Illumina 官方介绍不同机型index读取方式 indexed-sequencing-overview-guide-15057455-03
- 格式:pdf
- 大小:2.73 MB
- 文档页数:12
MiSeqDx儀器安全與合規指南ILLUMINA所屬財產文件#15034477v05CHT2021年11月此文件與其內容為Illumina,Inc.與其分支機構(「Illumina」)之專有財產,僅供客戶針對本文件所述之產品用途於契約規範內使用,不得移作他用。
此文件與其內容不得基於其他用途而使用或散播,和/或在未事先取得Illumina的書面同意下,以任何方式流通、揭露或複製。
Illumina並未藉由本文件傳遞其專利、商標、版權或任何普通法權利或任何第三方之類似權利的任何授權。
本文件的指示必須由受過適當訓練的合格人員嚴格且明確地遵守,以確保此處所述之產品的適當與安全使用。
在使用該產品之前,必須完整閱讀與了解文件的所有內容。
若未全文閱讀並明確遵守此處的所有指示,可能造成產品損壞、人員受傷(包括使用者或其他人),以及其他財產損壞,並導致產品保固失效。
對於不當使用本文所述產品(包括其零件或軟體)而造成的損失,Illumina不承擔任何責任。
©2021Illumina,Inc.保留一切權利。
所有商標均為Illumina,Inc.或其各自所有權人所擁有。
如需特定商標資訊,請參閱/company/legal.html。
修訂記錄目錄修訂記錄簡介1安全注意事項及標識2一般安全警告2電氣安全警告2高溫表面安全警告3重物安全警告3拆箱、安裝和移動儀器3環境限制4符號5產品合規與法規聲明6產品認證與合規6有害物質限制(RoHS)6無線電頻率的人體曝露量6 FCC合規6 IC合規7 ConformitéIC8巴西合規8韓國合規8 Españoles advertencia-Mexico9泰國合規9阿拉伯聯合大公國合規9技術協助10簡介本指南提供有關MiSeqDx安裝、維修和操作的重要安全資訊以及產品合規與法規聲明。
在MiSeqDx上執行任何程序之前,請先閱讀此文件。
MiSeqDx的原產國和製造日期皆印製在儀器標籤上。
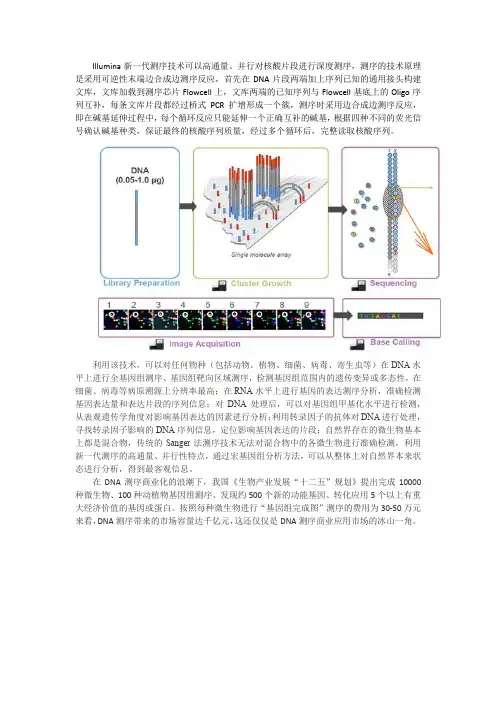
Illumina新一代测序技术可以高通量、并行对核酸片段进行深度测序,测序的技术原理是采用可逆性末端边合成边测序反应,首先在DNA片段两端加上序列已知的通用接头构建文库,文库加载到测序芯片Flowcell上,文库两端的已知序列与Flowcell基底上的Oligo序列互补,每条文库片段都经过桥式PCR扩增形成一个簇,测序时采用边合成边测序反应,即在碱基延伸过程中,每个循环反应只能延伸一个正确互补的碱基,根据四种不同的荧光信号确认碱基种类,保证最终的核酸序列质量,经过多个循环后,完整读取核酸序列。
利用该技术,可以对任何物种(包括动物、植物、细菌、病毒、寄生虫等)在DNA水平上进行全基因组测序、基因组靶向区域测序,检测基因组范围内的遗传变异或多态性,在细菌、病毒等病原溯源上分辨率最高;在RNA水平上进行基因的表达测序分析,准确检测基因表达量和表达片段的序列信息;对DNA处理后,可以对基因组甲基化水平进行检测,从表观遗传学角度对影响基因表达的因素进行分析;利用转录因子的抗体对DNA进行处理,寻找转录因子影响的DNA序列信息,定位影响基因表达的片段;自然界存在的微生物基本上都是混合物,传统的Sanger法测序技术无法对混合物中的各微生物进行准确检测,利用新一代测序的高通量、并行性特点,通过宏基因组分析方法,可以从整体上对自然界本来状态进行分析,得到最客观信息。
在DNA测序商业化的浪潮下,我国《生物产业发展“十二五”规划》提出完成10000种微生物、100种动植物基因组测序、发现约500个新的功能基因、转化应用5个以上有重大经济价值的基因或蛋白。
按照每种微生物进行“基因组完成图”测序的费用为30-50万元来看,DNA测序带来的市场容量达千亿元,这还仅仅是DNA测序商业应用市场的冰山一角。

illumina测序原理illumina测序是一种分子生物技术,是人类基因组测序领域最常用的序列技术。
Illumina测序可以帮助研究人员对物种基因组进行微观上的分析,以了解物种遗传和表型的变化,以及为药物开发和临床诊断提供技术支持。
本文将介绍illumina测序的原理、技术方法以及用于探索疾病的应用。
illumina测序的原理illumina测序以能够自动分辨双链DNA的激光荧光技术为基础。
它是一种借助微孔板的半结构化的、多芯片的平台,可以同时对多条双链DNA进行高通量测序。
