRICS AND INTELLIGENT LABO CHEMOMETR INTELL LAB0169-7439C HEMISTRY, A CHEMOMETRICS AND INTELLIGENT LABO CHEMOMETR INTELL LAB0169-7439C OMPUTER SC CHEMOMETRICS AND INTELLIGENT LABO CHEMOMETR INTELL LAB0169-7439M ATHEMATICCHEMOMETRICS AND INTELLIGENT LABO CHEMOMETR INTELL LAB0169-7439S TATISTICS & CHEMOMETRICS AND INTELLIGENT LABO CHEMOMETR INTELL LAB0169-7439I NSTRUMENT CHEMOMETRICS AND INTELLIGENT LABO CHEMOMETR INTELL LAB0169-7439A UTOMATIONCIRP ANN-MANUF TECHN0007-8506E NGINEERING CIRP ANNALS-MANUFACTURING TECHNOLCIRP ANN-MANUF TECHN0007-8506E NGINEERING CIRP ANNALS-MANUFACTURING TECHNOLCOMBUSTION AND FLAME COMBUST FLAME0010-2180E NGINEERING COMBUSTION AND FLAME COMBUST FLAME0010-2180E NGINEERING COMBUSTION AND FLAME COMBUST FLAME0010-2180E NGINEERING COMBUSTION AND FLAME COMBUST FLAME0010-2180E NERGY & FU COMBUSTION AND FLAME COMBUST FLAME0010-2180T HERMODYNA COMMUNICATIONS OF THE ACM COMMUN ACM0001-0782C OMPUTER SC COMMUNICATIONS OF THE ACM COMMUN ACM0001-0782C OMPUTER SCCOMMUNICATIONS OF THE ACM COMMUN ACM0001-0782C OMPUTER SCCOMPOS PART A-APPL S1359-835X M ATERIALS S COMPOSITES PART A-APPLIED SCIENCE ANCOMPOS PART A-APPL S1359-835X E NGINEERING COMPOSITES PART A-APPLIED SCIENCE ANCOMPOSITES PART B-ENGINEERING COMPOS PART B-ENG1359-8368M ATERIALS S COMPOSITES PART B-ENGINEERING COMPOS PART B-ENG1359-8368E NGINEERING COMPOSITES SCIENCE AND TECHNOLOGY COMPOS SCI TECHNOL0266-3538M ATERIALS SCOMPOSITE STRUCTURES COMPOS STRUCT0263-8223M ATERIALS S COMPUTERS & CHEMICAL ENGINEERING COMPUT CHEM ENG0098-1354E NGINEERINGCOMPUTERS & CHEMICAL ENGINEERING COMPUT CHEM ENG0098-1354C OMPUTER SC COMPUTERS & EDUCATION COMPUT EDUC0360-1315C OMPUTER SC COMPUTER METHODS IN APPLIED MECHA C OMPUT METHOD APPL M0045-7825E NGINEERING COMPUTER METHODS IN APPLIED MECHA C OMPUT METHOD APPL M0045-7825M ECHANICS COMPUTER METHODS IN APPLIED MECHA C OMPUT METHOD APPL M0045-7825M ATHEMATICCONSTR BUILD MATER0950-0618M ATERIALS S CONSTRUCTION AND BUILDING MATERIALCONSTR BUILD MATER0950-0618E NGINEERING CONSTRUCTION AND BUILDING MATERIALCONSTR BUILD MATER0950-0618C ONSTRUCTIO CONSTRUCTION AND BUILDING MATERIALCORROSION SCIENCE CORROS SCI0010-938X M ATERIALS S CORROSION SCIENCE CORROS SCI0010-938X M ETALLURGY CORROSION CORROSION0010-9312M ATERIALS S CORROSION CORROSION0010-9312M ETALLURGYDATA MIN KNOWL DISC1384-5810C OMPUTER SC DATA MINING AND KNOWLEDGE DISCOVEDATA MIN KNOWL DISC1384-5810C OMPUTER SC DATA MINING AND KNOWLEDGE DISCOVEDENTAL MATERIALS DENT MATER0109-5641M ATERIALS S DENTAL MATERIALS DENT MATER0109-5641D ENTISTRY, O DESALINATION DESALINATION0011-9164E NGINEERING DESALINATION DESALINATION0011-9164W ATER RESOU DYES AND PIGMENTS DYES PIGMENTS0143-7208M ATERIALS S DYES AND PIGMENTS DYES PIGMENTS0143-7208E NGINEERINGDYES AND PIGMENTS DYES PIGMENTS0143-7208C HEMISTRY, AELECTROCHIMICA ACTA ELECTROCHIM ACTA0013-4686E LECTROCHE Electronic Materials Letters ELECTRON MATER LETT1738-8090M ATERIALS S ENERGY AND BUILDINGS ENERG BUILDINGS0378-7788E NGINEERING ENERGY AND BUILDINGS ENERG BUILDINGS0378-7788C ONSTRUCTIOENERGY AND BUILDINGS ENERG BUILDINGS0378-7788E NERGY & FU ENERGY CONVERSION AND MANAGEMEN E NERG CONVERS MANAGE0196-8904M ECHANICS ENERGY CONVERSION AND MANAGEMEN E NERG CONVERS MANAGE0196-8904E NERGY & FUENERGY CONVERSION AND MANAGEMEN E NERG CONVERS MANAGE0196-8904T HERMODYNA ENERGY CONVERSION AND MANAGEMEN E NERG CONVERS MANAGE0196-8904P HYSICS, NUC ENERGY & FUELS ENERG FUEL0887-0624E NGINEERINGENERGY & FUELS ENERG FUEL0887-0624E NERGY & FU ENERGY JOURNAL ENERG J0195-6574E NERGY & FU ENERGY ENERGY0360-5442E NERGY & FU ENERGY ENERGY0360-5442T HERMODYNA ENVIRONMENTAL MODELLING & SOFTWA ENVIRON MODELL SOFTW1364-8152E NGINEERING ENVIRONMENTAL MODELLING & SOFTWA ENVIRON MODELL SOFTW1364-8152E NVIRONMEN ENVIRONMENTAL MODELLING & SOFTWA ENVIRON MODELL SOFTW1364-8152C OMPUTER SC EVOLUTIONARY COMPUTATION EVOL COMPUT1063-6560C OMPUTER SCEVOLUTIONARY COMPUTATION EVOL COMPUT1063-6560C OMPUTER SC FLUID PHASE EQUILIBRIA FLUID PHASE EQUILIBR0378-3812E NGINEERING FLUID PHASE EQUILIBRIA FLUID PHASE EQUILIBR0378-3812T HERMODYNAFLUID PHASE EQUILIBRIA FLUID PHASE EQUILIBR0378-3812C HEMISTRY, P FOOD ADDITIVES AND CONTAMINANTS FOOD ADDIT CONTAM A1944-0049T OXICOLOGY FOOD ADDITIVES AND CONTAMINANTS FOOD ADDIT CONTAM A1944-0049F OOD SCIENC FOOD ADDITIVES AND CONTAMINANTS FOOD ADDIT CONTAM A1944-0049C HEMISTRY, A FOOD AND BIOPRODUCTS PROCESSING FOOD BIOPROD PROCESS0960-3085E NGINEERING FOOD AND BIOPRODUCTS PROCESSING FOOD BIOPROD PROCESS0960-3085B IOTECHNOLO FOOD AND BIOPRODUCTS PROCESSING FOOD BIOPROD PROCESS0960-3085F OOD SCIENC FOOD CHEMISTRY FOOD CHEM0308-8146F OOD SCIENCFOOD CHEMISTRY FOOD CHEM0308-8146N UTRITION & FOOD CHEMISTRY FOOD CHEM0308-8146C HEMISTRY, A FOOD AND CHEMICAL TOXICOLOGY FOOD CHEM TOXICOL0278-6915T OXICOLOGY FOOD AND CHEMICAL TOXICOLOGY FOOD CHEM TOXICOL0278-6915F OOD SCIENC FOOD CONTROL FOOD CONTROL0956-7135F OOD SCIENC FOOD HYDROCOLLOIDS FOOD HYDROCOLLOID0268-005X F OOD SCIENC FOOD HYDROCOLLOIDS FOOD HYDROCOLLOID0268-005X C HEMISTRY, A FOOD MICROBIOLOGY FOOD MICROBIOL0740-0020B IOTECHNOLO FOOD MICROBIOLOGY FOOD MICROBIOL0740-0020F OOD SCIENC FOOD MICROBIOLOGY FOOD MICROBIOL0740-0020M ICROBIOLOG FOOD QUALITY AND PREFERENCE FOOD QUAL PREFER0950-3293F OOD SCIENC FOOD RESEARCH INTERNATIONAL FOOD RES INT0963-9969F OOD SCIENC FUEL FUEL0016-2361E NGINEERING FUEL FUEL0016-2361E NERGY & FU Fuel Cells FUEL CELLS1615-6846E LECTROCHE Fuel Cells FUEL CELLS1615-6846E NERGY & FUFUEL PROCESSING TECHNOLOGY FUEL PROCESS TECHNOL0378-3820E NGINEERINGFUEL PROCESSING TECHNOLOGY FUEL PROCESS TECHNOL0378-3820E NERGY & FUFUEL PROCESSING TECHNOLOGY FUEL PROCESS TECHNOL0378-3820C HEMISTRY, AFUTURE GENER COMP SY0167-739X C OMPUTER SC FUTURE GENERATION COMPUTER SYSTEMGEOTHERMICS GEOTHERMICS0375-6505G EOSCIENCES GEOTHERMICS GEOTHERMICS0375-6505E NERGY & FU GOLD BULLETIN GOLD BULL0017-1557M ATERIALS S GOLD BULLETIN GOLD BULL0017-1557C HEMISTRY, I GOLD BULLETIN GOLD BULL0017-1557C HEMISTRY, P HOLZFORSCHUNG HOLZFORSCHUNG0018-3830M ATERIALS S HOLZFORSCHUNG HOLZFORSCHUNG0018-3830F ORESTRY HUMAN-COMPUTER INTERACTION HUM-COMPUT INTERACT0737-0024C OMPUTER SC HUMAN-COMPUTER INTERACTION HUM-COMPUT INTERACT0737-0024C OMPUTER SC HYDROMETALLURGY HYDROMETALLURGY0304-386X M ETALLURGY IEEE COMMUNICATIONS MAGAZINE IEEE COMMUN MAG0163-6804T ELECOMMUNIEEE COMMUNICATIONS MAGAZINE IEEE COMMUN MAG0163-6804E NGINEERING IEEE Computational Intelligence Magazine IEEE COMPUT INTELL M1556-603X C OMPUTER SC IEEE CONTROL SYSTEMS MAGAZINE IEEE CONTR SYST MAG1066-033X A UTOMATION IEEE ELECTRON DEVICE LETTERS IEEE ELECTR DEVICE L0741-3106E NGINEERING IEEE Journal of Photovoltaics IEEE J PHOTOVOLT2156-3381M ATERIALS S IEEE Journal of Photovoltaics IEEE J PHOTOVOLT2156-3381E NERGY & FU IEEE Journal of Photovoltaics IEEE J PHOTOVOLT2156-3381P HYSICS, APPIEEE J SEL AREA COMM0733-8716T ELECOMMUN IEEE JOURNAL ON SELECTED AREAS IN COIEEE J SEL AREA COMM0733-8716E NGINEERING IEEE JOURNAL ON SELECTED AREAS IN COIEEE JOURNAL OF SELECTED TOPICS IN QUIEEE J SEL TOP QUANT1077-260X E NGINEERINGIEEE J SEL TOP QUANT1077-260X O PTICSIEEE JOURNAL OF SELECTED TOPICS IN QUIEEE J SEL TOP QUANT1077-260X P HYSICS, APP IEEE JOURNAL OF SELECTED TOPICS IN QUIEEE JOURNAL OF SOLID-STATE CIRCUITS IEEE J SOLID-ST CIRC0018-9200E NGINEERINGIEEE J-STSP1932-4553E NGINEERING IEEE Journal of Selected Topics in Signal ProcessIEEE NETWORK IEEE NETWORK0890-8044T ELECOMMUN IEEE NETWORK IEEE NETWORK0890-8044E NGINEERING IEEE NETWORK IEEE NETWORK0890-8044C OMPUTER SC IEEE NETWORK IEEE NETWORK0890-8044C OMPUTER SC IEEE PHOTONICS TECHNOLOGY LETTERS IEEE PHOTONIC TECH L1041-1135E NGINEERING IEEE PHOTONICS TECHNOLOGY LETTERS IEEE PHOTONIC TECH L1041-1135O PTICSIEEE PHOTONICS TECHNOLOGY LETTERS IEEE PHOTONIC TECH L1041-1135P HYSICS, APP IEEE Photonics Journal IEEE PHOTONICS J1943-0655E NGINEERING IEEE Photonics Journal IEEE PHOTONICS J1943-0655O PTICSIEEE Photonics Journal IEEE PHOTONICS J1943-0655P HYSICS, APPIEEE ROBOT AUTOM MAG1070-9932R OBOTICS IEEE ROBOTICS & AUTOMATION MAGAZINIEEE ROBOT AUTOM MAG1070-9932A UTOMATION IEEE ROBOTICS & AUTOMATION MAGAZINIEEE Transactions on Affective Computing IEEE T AFFECT COMPUT1949-3045C OMPUTER SC IEEE Transactions on Affective Computing IEEE T AFFECT COMPUT1949-3045C OMPUTER SC IEEE TRANSACTIONS ON ANTENNAS AND IEEE T ANTENN PROPAG0018-926X T ELECOMMUN IEEE TRANSACTIONS ON ANTENNAS AND IEEE T ANTENN PROPAG0018-926X E NGINEERINGIEEE T AUTOMAT CONTR0018-9286E NGINEERING IEEE TRANSACTIONS ON AUTOMATIC CONIEEE T AUTOMAT CONTR0018-9286A UTOMATION IEEE TRANSACTIONS ON AUTOMATIC CONIEEE Transactions on Biomedical Circuits and SyIEEE T BIOMED CIRC S1932-4545E NGINEERINGIEEE T BIOMED CIRC S1932-4545E NGINEERING IEEE Transactions on Biomedical Circuits and SyIEEE T BIO-MED ENG0018-9294E NGINEERING IEEE TRANSACTIONS ON BIOMEDICAL ENGIEEE TRANSACTIONS ON BROADCASTING IEEE T BROADCAST0018-9316T ELECOMMUN IEEE TRANSACTIONS ON BROADCASTING IEEE T BROADCAST0018-9316E NGINEERINGIEEE T CIRCUITS-I1549-8328E NGINEERING IEEE TRANSACTIONS ON CIRCUITS AND SYIEEE TRANSACTIONS ON CONTROL SYSTE IEEE T CONTR SYST T1063-6536E NGINEERING IEEE TRANSACTIONS ON CONTROL SYSTE IEEE T CONTR SYST T1063-6536A UTOMATION IEEE Transactions on Cybernetics IEEE T CYBERNETICS2168-2267C OMPUTER SC IEEE Transactions on Cybernetics IEEE T CYBERNETICS2168-2267C OMPUTER SC IEEE TRANSACTIONS ON ELECTRON DEVICIEEE T ELECTRON DEV0018-9383E NGINEERINGIEEE T ELECTRON DEV0018-9383P HYSICS, APP IEEE TRANSACTIONS ON ELECTRON DEVICIEEE T ENERGY CONVER0885-8969E NGINEERING IEEE TRANSACTIONS ON ENERGY CONVERIEEE T ENERGY CONVER0885-8969E NERGY & FU IEEE TRANSACTIONS ON ENERGY CONVERIEEE T GEOSCI REMOTE0196-2892I MAGING SCIE IEEE TRANSACTIONS ON GEOSCIENCE ANDIEEE T GEOSCI REMOTE0196-2892G EOCHEMIST IEEE TRANSACTIONS ON GEOSCIENCE ANDIEEE T GEOSCI REMOTE0196-2892E NGINEERING IEEE TRANSACTIONS ON GEOSCIENCE ANDIEEE T GEOSCI REMOTE0196-2892R EMOTE SENS IEEE TRANSACTIONS ON GEOSCIENCE ANDIEEE Transactions on Human-Machine Systems I EEE T HUM-MACH SYST2168-2291C OMPUTER SC IEEE Transactions on Human-Machine Systems I EEE T HUM-MACH SYST2168-2291C OMPUTER SC IEEE TRANSACTIONS ON IMAGE PROCESSI IEEE T IMAGE PROCESS1057-7149E NGINEERING IEEE TRANSACTIONS ON IMAGE PROCESSI IEEE T IMAGE PROCESS1057-7149C OMPUTER SCIEEE T INFORM THEORY0018-9448E NGINEERING IEEE TRANSACTIONS ON INFORMATION THIEEE TRANSACTIONS ON INFORMATION THIEEE T INFORM THEORY0018-9448C OMPUTER SC IEEE TRANSACTIONS ON INTELLIGENT TR IEEE T INTELL TRANSP1524-9050E NGINEERING。
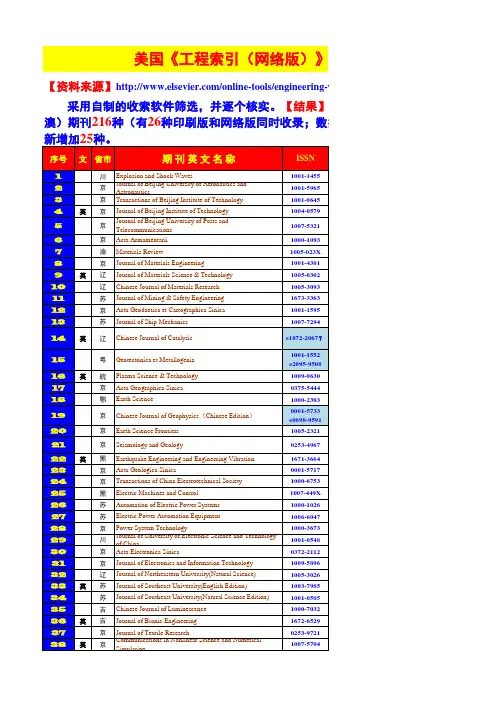
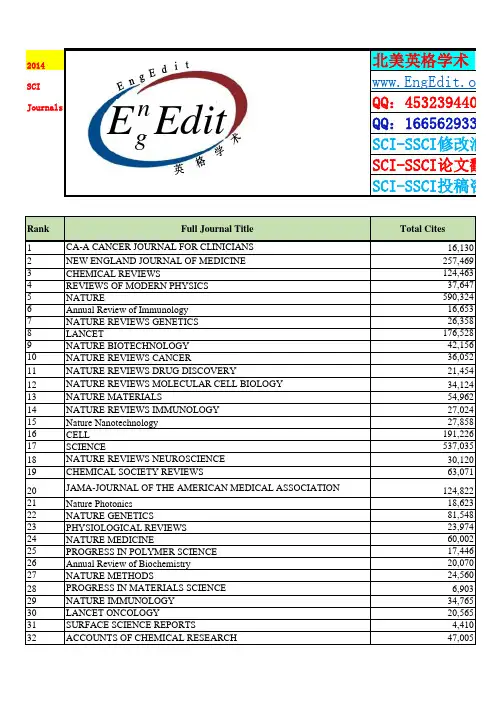

刊名全称BRAINJOURNAL OF HEAD TRAUMA REHABILITATIONLANCET NEUROLOGYNEUROLOGYNEUROREHABILITATION AND NEURAL REPAIRPain PhysicianSTROKEAMERICAN JOURNAL OF NEURORADIOLOGYBRAIN PATHOLOGY 病理Brain StimulationCurrent Alzheimer Research老年痴呆CURRENT OPINION IN NEUROLOGYEUROPEAN JOURNAL OF PAINJOURNAL OF NEUROPATHOLOGY AND EXPERIMENTAL NEUROLOGY JOURNAL OF NEUROTRAUMAJOURNAL OF PAINNEURO-ONCOLOGY 神经肿瘤 **NEUROPATHOLOGY AND APPLIED NEUROBIOLOGY NEUROSCIENTIST **PAINBMC Neurology 英国神经病学CEREBROVASCULAR DISEASES 脑血管CLINICAL JOURNAL OF PAINCurrent Neurology and Neuroscience Reports CURRENT NEUROVASCULAR RESEARCH 神经与血管EUROPEAN JOURNAL OF NEUROLOGY 欧洲神经病学HEADACHEJOURNAL OF NEUROLOGY 神经病学JOURNAL OF NEURO-ONCOLOGY 神经肿瘤JOURNAL OF NEUROSURGERY Neurodegenerative Diseases神经退行性病变NEUROGENETICS 神经遗传学NEUROLOGIC CLINICSNEURORADIOLOGY 神经放射学Neurosurgical FocusNEUROSURGERYSPINAL CORD 脊髓SPINE 脊柱Spine Journal 脊骨ACTA NEUROCHIRURGICA 欧亚神经外科学院Brain Tumor PathologyBRITISH JOURNAL OF NEUROSURGERY 英国神经外科CANADIAN JOURNAL OF NEUROLOGICAL SCIENCESCentral European NeurosurgeryCLINICAL NEUROLOGY AND NEUROSURGERYCLINICAL NEUROPATHOLOGYCurrent Pain and Headache ReportsCurrent Treatment Options in NeurologyDevelopmental NeurorehabilitationEUROPEAN JOURNAL OF PAEDIATRIC NEUROLOGY 小儿EUROPEAN NEUROLOGYEUROPEAN SPINE JOURNALJOURNAL OF CHILD NEUROLOGYJOURNAL OF CLINICAL AND EXPERIMENTAL NEUROPSYCHOLOGY Journal of Clinical NeurologyJOURNAL OF CLINICAL NEUROSCIENCEJOURNAL OF GERIATRIC PSYCHIATRY AND NEUROLOGY JOURNAL OF HEADACHE AND PAINJOURNAL OF NERVOUS AND MENTAL DISEASEJournal of Neurodevelopmental DisordersJOURNAL OF NEUROIMAGINGJOURNAL OF THE NEUROLOGICAL SCIENCESJOURNAL OF NEURO-OPHTHALMOLOGYJOURNAL OF NEUROPSYCHIATRY AND CLINICAL NEUROSCIENCES JOURNAL OF NEURORADIOLOGYJOURNAL OF NEUROSURGERY-SPINEJOURNAL OF SPINAL CORD MEDICINEJOURNAL OF SPINAL DISORDERS & TECHNIQUES NEUROCASENeurocritical CareNEUROLOGICAL RESEARCHNEUROLOGICAL SCIENCESNEUROLOGISTNEUROMODULATIONNEUROPATHOLOGYNEUROPHYSIOLOGIE CLINIQUE-CLINICAL NEUROPHYSIOLOGY NEUROSURGERY CLINICS OF NORTH AMERICANEUROSURGICAL REVIEWPain Research & ManagementPEDIATRIC NEUROLOGYPSYCHIATRY AND CLINICAL NEUROSCIENCESREVISTA DE NEUROLOGIASCHMERZSEIZURE-EUROPEAN JOURNAL OF EPILEPSYSEMINARS IN NEUROLOGYSeminars in Pediatric NeurologySURGICAL NEUROLOGYHUMAN BRAIN MAPPINGNEUROIMAGEAMERICAN JOURNAL OF NEURORADIOLOGYNEURORADIOLOGYPSYCHIATRY RESEARCH-NEUROIMAGINGSTEREOTACTIC AND FUNCTIONAL NEUROSURGERYCLINICAL EEG AND NEUROSCIENCEJOURNAL OF NEUROIMAGINGJournal of NeuroInterventional SurgeryJOURNAL OF NEURORADIOLOGYNEUROIMAGING CLINICS OF NORTH AMERICABrain Structure & FunctionFRONTIERS IN NEUROENDOCRINOLOGYHUMAN BRAIN MAPPINGJOURNAL OF COMPARATIVE NEUROLOGYJARO-JOURNAL OF THE ASSOCIATION FOR RESEARCH IN OTOLARYNGOLOGY MOLECULAR PSYCHIATRYNATURE NEUROSCIENCENATURE REVIEWS NEUROSCIENCENEUROIMAGENEURONNEUROPSYCHOPHARMACOLOGYPROGRESS IN NEUROBIOLOGYTRENDS IN COGNITIVE SCIENCESTRENDS IN NEUROSCIENCESAUDIOLOGY AND NEURO-OTOLOGYBIPOLAR DISORDERSBRAIN BEHAVIOR AND IMMUNITYBRAIN PATHOLOGYBRAIN RESEARCH REVIEWSBrain StimulationCEPHALALGIACurrent Alzheimer ResearchCURRENT OPINION IN NEUROBIOLOGYCURRENT OPINION IN NEUROLOGYEUROPEAN JOURNAL OF PAINHEARING RESEARCHHIPPOCAMPUSINTERNATIONAL JOURNAL OF NEUROPSYCHOPHARMACOLOGY JOURNAL OF ALZHEIMERS DISEASEJOURNAL OF CEREBRAL BLOOD FLOW AND METABOLISM JOURNAL OF COGNITIVE NEUROSCIENCEJOURNAL OF COMPARATIVE PHYSIOLOGY A-NEUROETHOLOGY Journal of Neural EngineeringJOURNAL OF NEUROCHEMISTRYJournal of NeuroinflammationJOURNAL OF NEUROPATHOLOGY AND EXPERIMENTAL NEUROLOGY JOURNAL OF NEUROSCIENCEJOURNAL OF NEUROTRAUMAJOURNAL OF PAINJOURNAL OF PINEAL RESEARCH 松果体NEURAL COMPUTATIONNEUROBIOLOGY OF AGINGNEUROBIOLOGY OF DISEASENEUROINFORMATICSNEUROPATHOLOGY AND APPLIED NEUROBIOLOGY NEUROSCIENCE AND BIOBEHAVIORAL REVIEWS NEUROSCIENTISTNeurotherapeuticsPAINPSYCHONEUROENDOCRINOLOGYPSYCHOPHARMACOLOGYSLEEPSLEEP MEDICINE REVIEWSSocial Cognitive and Affective NeuroscienceASN NeuroBEHAVIOURAL BRAIN RESEARCHBMC NEUROSCIENCEBRAIN INJURYBRAIN AND LANGUAGECEREBELLUMCLINICAL NEUROPHYSIOLOGYCNS & Neurological Disorders-Drug TargetsCNS Neuroscience & TherapeuticsCOGNITIVE AFFECTIVE & BEHAVIORAL NEUROSCIENCECurrent Neurology and Neuroscience ReportsCURRENT NEUROVASCULAR RESEARCHDevelopmental Disabilities Research Reviews DEVELOPMENTAL NEUROSCIENCEEUROPEAN JOURNAL OF NEUROLOGYEUROPEAN JOURNAL OF NEUROSCIENCEEUROPEAN NEUROPSYCHOPHARMACOLOGYEXPERIMENTAL NEUROLOGYFrontiers in Cellular NeuroscienceFrontiers in Neural CircuitsGENES BRAIN AND BEHAVIORINTERNATIONAL REVIEW OF NEUROBIOLOGYJOURNAL OF ELECTROMYOGRAPHY AND KINESIOLOGYJOURNAL OF THE INTERNATIONAL NEUROPSYCHOLOGICAL SOCIETY JOURNAL OF NEURAL TRANSMISSIONJOURNAL OF NEUROENDOCRINOLOGYJournal of Neuroimmune PharmacologyJOURNAL OF NEUROIMMUNOLOGYJOURNAL OF NEUROPHYSIOLOGYJOURNAL OF NEUROSCIENCE RESEARCHJOURNAL OF THE PERIPHERAL NERVOUS SYSTEMJOURNAL OF PSYCHOPHARMACOLOGYJOURNAL OF SLEEP RESEARCHLEARNING & MEMORYMOLECULAR AND CELLULAR NEUROSCIENCEMolecular PainMUSCLE & NERVENETWORK-COMPUTATION IN NEURAL SYSTEMSNeural DevelopmentNEURAL NETWORKSNEUROBIOLOGY OF LEARNING AND MEMORYNEUROCHEMISTRY INTERNATIONALNeurodegenerative DiseasesNEUROENDOCRINOLOGYNEUROGASTROENTEROLOGY AND MOTILITYNEUROLOGIC CLINICSNEUROMOLECULAR MEDICINENEUROMUSCULAR DISORDERSNEUROPHARMACOLOGYNEUROPSYCHOLOGY REVIEWNEUROPSYCHOLOGIANEUROPSYCHOLOGYNEUROSCIENCENEUROSIGNALSNEUROTOXICOLOGY AND TERATOLOGYNEUROTOXICOLOGYPROGRESS IN BRAIN RESEARCHPROGRESS IN NEURO-PSYCHOPHARMACOLOGY & BIOLOGICAL PSYCHIATRY PSYCHOPHYSIOLOGYPurinergic SignallingRESTORATIVE NEUROLOGY AND NEUROSCIENCESocial NeuroscienceSTEREOTACTIC AND FUNCTIONAL NEUROSURGERYSTRESS-THE INTERNATIONAL JOURNAL ON THE BIOLOGY OF STRESS ACTA NEUROBIOLOGIAE EXPERIMENTALISACUPUNCTURE & ELECTRO-THERAPEUTICS RESEARCHAUTONOMIC NEUROSCIENCE-BASIC & CLINICALBehavioral and Brain FunctionsBEHAVIORAL NEUROSCIENCEBEHAVIOURAL PHARMACOLOGYBRAIN BEHAVIOR AND EVOLUTIONBRAIN AND COGNITIONBRAIN RESEARCHBRAIN RESEARCH BULLETINBRAIN TOPOGRAPHYCELLULAR AND MOLECULAR NEUROBIOLOGYCHEMICAL SENSESChemosensory PerceptionCLINICAL AUTONOMIC RESEARCHCLINICAL EEG AND NEUROSCIENCECognitive NeurodynamicsCognitive Systems ResearchCurrent NeuropharmacologyDevelopmental NeurobiologyEUROPEAN NEUROLOGYEXPERIMENTAL BRAIN RESEARCHFOLIA NEUROPATHOLOGICAFrontiers in Computational NeuroscienceFrontiers in Human NeuroscienceFUNCTIONAL NEUROLOGYGAIT & POSTUREHUMAN MOVEMENT SCIENCEINTERNATIONAL JOURNAL OF DEVELOPMENTAL NEUROSCIENCE INTERNATIONAL JOURNAL OF PSYCHOPHYSIOLOGY JOURNAL OF CHEMICAL NEUROANATOMYJOURNAL OF CLINICAL NEUROPHYSIOLOGYJOURNAL OF CLINICAL NEUROSCIENCEJOURNAL OF COMPUTATIONAL NEUROSCIENCEJOURNAL OF HEADACHE AND PAINJournal of Integrative NeuroscienceJOURNAL OF MOLECULAR NEUROSCIENCEJOURNAL OF MOTOR BEHAVIORJournal of Neurodevelopmental DisordersJOURNAL OF NEUROGENETICSJOURNAL OF THE NEUROLOGICAL SCIENCESJOURNAL OF NEUROLINGUISTICSJOURNAL OF NEUROPSYCHIATRY AND CLINICAL NEUROSCIENCES JOURNAL OF NEUROSCIENCE METHODSJOURNAL OF NEUROVIROLOGYJOURNAL OF PHYSIOLOGY-PARISMETABOLIC BRAIN DISEASEMOTOR CONTROLNEUROCHEMICAL RESEARCHNEUROENDOCRINOLOGY LETTERSNEUROIMAGING CLINICS OF NORTH AMERICA NEUROIMMUNOMODULATIONNEUROLOGICAL RESEARCHNEUROLOGICAL SCIENCESNeuron Glia BiologyNEUROPATHOLOGYNEUROPEPTIDESNEUROPHYSIOLOGIE CLINIQUE-CLINICAL NEUROPHYSIOLOGY NEUROPSYCHOBIOLOGYNEUROPSYCHOLOGICAL REHABILITATIONNEUROREPORTNEUROSCIENCE LETTERSNEUROSCIENCE RESEARCHNEUROTOXICITY RESEARCHNUTRITIONAL NEUROSCIENCEPHARMACOLOGY BIOCHEMISTRY AND BEHAVIORPSYCHIATRY AND CLINICAL NEUROSCIENCESPSYCHIATRIC GENETICSREVIEWS IN THE NEUROSCIENCESSEIZURE-EUROPEAN JOURNAL OF EPILEPSY SOMATOSENSORY AND MOTOR RESEARCHISSN名称小区大区0006-8950临床神经学11 Journal Of Head Trauma Rehabilitation0885-9701临床神经学131474-4422临床神经学110028-3878临床神经学11 Neurorehabilitation And Neural Repair1545-9683临床神经学121533-3159临床神经学110039-2499临床神经学110195-6108临床神经学231015-6305临床神经学221935-861X临床神经学221567-2050临床神经学22 current opinion in neurology1350-7540临床神经学22european journal of pain1090-3801临床神经学22 AL NEUROLOGY0022-3069临床神经学220897-7151临床神经学221526-5900临床神经学22 neuro-oncology 神经肿瘤 **1522-8517临床神经学220305-1846临床神经学22 Neuroscientist **1073-8584临床神经学220304-3959临床神经学221471-2377临床神经学331015-9770临床神经学330749-8047临床神经学331528-4042临床神经学331567-2026临床神经学331351-5101临床神经学33 Headache0017-8748临床神经学330340-5354临床神经学330167-594X临床神经学330022-3085临床神经学331660-2854临床神经学33 neurogenetics 神经遗传学1364-6745临床神经学33neurologic clinics0733-8619临床神经学33neuroradiology 神经放射学0028-3940临床神经学331092-0684临床神经学33 neurosurgery0148-396X临床神经学331362-4393临床神经学340362-2436临床神经学331529-9430临床神经学330001-6268临床神经学441433-7398临床神经学440268-8697临床神经学440317-1671临床神经学440044-4251临床神经学44 Clinical Neurology And Neurosurgery0303-8467临床神经学440722-5091临床神经学441531-3433临床神经学441092-8480临床神经学441751-8423临床神经学441090-3798临床神经学440014-3022临床神经学440940-6719临床神经学430883-0738临床神经学44 ROPSYCHOLOGY1380-3395临床神经学441738-6586临床神经学44 Journal Of Clinical Neuroscience0967-5868临床神经学440891-9887临床神经学441129-2369临床神经学440022-3018临床神经学441866-1947临床神经学441051-2284临床神经学440022-510X临床神经学431070-8022临床神经学44 NEUROSCIENCES0895-0172临床神经学430150-9861临床神经学441547-5654临床神经学441079-0268临床神经学441536-0652临床神经学44 Neurocase1355-4794临床神经学441541-6933临床神经学440161-6412临床神经学441590-1874临床神经学441074-7931临床神经学441094-7159临床神经学440919-6544临床神经学44 PHYSIOLOGY0987-7053临床神经学441042-3680临床神经学44 Neurosurgical Review0344-5607临床神经学441203-6765临床神经学44 Pediatric Neurology0887-8994临床神经学441323-1316临床神经学440210-0010临床神经学440932-433X临床神经学441059-1311临床神经学440271-8235临床神经学431071-9091临床神经学440090-3019临床神经学441065-9471神经成像121053-8119神经成像120195-6108神经成像230028-3940神经成像330925-4927神经成像331011-6125神经成像341550-0594神经成像441051-2284神经成像441759-8478神经成像440150-9861神经成像441052-5149神经成像441863-2653神经科学120091-3022神经科学111065-9471神经科学120021-9967神经科学12 ARCH IN OTOLARYNGOLOGY1525-3961神经科学131359-4184神经科学111097-6256神经科学111471-0048神经科学111053-8119神经科学120896-6273神经科学110166-2236神经科学111420-3030神经科学231398-5647神经科学220889-1591神经科学221015-6305神经科学220165-0173神经科学211935-861X神经科学220333-1024神经科学221567-2050神经科学220959-4388神经科学211350-7540神经科学221090-3801神经科学220378-5955神经科学231050-9631神经科学22 MACOLOGY1461-1457神经科学221387-2877神经科学220271-678X神经科学220898-929X神经科学22 LOGY SENSORY NEURAL AND BEHAVIORAL PHYSIOLO0340-7594神经科学241741-2560神经科学220022-3042神经科学221742-2094神经科学22 AL NEUROLOGY0022-3069神经科学22 Journal Of Neuroscience0270-6474神经科学210897-7151神经科学221526-5900神经科学220742-3098神经科学220899-7667神经科学220197-4580神经科学220969-9961神经科学22 neuroinformatics1539-2791神经科学230305-1846神经科学220149-7634神经科学21 Neuroscientist1073-8584神经科学220033-3158神经科学220161-8105神经科学221087-0792神经科学221749-5016神经科学221759-0914神经科学320166-4328神经科学331471-2202神经科学330269-9052神经科学340093-934X神经科学331473-4222神经科学32 Clinical Neurophysiology1388-2457神经科学331871-5273神经科学321755-5930神经科学331530-7026神经科学321528-4042神经科学331567-2026神经科学331940-5510神经科学330378-5866神经科学331351-5101神经科学330953-816X神经科学320924-977X神经科学320014-4886神经科学321662-5102神经科学321662-5110神经科学321601-1848神经科学320074-7742神经科学331050-6411神经科学34 LOGICAL SOCIETY1355-6177神经科学330300-9564神经科学330953-8194神经科学321557-1890神经科学330165-5728神经科学330022-3077神经科学330360-4012神经科学331072-0502神经科学321044-7431神经科学321744-8069神经科学320148-639X神经科学330954-898X神经科学331749-8104神经科学330893-6080神经科学321074-7427神经科学320197-0186神经科学321660-2854神经科学330028-3835神经科学331350-1925神经科学32 Neurologic Clinics0733-8619神经科学331535-1084神经科学330960-8966神经科学330028-3908神经科学321040-7308神经科学320028-3932神经科学320894-4105神经科学330306-4522神经科学331424-862X神经科学320892-0362神经科学330161-813X神经科学330079-6123神经科学33 IOLOGICAL PSYCHIATRY0278-5846神经科学330048-5772神经科学321573-9538神经科学330922-6028神经科学331747-0919神经科学321011-6125神经科学34 BIOLOGY OF STRESS1025-3890神经科学330065-1400神经科学440360-1293神经科学441566-0702神经科学440006-8977神经科学430278-2626神经科学430006-8993神经科学430361-9230神经科学430896-0267神经科学430272-4340神经科学430379-864X神经科学431936-5802神经科学430959-9851神经科学441550-0594神经科学441871-4080神经科学441389-0417神经科学441570-159X神经科学441932-8451神经科学430014-3022神经科学440014-4819神经科学431641-4640神经科学441662-5188神经科学431662-5161神经科学440393-5264神经科学440966-6362神经科学430167-9457神经科学44 EUROSCIENCE0736-5748神经科学440167-8760神经科学430891-0618神经科学430736-0258神经科学440967-5868神经科学440929-5313神经科学431129-2369神经科学440219-6352神经科学440895-8696神经科学430022-2895神经科学441866-1947神经科学440167-7063神经科学440911-6044神经科学44 NEUROSCIENCES0895-0172神经科学430165-0270神经科学431355-0284神经科学430928-4257神经科学430885-7490神经科学441087-1640神经科学440364-3190神经科学430172-780X神经科学441052-5149神经科学441021-7401神经科学440161-6412神经科学441590-1874神经科学441740-925X神经科学440919-6544神经科学440143-4179神经科学43 PHYSIOLOGY0987-7053神经科学440302-282X神经科学430960-2011神经科学440959-4965神经科学440304-3940神经科学440168-0102神经科学431029-8428神经科学431028-415X神经科学440091-3057神经科学431323-1316神经科学440955-8829神经科学430334-1763神经科学431059-1311神经科学440899-0220神经科学4410年因子-2010年平均影响备注9.2329.4422.779 2.58521.65918.0188.0177.7443.7724.2637.7937.7935.7566.4323.464 3.1684.7415.4074.964 3.9824.953 4.6855.021 5.3343.819 3.7444.190 4.6313.426 3.7364.851 4.0055.483 5.1563.625 3.5934.5775.5175.355 5.5852.797 2.5632.9873.1883.114 3.0032.697 2.4673.047 3.2833.765 3.0022.642 2.8363.853 3.0972.929 2.6682.739 2.4863.791 3.4253.488 3.3252.533 2.5342.870 2.5052.348 2.3483.298 3.1862.510 2.6423.024 2.9631.329 1.4781.1290.8780.9660.9141.179 1.1530.7160.6362009更名,原名为ZENTRALBLATT FUR NEUROCHIRURGIE1.636 1.4211.067 1.1011.450 1.4501.810 1.8101.384 1.3841.994 1.8071.760 1.7171.9942.1151.668 1.5641.805 1.9581.0970.7711.165 1.1752.131 1.9472.015 2.0091.796 1.7921.269 1.2691.287 1.6062.167 2.2831.059 1.1351.9812.2261.203 1.2621.594 1.5371.442 1.3881.333 1.3011.0690.9862.353 2.0611.621 1.5111.220 1.2581.8812.0581.605 1.731 1.693 1.3411.477 1.4562.259 1.961 1.515 1.515 1.513 1.505 1.559 1.426 1.218 1.178 1.170 1.2031.6492.0202.000 2.209 1.862 1.862 1.260 1.399 5.107 5.586 5.937 5.7903.464 3.168 2.870 2.505 2.064 2.712 1.882 1.727 1.325 1.199 1.287 1.606 1.069 1.069 1.203 1.262 1.420 1.4974.982 3.938 12.75011.1635.107 5.586 3.774 3.745 3.038 2.634 15.47014.352 14.19114.233 29.51027.311 5.937 5.790 14.02713.8199.9669.412 9.68610.777 13.32012.977 2.228 2.162 5.221 4.8943.9564.6424.7415.407 8.8427.489 4.964 3.982 4.265 3.805 4.953 4.6856.8917.401 5.021 5.334 3.819 3.744 2.428 2.313 4.609 4.584 4.699 4.650 4.261 4.3984.5225.2405.357 5.202 2.000 1.955 2.628 3.0354.337 4.2795.785 4.555 4.190 4.631 7.2717.3003.426 3.7364.851 4.0055.855 5.373 2.290 2.2816.634 6.177 5.121 4.830 3.027 2.990 3.625 3.593 9.0158.203 4.577 5.5175.355 5.585 5.168 4.383 3.817 3.8655.486 5.1216.338 6.149 4.482 4.343 3.833 3.833 3.393 3.261 3.091 2.895 1.750 1.466 3.162 3.021 3.288 3.4722.786 2.9603.618 3.962 3.492 3.091 3.512 3.5652.697 2.4673.047 3.283 3.436 2.7182.707 2.7303.765 3.0023.658 3.4874.201 3.849 4.436 4.108 3.588 3.5883.960 3.9604.061 3.915 2.183 2.892 2.372 2.084 2.910 2.767 2.597 2.457 4.650 3.867 3.528 3.1012.901 2.9673.114 3.4152.9583.0103.032 3.1523.801 3.8373.361 3.3724.607 4.2713.861 3.7884.148 4.013 2.302 2.394 0.957 1.275 3.395 3.411 1.972 2.169 3.701 3.646 3.601 3.457 3.791 3.425 3.272 3.086 3.349 3.466 2.533 2.534 4.657 3.373 2.764 2.891 4.677 3.990 4.231 4.270 3.949 4.123 3.176 3.109 3.215 3.354 3.967 3.997 2.622 2.8792.921 2.7493.134 2.9592.877 2.7793.263 3.5022.975 2.9233.349 3.014 2.823 3.4491.882 1.7272.553 2.903 1.533 1.320 1.0000.6471.671 1.8722.305 2.3212.481 2.650 2.530 2.6511.9682.4722.838 2.609 2.623 2.5272.498 2.3213.288 2.182 2.423 2.360 2.327 2.800 1.091 1.046 1.333 1.376 1.325 1.199 1.625 1.9441.000 1.0512.783 1.888 2.855 2.6401.760 1.7172.296 2.2491.222 1.1532.586 2.586 1.940 1.7441.481 1.2112.313 2.544 1.967 1.9471.938 1.9442.378 2.562 2.121 2.151 1.154 1.4551.165 1.1752.325 2.432 2.015 2.0091.216 1.2552.922 2.568 1.650 1.428 1.269 1.2691.950 1.0152.167 2.2831.742 1.6161.9812.2262.100 2.1622.243 2.1463.030 2.401 2.343 2.0421.204 1.4112.608 2.530 1.621 1.3421.420 1.4972.642 1.963 1.621 1.5111.220 1.2582.611 1.829 1.605 1.731 1.917 2.1301.693 1.3412.567 2.155 1.731 1.7591.822 1.8442.055 2.0602.096 2.2383.015 2.7611.301 1.1792.624 2.7811.559 1.4262.061 2.361 2.047 2.612 1.649 2.020 1.115 1.011。

刊名简称刊名全称CHEM REV=L CHEMICAL REVIEWS 化学评论 美国ACCOUNTS CHEM RES ac ACCOUNTS OF CHEMICAL RESEARCH 化学研究述评 美国PROG POLYM SCI pr PROGRESS IN POLYMER SCIENCECHEM SOC REV ch CHEMICAL SOCIETY REVIEWS 化学会评论 英国ALDRICHIM ACTA al ALDRICHIMICA ACTAANNU REV PHYS CHEM an ANNUAL REVIEW OF PHYSICAL CHEMISTRYSURF SCI REP su SURFACE SCIENCE REPORTSSURF SCI REP su SURFACE SCIENCE REPORTSANGEW CHEM INT EDIT an ANGEWANDTE CHEMIE-INTERNATIONAL EDITION 德国应用化学COORDIN CHEM REV co COORDINATION CHEMISTRY REVIEWSNAT PROD REP na NATURAL PRODUCT REPORTSNAT PROD REP na NATURAL PRODUCT REPORTSNAT PROD REP na NATURAL PRODUCT REPORTSADV CATAL ad ADVANCES IN CATALYSISJ AM CHEM SOC jo JOURNAL OF THE AMERICAN CHEMICAL SOCIETY 美国化学会志CATAL REV ca CATALYSIS REVIEWS-SCIENCE AND ENGINEERINGINT REV PHYS CHEM in INTERNATIONAL REVIEWS IN PHYSICAL CHEMISTRYJ PHOTOCH PHOTOBIO C jo JOURNAL OF PHOTOCHEMISTRY AND PHOTOBIOLOGY C-PHOTOCHEM ADV POLYM SCI ad ADVANCES IN POLYMER SCIENCEANAL CHEM an ANALYTICAL CHEMISTRYTOP CURR CHEM to TOPICS IN CURRENT CHEMISTRYTRAC-TREND ANAL CHEM tr TRAC-TRENDS IN ANALYTICAL CHEMISTRYCHEM-EUR J ch CHEMISTRY-A EUROPEAN JOURNAL 化学 德国ADV SYNTH CATAL ad ADVANCED SYNTHESIS & CATALYSISADV SYNTH CATAL ad ADVANCED SYNTHESIS & CATALYSISCHEM COMMUN ch CHEMICAL COMMUNICATIONS 化学通讯 英国ADV ORGANOMET CHEM ad ADVANCES IN ORGANOMETALLIC CHEMISTRYADV ORGANOMET CHEM ad ADVANCES IN ORGANOMETALLIC CHEMISTRY 有机金属化学进展 ORG LETT or ORGANIC LETTERSCURR OPIN COLLOID IN cu CURRENT OPINION IN COLLOID & INTERFACE SCIENCE FARADAY DISCUSS fa FARADAY DISCUSSIONSGREEN CHEM gr GREEN CHEMISTRYSTRUCT BOND st STRUCTURE AND BONDINGSTRUCT BOND st STRUCTURE AND BONDINGADV INORG CHEM ad ADVANCES IN INORGANIC CHEMISTRYCRYST GROWTH DES cr CRYSTAL GROWTH & DESIGNCRYST GROWTH DES cr CRYSTAL GROWTH & DESIGNCRYST GROWTH DES cr CRYSTAL GROWTH & DESIGNCHEM-ASIAN J ch Chemistry-An Asian JournalJ COMPUT CHEM jo JOURNAL OF COMPUTATIONAL CHEMISTRYJ PHYS CHEM B jo JOURNAL OF PHYSICAL CHEMISTRY BJ CHEM THEORY COMPUT jo Journal of Chemical Theory and ComputationADV COLLOID INTERFAC ad ADVANCES IN COLLOID AND INTERFACE SCIENCEINORG CHEM in INORGANIC CHEMISTRYLANGMUIR la LANGMUIR 兰格缪尔 美国BIOMACROMOLECULES bi BIOMACROMOLECULESBIOMACROMOLECULES bi BIOMACROMOLECULESBIOMACROMOLECULES bi BIOMACROMOLECULESJ ORG CHEM jo JOURNAL OF ORGANIC CHEMISTRY 有机化学杂志 美国ORGANOMETALLICS or ORGANOMETALLICSORGANOMETALLICS or ORGANOMETALLICSJ CHROMATOGR A jo JOURNAL OF CHROMATOGRAPHY AJ CHROMATOGR A jo JOURNAL OF CHROMATOGRAPHY AJ ANAL ATOM SPECTROM jo JOURNAL OF ANALYTICAL ATOMIC SPECTROMETRYJ ANAL ATOM SPECTROM jo JOURNAL OF ANALYTICAL ATOMIC SPECTROMETRYJ POLYM SCI POL CHEM jo JOURNAL OF POLYMER SCIENCE PART A-POLYMER CHEMISTRY CRYSTENGCOMM cr CRYSTENGCOMMCRYSTENGCOMM cr CRYSTENGCOMMCHEMPHYSCHEM ch CHEMPHYSCHEMCHEMPHYSCHEM ch CHEMPHYSCHEMANALYST an ANALYSTCURR ORG CHEM cu CURRENT ORGANIC CHEMISTRYPHYS CHEM CHEM PHYS ph PHYSICAL CHEMISTRY CHEMICAL PHYSICSPHYS CHEM CHEM PHYS ph PHYSICAL CHEMISTRY CHEMICAL PHYSICSJ BIOL INORG CHEM jo JOURNAL OF BIOLOGICAL INORGANIC CHEMISTRYJ BIOL INORG CHEM jo JOURNAL OF BIOLOGICAL INORGANIC CHEMISTRYJ PHYS CHEM C jo Journal of Physical Chemistry CJ PHYS CHEM C jo Journal of Physical Chemistry CJ PHYS CHEM C jo Journal of Physical Chemistry CJ AM SOC MASS SPECTR jo JOURNAL OF THE AMERICAN SOCIETY FOR MASS SPECTROMETRY J AM SOC MASS SPECTR jo JOURNAL OF THE AMERICAN SOCIETY FOR MASS SPECTROMETRY J AM SOC MASS SPECTR jo JOURNAL OF THE AMERICAN SOCIETY FOR MASS SPECTROMETRY J CHEM INF MODEL jo Journal of Chemical Information and ModelingJ CHEM INF MODEL jo Journal of Chemical Information and ModelingJ CHEM INF MODEL jo Journal of Chemical Information and Modeling DALTON T da DALTON TRANSACTIONSCHEM REC ch CHEMICAL RECORDORG BIOMOL CHEM or ORGANIC & BIOMOLECULAR CHEMISTRYTALANTA ta TALANTAJ COMB CHEM jo JOURNAL OF COMBINATORIAL CHEMISTRYJ COMB CHEM jo JOURNAL OF COMBINATORIAL CHEMISTRYJ COMB CHEM jo JOURNAL OF COMBINATORIAL CHEMISTRYANAL CHIM ACTA an ANALYTICA CHIMICA ACTAPOLYMER po POLYMERAPPL CATAL A-GEN ap APPLIED CATALYSIS A-GENERALAPPL CATAL A-GEN ap APPLIED CATALYSIS A-GENERALJ MASS SPECTROM jo JOURNAL OF MASS SPECTROMETRYJ MASS SPECTROM jo JOURNAL OF MASS SPECTROMETRYJ MASS SPECTROM jo JOURNAL OF MASS SPECTROMETRYJ PHYS CHEM REF DATA jo JOURNAL OF PHYSICAL AND CHEMICAL REFERENCE DATAJ PHYS CHEM REF DATA jo JOURNAL OF PHYSICAL AND CHEMICAL REFERENCE DATAJ PHYS CHEM REF DATA jo JOURNAL OF PHYSICAL AND CHEMICAL REFERENCE DATAJ PHYS CHEM A jo JOURNAL OF PHYSICAL CHEMISTRY AJ PHYS CHEM A jo JOURNAL OF PHYSICAL CHEMISTRY AANAL BIOANAL CHEM an ANALYTICAL AND BIOANALYTICAL CHEMISTRYANAL BIOANAL CHEM an ANALYTICAL AND BIOANALYTICAL CHEMISTRYMAR CHEM ma MARINE CHEMISTRYMAR CHEM ma MARINE CHEMISTRYEUR J ORG CHEM eu EUROPEAN JOURNAL OF ORGANIC CHEMISTRYTETRAHEDRON te TETRAHEDRONCURR ORG SYNTH cu CURRENT ORGANIC SYNTHESISRAPID COMMUN MASS SP ra RAPID COMMUNICATIONS IN MASS SPECTROMETRYRAPID COMMUN MASS SP ra RAPID COMMUNICATIONS IN MASS SPECTROMETRY ELECTROANAL el ELECTROANALYSISELECTROANAL el ELECTROANALYSISSYNLETT sy SYNLETTNEW J CHEM ne NEW JOURNAL OF CHEMISTRYCRIT REV ANAL CHEM cr CRITICAL REVIEWS IN ANALYTICAL CHEMISTRYCOMMENT INORG CHEM co COMMENTS ON INORGANIC CHEMISTRYMATCH-COMMUN MATH CO ma MATCH-COMMUNICATIONS IN MATHEMATICAL AND IN COMPUTER C MATCH-COMMUN MATH CO ma MATCH-COMMUNICATIONS IN MATHEMATICAL AND IN COMPUTER C MATCH-COMMUN MATH CO ma MATCH-COMMUNICATIONS IN MATHEMATICAL AND IN COMPUTER C J MOL CATAL A-CHEM jo JOURNAL OF MOLECULAR CATALYSIS A-CHEMICALEUR J INORG CHEM eu EUROPEAN JOURNAL OF INORGANIC CHEMISTRYCATAL TODAY ca CATALYSIS TODAYCATAL TODAY ca CATALYSIS TODAYCATAL TODAY ca CATALYSIS TODAYJ SEP SCI jo JOURNAL OF SEPARATION SCIENCETETRAHEDRON-ASYMMETR te TETRAHEDRON-ASYMMETRYTETRAHEDRON-ASYMMETR te TETRAHEDRON-ASYMMETRYTETRAHEDRON-ASYMMETR te TETRAHEDRON-ASYMMETRY 四面体 英国TETRAHEDRON LETT te TETRAHEDRON LETTERS 四面体通讯 英国MICROPOR MESOPOR MAT mi MICROPOROUS AND MESOPOROUS MATERIALSMICROPOR MESOPOR MAT mi MICROPOROUS AND MESOPOROUS MATERIALSMICROPOR MESOPOR MAT mi MICROPOROUS AND MESOPOROUS MATERIALSMICROPOR MESOPOR MAT mi MICROPOROUS AND MESOPOROUS MATERIALSADV PHYS ORG CHEM ad ADVANCES IN PHYSICAL ORGANIC CHEMISTRYADV PHYS ORG CHEM ad ADVANCES IN PHYSICAL ORGANIC CHEMISTRYJ ELECTROANAL CHEM jo JOURNAL OF ELECTROANALYTICAL CHEMISTRYEUR J MED CHEM eu EUROPEAN JOURNAL OF MEDICINAL CHEMISTRYTHEOR CHEM ACC th THEORETICAL CHEMISTRY ACCOUNTSPROG SOLID STATE CH pr PROGRESS IN SOLID STATE CHEMISTRYULTRASON SONOCHEM ul ULTRASONICS SONOCHEMISTRYULTRASON SONOCHEM ul ULTRASONICS SONOCHEMISTRYCATAL COMMUN ca CATALYSIS COMMUNICATIONSSYNTHESIS-STUTTGART sy SYNTHESIS-STUTTGARTJ COLLOID INTERF SCI jo JOURNAL OF COLLOID AND INTERFACE SCIENCETOP CATAL to TOPICS IN CATALYSISTOP CATAL to TOPICS IN CATALYSISCHEM PHYS LETT ch CHEMICAL PHYSICS LETTERSCHEM PHYS LETT ch CHEMICAL PHYSICS LETTERSADV CHROMATOGR ad ADVANCES IN CHROMATOGRAPHYAUST J CHEM au AUSTRALIAN JOURNAL OF CHEMISTRYCOMB CHEM HIGH T SCR co COMBINATORIAL CHEMISTRY & HIGH THROUGHPUT SCREENING COMB CHEM HIGH T SCR co COMBINATORIAL CHEMISTRY & HIGH THROUGHPUT SCREENING COMB CHEM HIGH T SCR co COMBINATORIAL CHEMISTRY & HIGH THROUGHPUT SCREENINGJ FLUORESC jo JOURNAL OF FLUORESCENCEJ FLUORESC jo JOURNAL OF FLUORESCENCEPOLYM DEGRAD STABIL po POLYMER DEGRADATION AND STABILITYEUR POLYM J eu EUROPEAN POLYMER JOURNALPURE APPL CHEM pu PURE AND APPLIED CHEMISTRY 理论化学与应用化学 美国J PHOTOCH PHOTOBIO A jo JOURNAL OF PHOTOCHEMISTRY AND PHOTOBIOLOGY A-CHEMISTRY J ORGANOMET CHEM jo JOURNAL OF ORGANOMETALLIC CHEMISTRYJ ORGANOMET CHEM jo JOURNAL OF ORGANOMETALLIC CHEMISTRYMACROMOL CHEM PHYS ma MACROMOLECULAR CHEMISTRY AND PHYSICSCARBOHYD POLYM ca CARBOHYDRATE POLYMERSCARBOHYD POLYM ca CARBOHYDRATE POLYMERSCARBOHYD POLYM ca CARBOHYDRATE POLYMERSJ SOLID STATE CHEM jo JOURNAL OF SOLID STATE CHEMISTRYJ SOLID STATE CHEM jo JOURNAL OF SOLID STATE CHEMISTRYADV HETEROCYCL CHEM ad ADVANCES IN HETEROCYCLIC CHEMISTRYMICROCHEM J mi MICROCHEMICAL JOURNALCHEM PHYS ch CHEMICAL PHYSICSCHEM PHYS ch CHEMICAL PHYSICSORG PROCESS RES DEV or ORGANIC PROCESS RESEARCH & DEVELOPMENTORG PROCESS RES DEV or ORGANIC PROCESS RESEARCH & DEVELOPMENTCATAL LETT ca CATALYSIS LETTERSINORG CHEM COMMUN in INORGANIC CHEMISTRY COMMUNICATIONSVIB SPECTROSC vi VIBRATIONAL SPECTROSCOPYVIB SPECTROSC vi VIBRATIONAL SPECTROSCOPYVIB SPECTROSC vi VIBRATIONAL SPECTROSCOPYSURF SCI su SURFACE SCIENCESURF SCI su SURFACE SCIENCEJ ANAL APPL PYROL jo JOURNAL OF ANALYTICAL AND APPLIED PYROLYSISJ ANAL APPL PYROL jo JOURNAL OF ANALYTICAL AND APPLIED PYROLYSISUSP KHIM+us USPEKHI KHIMIIPOLYHEDRON po POLYHEDRONPOLYHEDRON po POLYHEDRONCARBOHYD RES ca CARBOHYDRATE RESEARCHCARBOHYD RES ca CARBOHYDRATE RESEARCHCARBOHYD RES ca CARBOHYDRATE RESEARCHSUPRAMOL CHEM su SUPRAMOLECULAR CHEMISTRYINORG CHIM ACTA in INORGANICA CHIMICA ACTAMINI-REV ORG CHEM mi MINI-REVIEWS IN ORGANIC CHEMISTRYSOLID STATE SCI so SOLID STATE SCIENCESSOLID STATE SCI so SOLID STATE SCIENCESSOLID STATE SCI so SOLID STATE SCIENCESCOLLOID SURFACE A co COLLOIDS AND SURFACES A-PHYSICOCHEMICAL AND ENGINEERIN MICROCHIM ACTA mi MICROCHIMICA ACTAADV QUANTUM CHEM ad ADVANCES IN QUANTUM CHEMISTRYJ MOL MODEL jo JOURNAL OF MOLECULAR MODELINGJ MOL MODEL jo JOURNAL OF MOLECULAR MODELINGJ MOL MODEL jo JOURNAL OF MOLECULAR MODELINGJ MOL MODEL jo JOURNAL OF MOLECULAR MODELINGPOLYM INT po POLYMER INTERNATIONALCURR ANAL CHEM cu Current Analytical ChemistryANAL SCI an ANALYTICAL SCIENCESCHEM LETT ch CHEMISTRY LETTERSSEP PURIF REV se SEPARATION AND PURIFICATION REVIEWSSEP PURIF REV se SEPARATION AND PURIFICATION REVIEWSSEP PURIF REV se SEPARATION AND PURIFICATION REVIEWSTHERMOCHIM ACTA th THERMOCHIMICA ACTATHERMOCHIM ACTA th THERMOCHIMICA ACTAJ FLUORINE CHEM jo JOURNAL OF FLUORINE CHEMISTRYJ FLUORINE CHEM jo JOURNAL OF FLUORINE CHEMISTRYCOLLOID POLYM SCI co COLLOID AND POLYMER SCIENCECOLLOID POLYM SCI co COLLOID AND POLYMER SCIENCEJ PHYS ORG CHEM jo JOURNAL OF PHYSICAL ORGANIC CHEMISTRYJ PHYS ORG CHEM jo JOURNAL OF PHYSICAL ORGANIC CHEMISTRYB CHEM SOC JPN bu BULLETIN OF THE CHEMICAL SOCIETY OF JAPAN 日本化学会通J MOL STRUCT jo JOURNAL OF MOLECULAR STRUCTUREJ THERM ANAL CALORIM jo JOURNAL OF THERMAL ANALYSIS AND CALORIMETRYJ THERM ANAL CALORIM jo JOURNAL OF THERMAL ANALYSIS AND CALORIMETRYMAGN RESON CHEM ma MAGNETIC RESONANCE IN CHEMISTRYMAGN RESON CHEM ma MAGNETIC RESONANCE IN CHEMISTRYMAGN RESON CHEM ma MAGNETIC RESONANCE IN CHEMISTRYHELV CHIM ACTA he HELVETICA CHIMICA ACTACALPHAD ca CALPHAD-COMPUTER COUPLING OF PHASE DIAGRAMS AND THERMO CALPHAD ca CALPHAD-COMPUTER COUPLING OF PHASE DIAGRAMS AND THERMO J INORG ORGANOMET P jo JOURNAL OF INORGANIC AND ORGANOMETALLIC POLYMERSJ IRAN CHEM SOC jo Journal of the Iranian Chemical SocietyJ CHEMOMETR jo JOURNAL OF CHEMOMETRICSJ CHEMOMETR jo JOURNAL OF CHEMOMETRICSJ CHEMOMETR jo JOURNAL OF CHEMOMETRICSJ CHEMOMETR jo JOURNAL OF CHEMOMETRICSJ CHEMOMETR jo JOURNAL OF CHEMOMETRICSJ CHEMOMETR jo JOURNAL OF CHEMOMETRICSPOLYM J po POLYMER JOURNALCR CHIM co COMPTES RENDUS CHIMIEJ BRAZIL CHEM SOC jo JOURNAL OF THE BRAZILIAN CHEMICAL SOCIETYINT J QUANTUM CHEM in INTERNATIONAL JOURNAL OF QUANTUM CHEMISTRYINT J QUANTUM CHEM in INTERNATIONAL JOURNAL OF QUANTUM CHEMISTRYINT J QUANTUM CHEM in INTERNATIONAL JOURNAL OF QUANTUM CHEMISTRYSTRUCT CHEM st STRUCTURAL CHEMISTRYSTRUCT CHEM st STRUCTURAL CHEMISTRYSTRUCT CHEM st STRUCTURAL CHEMISTRYSURF INTERFACE ANAL su SURFACE AND INTERFACE ANALYSISAPPL ORGANOMET CHEM ap APPLIED ORGANOMETALLIC CHEMISTRYAPPL ORGANOMET CHEM ap APPLIED ORGANOMETALLIC CHEMISTRYSOLVENT EXTR ION EXC so SOLVENT EXTRACTION AND ION EXCHANGEANAL LETT an ANALYTICAL LETTERSCHROMATOGRAPHIA ch CHROMATOGRAPHIACHROMATOGRAPHIA ch CHROMATOGRAPHIAZ ANORG ALLG CHEM ze ZEITSCHRIFT FUR ANORGANISCHE UND ALLGEMEINE CHEMIE CAN J CHEM ca CANADIAN JOURNAL OF CHEMISTRY-REVUE CANADIENNE DE CHIM CATAL SURV ASIA ca CATALYSIS SURVEYS FROM ASIAMOL SIMULAT mo MOLECULAR SIMULATIONMOL SIMULAT mo MOLECULAR SIMULATIONJ INCL PHENOM MACRO jo JOURNAL OF INCLUSION PHENOMENA AND MACROCYCLIC CHEMIST J INCL PHENOM MACRO jo JOURNAL OF INCLUSION PHENOMENA AND MACROCYCLIC CHEMIST J APPL POLYM SCI jo JOURNAL OF APPLIED POLYMER SCIENCEINT J CHEM KINET in INTERNATIONAL JOURNAL OF CHEMICAL KINETICSJ MATH CHEM jo JOURNAL OF MATHEMATICAL CHEMISTRYJ MATH CHEM jo JOURNAL OF MATHEMATICAL CHEMISTRYARKIVOC ar ARKIVOCJ SOLUTION CHEM jo JOURNAL OF SOLUTION CHEMISTRYB KOR CHEM SOC bu BULLETIN OF THE KOREAN CHEMICAL SOCIETYRADIOCHIM ACTA ra RADIOCHIMICA ACTARADIOCHIM ACTA ra RADIOCHIMICA ACTAJ PORPHYR PHTHALOCYA jo JOURNAL OF PORPHYRINS AND PHTHALOCYANINESMONATSH CHEM mo MONATSHEFTE FUR CHEMIEJ MOL STRUC-THEOCHEM jo JOURNAL OF MOLECULAR STRUCTURE-THEOCHEMJ MOL LIQ jo JOURNAL OF MOLECULAR LIQUIDSJ MOL LIQ jo JOURNAL OF MOLECULAR LIQUIDSJ COAT TECHNOL RES jo Journal of Coatings Technology and ResearchJ PHOTOPOLYM SCI TEC jo JOURNAL OF PHOTOPOLYMER SCIENCE AND TECHNOLOGY HETEROCYCLES he HETEROCYCLESPOLYM BULL po POLYMER BULLETINMOLECULES mo MOLECULESBIOINORG CHEM APPL bi BIOINORGANIC CHEMISTRY AND APPLICATIONSBIOINORG CHEM APPL bi BIOINORGANIC CHEMISTRY AND APPLICATIONSBIOINORG CHEM APPL bi BIOINORGANIC CHEMISTRY AND APPLICATIONS HETEROATOM CHEM he HETEROATOM CHEMISTRYSYNTHETIC COMMUN sy SYNTHETIC COMMUNICATIONSLETT ORG CHEM le LETTERS IN ORGANIC CHEMISTRYJ CHEM SCI jo JOURNAL OF CHEMICAL SCIENCESJ CHROMATOGR SCI jo JOURNAL OF CHROMATOGRAPHIC SCIENCEJ CHROMATOGR SCI jo JOURNAL OF CHROMATOGRAPHIC SCIENCEJ CLUST SCI jo JOURNAL OF CLUSTER SCIENCEJ LIQ CHROMATOGR R T jo JOURNAL OF LIQUID CHROMATOGRAPHY & RELATED TECHNOLOGIE J LIQ CHROMATOGR R T jo JOURNAL OF LIQUID CHROMATOGRAPHY & RELATED TECHNOLOGIECHIMIA ch CHIMIAJPC-J PLANAR CHROMAT jp JPC-JOURNAL OF PLANAR CHROMATOGRAPHY-MODERN TLC TRANSIT METAL CHEM tr TRANSITION METAL CHEMISTRYACTA CHIM SLOV ac ACTA CHIMICA SLOVENICARADIAT PHYS CHEM ra RADIATION PHYSICS AND CHEMISTRYRADIAT PHYS CHEM ra RADIATION PHYSICS AND CHEMISTRYRADIAT PHYS CHEM ra RADIATION PHYSICS AND CHEMISTRYJ CARBOHYD CHEM jo JOURNAL OF CARBOHYDRATE CHEMISTRYJ CARBOHYD CHEM jo JOURNAL OF CARBOHYDRATE CHEMISTRYJ COORD CHEM jo JOURNAL OF COORDINATION CHEMISTRYJ THEOR COMPUT CHEM jo JOURNAL OF THEORETICAL & COMPUTATIONAL CHEMISTRY COLLECT CZECH CHEM C co COLLECTION OF CZECHOSLOVAK CHEMICAL COMMUNICATIONS QUIM NOVA qu QUIMICA NOVAE-POLYMERS e-E-POLYMERSJ POLYM RES jo JOURNAL OF POLYMER RESEARCHJ HETEROCYCLIC CHEM jo JOURNAL OF HETEROCYCLIC CHEMISTRYJ DISPER SCI TECHNOL jo JOURNAL OF DISPERSION SCIENCE AND TECHNOLOGYACTA CHROMATOGR ac ACTA CHROMATOGRAPHICAZ NATURFORSCH B ze ZEITSCHRIFT FUR NATURFORSCHUNG SECTION B-A JOURNAL OF Z NATURFORSCH B ze ZEITSCHRIFT FUR NATURFORSCHUNG SECTION B-A JOURNAL OF INT J MOL SCI in INTERNATIONAL JOURNAL OF MOLECULAR SCIENCESCHINESE J CHEM ch CHINESE JOURNAL OF CHEMISTRYACTA CHIM SINICA ac ACTA CHIMICA SINICAJ MACROMOL SCI A jo JOURNAL OF MACROMOLECULAR SCIENCE-PURE AND APPLIED CHE NAT PROD RES na NATURAL PRODUCT RESEARCHNAT PROD RES na NATURAL PRODUCT RESEARCHHIGH PERFORM POLYM hi HIGH PERFORMANCE POLYMERSPHYS CHEM LIQ ph PHYSICS AND CHEMISTRY OF LIQUIDSPHYS CHEM LIQ ph PHYSICS AND CHEMISTRY OF LIQUIDSORG PREP PROCED INT or ORGANIC PREPARATIONS AND PROCEDURES INTERNATIONAL CROAT CHEM ACTA cr CROATICA CHEMICA ACTACHINESE J ORG CHEM ch CHINESE JOURNAL OF ORGANIC CHEMISTRYPOLYCYCL AROMAT COMP po POLYCYCLIC AROMATIC COMPOUNDSDES MONOMERS POLYM de DESIGNED MONOMERS AND POLYMERSCHINESE J STRUC CHEM ch CHINESE JOURNAL OF STRUCTURAL CHEMISTRYCHINESE J STRUC CHEM ch CHINESE JOURNAL OF STRUCTURAL CHEMISTRYJ ADV OXID TECHNOL jo JOURNAL OF ADVANCED OXIDATION TECHNOLOGIESCENT EUR J CHEM ce CENTRAL EUROPEAN JOURNAL OF CHEMISTRYMENDELEEV COMMUN me MENDELEEV COMMUNICATIONSJ SYN ORG CHEM JPN jo JOURNAL OF SYNTHETIC ORGANIC CHEMISTRY JAPANIRAN POLYM J ir IRANIAN POLYMER JOURNALTURK J CHEM tu TURKISH JOURNAL OF CHEMISTRYTURK J CHEM tu TURKISH JOURNAL OF CHEMISTRYISR J CHEM is ISRAEL JOURNAL OF CHEMISTRYCHEM J CHINESE U ch CHEMICAL JOURNAL OF CHINESE UNIVERSITIES-CHINESEJ CHIN CHEM SOC-TAIP jo JOURNAL OF THE CHINESE CHEMICAL SOCIETYBEILSTEIN J ORG CHEM be Beilstein Journal of Organic ChemistryCHINESE J POLYM SCI ch CHINESE JOURNAL OF POLYMER SCIENCECOLLOID J+co COLLOID JOURNALINDIAN J CHEM A in INDIAN JOURNAL OF CHEMISTRY SECTION A-INORGANIC BIO-IN PHOSPHORUS SULFUR ph PHOSPHORUS SULFUR AND SILICON AND THE RELATED ELEMENTS KINET CATAL+ki KINETICS AND CATALYSISREV ANAL CHEM re REVIEWS IN ANALYTICAL CHEMISTRYACTA PHYS-CHIM SIN ac ACTA PHYSICO-CHIMICA SINICAJ CHEM CRYSTALLOGR jo JOURNAL OF CHEMICAL CRYSTALLOGRAPHYJ CHEM CRYSTALLOGR jo JOURNAL OF CHEMICAL CRYSTALLOGRAPHYANN CHIM-ROME an ANNALI DI CHIMICAANN CHIM-ROME an ANNALI DI CHIMICAINORG REACT MECH in INORGANIC REACTION MECHANISMSINT J POLYM ANAL CH in INTERNATIONAL JOURNAL OF POLYMER ANALYSIS AND CHARACTE SCI CHINA SER B sc SCIENCE IN CHINA SERIES B-CHEMISTRYRES CHEM INTERMEDIAT re RESEARCH ON CHEMICAL INTERMEDIATESJ ANAL CHEM+jo JOURNAL OF ANALYTICAL CHEMISTRYREACT KINET CATAL L re REACTION KINETICS AND CATALYSIS LETTERSCHEM LISTY ch CHEMICKE LISTYCHINESE J INORG CHEM ch CHINESE JOURNAL OF INORGANIC CHEMISTRYJ RADIOANAL NUCL CH jo JOURNAL OF RADIOANALYTICAL AND NUCLEAR CHEMISTRYJ RADIOANAL NUCL CH jo JOURNAL OF RADIOANALYTICAL AND NUCLEAR CHEMISTRYJ RADIOANAL NUCL CH jo JOURNAL OF RADIOANALYTICAL AND NUCLEAR CHEMISTRY CHEM ANAL-WARSAW ch CHEMIA ANALITYCZNAJ CHIL CHEM SOC jo JOURNAL OF THE CHILEAN CHEMICAL SOCIETYPOLYM SCI SER A+po POLYMER SCIENCE SERIES AACTA POLYM SIN ac ACTA POLYMERICA SINICAJ SERB CHEM SOC jo JOURNAL OF THE SERBIAN CHEMICAL SOCIETYIONICS io IONICSIONICS io IONICSIONICS io IONICSRUSS J ORG CHEM+ru RUSSIAN JOURNAL OF ORGANIC CHEMISTRYPROG CHEM pr PROGRESS IN CHEMISTRYHIGH ENERG CHEM+hi HIGH ENERGY CHEMISTRYJ CHEM EDUC jo JOURNAL OF CHEMICAL EDUCATIONJ CHEM EDUC jo JOURNAL OF CHEMICAL EDUCATIONRUSS CHEM B+ru RUSSIAN CHEMICAL BULLETINCHINESE J ANAL CHEM ch CHINESE JOURNAL OF ANALYTICAL CHEMISTRYPOL J CHEM po POLISH JOURNAL OF CHEMISTRYRUSS J COORD CHEM+ru RUSSIAN JOURNAL OF COORDINATION CHEMISTRYCHEM PAP ch CHEMICAL PAPERS-CHEMICKE ZVESTICAN J ANAL SCI SPECT ca CANADIAN JOURNAL OF ANALYTICAL SCIENCES AND SPECTROSCO CAN J ANAL SCI SPECT ca CANADIAN JOURNAL OF ANALYTICAL SCIENCES AND SPECTROSCO STUD CONSERV st STUDIES IN CONSERVATIONSTUD CONSERV st STUDIES IN CONSERVATIONSTUD CONSERV st STUDIES IN CONSERVATIONHETEROCYCL COMMUN he HETEROCYCLIC COMMUNICATIONSREV INORG CHEM re REVIEWS IN INORGANIC CHEMISTRYJ STRUCT CHEM+jo JOURNAL OF STRUCTURAL CHEMISTRYJ STRUCT CHEM+jo JOURNAL OF STRUCTURAL CHEMISTRYLC GC EUR lc LC GC EUROPEPOLYM-KOREA po POLYMER-KOREADOKL PHYS CHEM do DOKLADY PHYSICAL CHEMISTRYCHEM WORLD-UK ch CHEMISTRY WORLDINDIAN J CHEM B in INDIAN JOURNAL OF CHEMISTRY SECTION B-ORGANIC CHEMISTR RUSS J GEN CHEM+ru RUSSIAN JOURNAL OF GENERAL CHEMISTRYCHEM NAT COMPD+ch CHEMISTRY OF NATURAL COMPOUNDSJ RARE EARTH jo JOURNAL OF RARE EARTHSS AFR J CHEM-S-AFR T so SOUTH AFRICAN JOURNAL OF CHEMISTRY-SUID-AFRIKAANSE TYD CHIM OGGI ch CHIMICA OGGI-CHEMISTRY TODAYCHIM OGGI ch CHIMICA OGGI-CHEMISTRY TODAYRUSS J PHYS CHEM A+ru RUSSIAN JOURNAL OF PHYSICAL Chemistry AJ MED PLANTS RES jo Journal of Medicinal Plants ResearchRUSS J INORG CHEM+ru RUSSIAN JOURNAL OF INORGANIC CHEMISTRYCHEM RES CHINESE U ch CHEMICAL RESEARCH IN CHINESE UNIVERSITIESCHINESE CHEM LETT ch CHINESE CHEMICAL LETTERSDOKL CHEM do DOKLADY CHEMISTRYMAIN GROUP MET CHEM ma MAIN GROUP METAL CHEMISTRYMAIN GROUP MET CHEM ma MAIN GROUP METAL CHEMISTRYCHEM UNSERER ZEIT ch CHEMIE IN UNSERER ZEITPROG REACT KINET MEC pr PROGRESS IN REACTION KINETICS AND MECHANISMJ INDIAN CHEM SOC jo JOURNAL OF THE INDIAN CHEMICAL SOCIETYLC GC N AM lc LC GC NORTH AMERICABUNSEKI KAGAKU bu BUNSEKI KAGAKUREV CHIM-BUCHAREST re REVISTA DE CHIMIEREV CHIM-BUCHAREST re REVISTA DE CHIMIEPOLYM SCI SER B+po POLYMER SCIENCE SERIES BINDIAN J HETEROCY CH in INDIAN JOURNAL OF HETEROCYCLIC CHEMISTRYCHEM IND-LONDON ch CHEMISTRY & INDUSTRYOXID COMMUN ox OXIDATION COMMUNICATIONSREV ROUM CHIM re REVUE ROUMAINE DE CHIMIERUSS J APPL CHEM+ru RUSSIAN JOURNAL OF APPLIED CHEMISTRYASIAN J CHEM as ASIAN JOURNAL OF CHEMISTRYB CHEM SOC ETHIOPIA bu BULLETIN OF THE CHEMICAL SOCIETY OF ETHIOPIA AFINIDAD af AFINIDADJ AUTOM METHOD MANAG jo JOURNAL OF AUTOMATED METHODS & MANAGEMENT IN CHEMISTRY J AUTOM METHOD MANAG jo JOURNAL OF AUTOMATED METHODS & MANAGEMENT IN CHEMISTRY KOBUNSHI RONBUNSHU ko KOBUNSHI RONBUNSHUJ CHEM SOC PAKISTAN jo JOURNAL OF THE CHEMICAL SOCIETY OF PAKISTANJ CHEM RES-S jo JOURNAL OF CHEMICAL RESEARCH-SACTUAL CHIMIQUE ac ACTUALITE CHIMIQUERUSS J PHYS CHEM B+ru Russian Journal of Physical Chemistry BCHEM PHYS CARBON ch CHEMISTRY AND PHYSICS OF CARBONCHEM PHYS CARBON ch CHEMISTRY AND PHYSICS OF CARBONCHEM PHYS CARBON ch CHEMISTRY AND PHYSICS OF CARBONJ APPL CRYSTALLOGR jo JOURNAL OF APPLIED CRYSTALLOGRAPHYACTA CRYSTALLOGR B ac ACTA CRYSTALLOGRAPHICA SECTION B-STRUCTURAL SCIENCE ACTA CRYSTALLOGR A ac ACTA CRYSTALLOGRAPHICA SECTION AJ CRYST GROWTH jo JOURNAL OF CRYSTAL GROWTHLIQ CRYST li LIQUID CRYSTALSCRYST RES TECHNOL cr CRYSTAL RESEARCH AND TECHNOLOGYACTA CRYSTALLOGR C ac ACTA CRYSTALLOGRAPHICA SECTION C-CRYSTAL STRUCTURE COM MOL CRYST LIQ CRYST mo MOLECULAR CRYSTALS AND LIQUID CRYSTALSACTA CRYSTALLOGR E ac ACTA CRYSTALLOGRAPHICA SECTION E-STRUCTURE REPORTS ONL CRYSTALLOGR REP+cr CRYSTALLOGRAPHY REPORTSZ KRIST-NEW CRYST ST ze ZEITSCHRIFT FUR KRISTALLOGRAPHIE-NEW CRYSTAL STRUCTURE小类名称(英文)小类分区大类名称大类分区2008年影响因ISSN小类名称(中文0009-2665化学综合CHEMISTRY, MULTIDISCIPLINA1化学123.592 0001-4842化学综合CHEMISTRY, MULTIDISCIPLINA1化学112.176 0079-6700高分子科学POLYMER SCIENCE1化学116.819 0306-0012化学综合CHEMISTRY, MULTIDISCIPLINA1化学117.419 0002-5100有机化学CHEMISTRY, ORGANIC1化学116.733 0066-426X物理化学CHEMISTRY, PHYSICAL1化学114.688 0167-5729物理化学CHEMISTRY, PHYSICAL1化学112.808 0167-5729物理:凝聚态物PHYSICS, CONDENSED MATTER1化学112.808 1433-7851化学综合CHEMISTRY, MULTIDISCIPLINA1化学110.879CHEMISTRY, INORGANIC & NUC1化学110.566 0010-8545无机化学与核化0265-0568医药化学CHEMISTRY, MEDICINAL1化学17.45 0265-0568有机化学CHEMISTRY, ORGANIC1化学17.45 0265-0568生化与分子生物BIOCHEMISTRY & MOLECULAR B2化学17.45 0360-0564物理化学CHEMISTRY, PHYSICAL1化学1 4.812 0002-7863化学综合CHEMISTRY, MULTIDISCIPLINA2化学18.091 0161-4940物理化学CHEMISTRY, PHYSICAL1化学1 5.625 0144-235X物理化学CHEMISTRY, PHYSICAL2化学1 6.892 1389-5567物理化学CHEMISTRY, PHYSICAL2化学1 5.36 0065-3195高分子科学POLYMER SCIENCE1化学2 6.802 0003-2700分析化学CHEMISTRY, ANALYTICAL1化学2 5.712 0340-1022化学综合CHEMISTRY, MULTIDISCIPLINA2化学2 5.27 0165-9936分析化学CHEMISTRY, ANALYTICAL1化学2 5.485 0947-6539化学综合CHEMISTRY, MULTIDISCIPLINA2化学2 5.454 1615-4150应用化学CHEMISTRY, APPLIED1化学2 5.619 1615-4150有机化学CHEMISTRY, ORGANIC1化学2 5.619 1359-7345化学综合CHEMISTRY, MULTIDISCIPLINA2化学2 5.34 0065-3055无机化学与核化CHEMISTRY, INORGANIC & NUC1化学2 3.571 0065-3055有机化学CHEMISTRY, ORGANIC2化学2 3.571 1523-7060有机化学CHEMISTRY, ORGANIC2化学2 5.128 1359-0294物理化学CHEMISTRY, PHYSICAL2化学2 5.493 1364-5498物理化学CHEMISTRY, PHYSICAL2化学2 4.604 1463-9262化学综合CHEMISTRY, MULTIDISCIPLINA2化学2 4.542 0081-5993物理化学CHEMISTRY, PHYSICAL2化学2 6.511 0081-5993无机化学与核化CHEMISTRY, INORGANIC & NUC2化学2 6.511CHEMISTRY, INORGANIC & NUC2化学2 4.214 0898-8838无机化学与核化1528-7483晶体学CRYSTALLOGRAPHY1化学2 4.215MATERIALS SCIENCE, MULTIDI2化学2 4.215 1528-7483材料科学:综合1528-7483化学综合CHEMISTRY, MULTIDISCIPLINA2化学2 4.215 1861-4728化学综合CHEMISTRY, MULTIDISCIPLINA2化学2 4.197 0192-8651化学综合CHEMISTRY, MULTIDISCIPLINA3化学2 3.39 1520-6106物理化学CHEMISTRY, PHYSICAL2化学2 4.189 1549-9618化学综合CHEMISTRY, MULTIDISCIPLINA3化学2 4.274 0001-8686物理化学CHEMISTRY, PHYSICAL3化学2 5.333CHEMISTRY, INORGANIC & NUC2化学2 4.147 0020-1669无机化学与核化0743-7463物理化学CHEMISTRY, PHYSICAL3化学2 4.097 1525-7797高分子科学POLYMER SCIENCE2化学2 4.1461525-7797有机化学CHEMISTRY, ORGANIC2化学2 4.146BIOCHEMISTRY & MOLECULAR B3化学2 4.146 1525-7797生化与分子生物0022-3263有机化学CHEMISTRY, ORGANIC2化学2 3.952CHEMISTRY, INORGANIC & NUC2化学2 3.815 0276-7333无机化学与核化0276-7333有机化学CHEMISTRY, ORGANIC2化学2 3.815 0021-9673分析化学CHEMISTRY, ANALYTICAL2化学2 3.756 0021-9673生化研究方法BIOCHEMICAL RESEARCH METHO2化学2 3.756 0267-9477分析化学CHEMISTRY, ANALYTICAL2化学2 4.028 0267-9477光谱学SPECTROSCOPY2化学2 4.028 0887-624X高分子科学POLYMER SCIENCE2化学2 3.821 1466-8033晶体学CRYSTALLOGRAPHY2化学2 3.535 1466-8033化学综合CHEMISTRY, MULTIDISCIPLINA3化学2 3.535PHYSICS, ATOMIC, MOLECULAR1化学2 3.636 1439-4235物理:原子、分1439-4235物理化学CHEMISTRY, PHYSICAL3化学2 3.636 0003-2654分析化学CHEMISTRY, ANALYTICAL2化学2 3.761 1385-2728有机化学CHEMISTRY, ORGANIC2化学2 3.184PHYSICS, ATOMIC, MOLECULAR2化学2 4.064 1463-9076物理:原子、分1463-9076物理化学CHEMISTRY, PHYSICAL3化学2 4.064CHEMISTRY, INORGANIC & NUC2化学2 3.6 0949-8257无机化学与核化BIOCHEMISTRY & MOLECULAR B3化学2 3.6 0949-8257生化与分子生物MATERIALS SCIENCE, MULTIDI2化学2 3.396 1932-7447材料科学:综合1932-7447物理化学CHEMISTRY, PHYSICAL3化学2 3.396 1932-7447纳米科技NANOSCIENCE & NANOTECHNOLO3化学2 3.396 1044-0305分析化学CHEMISTRY, ANALYTICAL2化学2 3.181 1044-0305光谱学SPECTROSCOPY2化学2 3.181 1044-0305物理化学CHEMISTRY, PHYSICAL3化学2 3.181 1549-9596计算机:信息系COMPUTER SCIENCE, INFORMAT1化学2 3.643COMPUTER SCIENCE, INTERDIS2化学2 3.643 1549-9596计算机:跨学科1549-9596化学综合CHEMISTRY, MULTIDISCIPLINA3化学2 3.643CHEMISTRY, INORGANIC & NUC2化学2 3.58 1477-9226无机化学与核化1527-8999化学综合CHEMISTRY, MULTIDISCIPLINA3化学3 3.477 1477-0520有机化学CHEMISTRY, ORGANIC2化学3 3.55 0039-9140分析化学CHEMISTRY, ANALYTICAL2化学3 3.206 1520-4766应用化学CHEMISTRY, APPLIED1化学3 3.011 1520-4766化学综合CHEMISTRY, MULTIDISCIPLINA3化学3 3.011 1520-4766医药化学CHEMISTRY, MEDICINAL3化学3 3.011 0003-2670分析化学CHEMISTRY, ANALYTICAL2化学3 3.146 0032-3861高分子科学POLYMER SCIENCE2化学3 3.331 0926-860X环境科学ENVIRONMENTAL SCIENCES2化学3 3.19 0926-860X物理化学CHEMISTRY, PHYSICAL3化学3 3.19 1076-5174光谱学SPECTROSCOPY2化学3 2.94 1076-5174生物物理BIOPHYSICS3化学3 2.94 1076-5174有机化学CHEMISTRY, ORGANIC3化学3 2.94 0047-2689物理化学CHEMISTRY, PHYSICAL3化学3 2.424 0047-2689化学综合CHEMISTRY, MULTIDISCIPLINA3化学3 2.424 0047-2689物理:综合PHYSICS, MULTIDISCIPLINARY3化学3 2.424PHYSICS, ATOMIC, MOLECULAR2化学3 2.871 1089-5639物理:原子、分1089-5639物理化学CHEMISTRY, PHYSICAL3化学3 2.871 1618-2642分析化学CHEMISTRY, ANALYTICAL2化学3 3.328 1618-2642生化研究方法BIOCHEMICAL RESEARCH METHO3化学3 3.328 0304-4203海洋学OCEANOGRAPHY2化学3 2.977 0304-4203化学综合CHEMISTRY, MULTIDISCIPLINA3化学3 2.977 1434-193X有机化学CHEMISTRY, ORGANIC3化学3 3.016 0040-4020有机化学CHEMISTRY, ORGANIC3化学3 2.897 1570-1794有机化学CHEMISTRY, ORGANIC3化学3 2.61 0951-4198分析化学CHEMISTRY, ANALYTICAL2化学3 2.772 0951-4198光谱学SPECTROSCOPY3化学3 2.772 1040-0397分析化学CHEMISTRY, ANALYTICAL2化学3 2.901 1040-0397电化学ELECTROCHEMISTRY3化学3 2.901 0936-5214有机化学CHEMISTRY, ORGANIC3化学3 2.659 1144-0546化学综合CHEMISTRY, MULTIDISCIPLINA3化学3 2.942 1040-8347分析化学CHEMISTRY, ANALYTICAL3化学3 3.5CHEMISTRY, INORGANIC & NUC3化学32 0260-3594无机化学与核化MATHEMATICS, INTERDISCIPLI1化学3 3.5 0340-6253数学跨学科应用COMPUTER SCIENCE, INTERDIS2化学3 3.5 0340-6253计算机:跨学科0340-6253化学综合CHEMISTRY, MULTIDISCIPLINA3化学3 3.5 1381-1169物理化学CHEMISTRY, PHYSICAL3化学3 2.814CHEMISTRY, INORGANIC & NUC3化学3 2.694 1434-1948无机化学与核化0920-5861工程:化工ENGINEERING, CHEMICAL1化学3 3.004 0920-5861应用化学CHEMISTRY, APPLIED2化学3 3.004 0920-5861物理化学CHEMISTRY, PHYSICAL3化学3 3.004 1615-9306分析化学CHEMISTRY, ANALYTICAL3化学3 2.746 0957-4166物理化学CHEMISTRY, PHYSICAL3化学3 2.796 0957-4166无机化学与核化CHEMISTRY, INORGANIC & NUC3化学3 2.796 0957-4166有机化学CHEMISTRY, ORGANIC3化学3 2.796 0040-4039有机化学CHEMISTRY, ORGANIC3化学3 2.538MATERIALS SCIENCE, MULTIDI2化学3 2.555 1387-1811材料科学:综合1387-1811应用化学CHEMISTRY, APPLIED2化学3 2.555 1387-1811物理化学CHEMISTRY, PHYSICAL3化学3 2.555 1387-1811纳米科技NANOSCIENCE & NANOTECHNOLO3化学3 2.555 0065-3160物理化学CHEMISTRY, PHYSICAL3化学3 1.833 0065-3160有机化学CHEMISTRY, ORGANIC3化学3 1.833 1572-6657分析化学CHEMISTRY, ANALYTICAL3化学3 2.484 0223-5234医药化学CHEMISTRY, MEDICINAL3化学3 2.882 1432-881X物理化学CHEMISTRY, PHYSICAL3化学3 2.37 0079-6786无机化学与核化CHEMISTRY, INORGANIC & NUC3化学3 2.938 1350-4177声学ACOUSTICS2化学3 2.796 1350-4177化学综合CHEMISTRY, MULTIDISCIPLINA3化学3 2.796 1566-7367物理化学CHEMISTRY, PHYSICAL3化学3 2.791 0039-7881有机化学CHEMISTRY, ORGANIC3化学3 2.47 0021-9797物理化学CHEMISTRY, PHYSICAL3化学3 2.443 1022-5528应用化学CHEMISTRY, APPLIED2化学3 2.212 1022-5528物理化学CHEMISTRY, PHYSICAL3化学3 2.212 0009-2614物理化学CHEMISTRY, PHYSICAL3化学3 2.169。
补充硫酸亚铁对运动训练大鼠心肌线粒体呼吸链酶复合物活性及自由基代谢的影响The Effect of Supplemented Ferons Sulfate on Activities of Respiratory Chain Complexes in Mitochondria and Free Radicals Metabolism from Mice’s Myocardium * 分页下载* 分章下载* 整本下载* 在线阅读* 不支持迅雷等下载工具。
如阅读文献显示异常,请下载并安装新版CAJ阅读器。
【作者】张军;【导师】李洁【作者基本信息】西北师范大学,运动人体科学, 2010,硕士【摘要】人体在长期长时间大强度的运动下会导致缺铁性贫血。
本研究主要是通过对缺铁性贫血的运动大鼠补充硫酸亚铁,运用运动生物化学理论和细胞生物学研究方法对大鼠心肌线粒体呼吸链酶复合物活性和自由基含量进行比较,在此基础上探讨补铁对大鼠心脏代谢具有积极意义的可能机制,以至为进一步的科学研究提供参考资料。
实验方法:健康雄性Whistar 大鼠60只,体重在100g左右,所有大鼠在建模成功后按要求随机分为:安静对照组(C,n=10),小剂量补铁+运动组(S,n=10),中剂量补铁+运动组(M,n=10)。
大剂量补铁+运动组(L,n=10),运动组(T,n=10),补铁组每天采用灌胃方式补充硫酸亚铁溶液,补铁的剂量为:小剂量补铁组每鼠每天28.85mg/kg大鼠体重,中剂量补铁组每鼠每天57.7mg/kg大鼠体重,大剂量补铁组每鼠每天115.4mg/kg大鼠体重。
运动组和补铁+运动组大鼠进行递增负荷跑台训练8周,每周训练6天,周日休息,动物跑台坡度为0度,跑台速度为30米/分。
前4周运动性贫血模型建立时的训练安排计划为:前两周每天1次进行训练,其余时间为每天早晚各1次;第1次训练时间为5min,之后2min/...?更多还原【Abstract】 The human body will lead to iron-deficiency anemia under the condition oflong time and high-intensity exercise, This study is mainly through supplementferrous sulfate to the rats of iron-deficiency anemia. Applied theory and method ofsports biochemistry and cell biology to find the differences of mitochondrial enzymecomplex’activity and oxygen free radicals content in the rats’myocardium,and todiscuss the Mechanism of the added ferrous sulfate has positive influence on cardiacmetabolism of the b...?更多还原【关键词】硫酸亚铁;心肌;线粒体;呼吸链酶活性; MDA; SOD; GSH-PX;【Key words】 ferrous sulfate; myocardium; mitochondria; respiratory chain enzymeactivities; MDA; SOD; GSH-PX;* 【网络出版投稿人】西北师范大学【网络出版年期】2011年 05期* 【分类号】G804.7我国优秀花样游泳运动员身体机能的生理生化评定* 推荐 CAJ下载* PDF下载* 不支持迅雷等下载工具,请取消加速工具后下载。