其基本原理是,在介质中悬浮的DNA片段会在illumina仪器上被电场热像(piezo-electric heating)做成微型沉积,然后由四种激光激发激光(excitation laser)、发射激光(emission laser)、紫外线激光(UV laser)、紫外线破坏激光(UV destruction laser)对每个沉积的微粒进行精确的检测。
illumina测序的技术方法illumina测序通常由四个步骤组成:DNA片段制备、多聚合酶链反应(PCR)、样品分配和测序读取。
1、DNA片段制备:这一步是首先利用核酸在体外产生固定大小的DNA片段,然后加入抑制剂阻止PCR反应,最后将数据存储在微孔板中。
2、多聚合酶链反应(PCR):多聚合酶链反应(PCR)使得每个DNA 片段可以得到大量的复制,以便illumina仪器可以测试它们。
3、样品分配:在这个步骤中,借助芯片的能力,illumina测序仪可以将样品放置在固定的位置上,并精确地控制它们的移动速度和沉积位置,以实现多个样品的定位和分配。
4、测序读取:这是最后一步,也是最重要的步骤,即对每一个沉积的DNA片段序列进行精确的测序,以获得每一条双链DNA的完整序列。
illumina测序用于疾病探索illumina测序不仅应用于普通的基因组测序,还可以用于探索疾病机制,鉴定敏感性突变,并检测基因组结构变异。

illumina的read 读段Illumina, Inc. is an innovative company revolutionizing the field of genomics and genetic analysis. Their groundbreaking DNA sequencing technology has opened up new possibilities in medical research, agriculture, and personalized medicine. Illumina's "reads" refer to the process of analyzing the data generated from DNA sequencing. In this article, we will delve into the intricacies of Illumina reads and explore their significance in advancing genomics research.DNA sequencing is the process of determining the exact order of nucleotides in a DNA molecule. Illumina sequencing, specifically, is based on a method called "sequencing by synthesis." This technique fragments the DNA sample into smaller pieces and attaches short DNA fragments called adapters to each end. These adapters allow the DNA fragments to bind to a solid surface, creating a DNA template for sequencing.Illumina sequencing utilizes a complex system that involves multiple steps. First, the DNA fragments are amplified through a process called polymerase chain reaction (PCR), creating millions of identical copies of each fragment. This step is crucial because it enables the detection of weak signals and reduces errors in the sequencing process.After amplification, the DNA fragments are loaded onto a specially designed flow cell, where they are annealed to complementary oligonucleotides known as primers. These primers act as starting points for DNA synthesis. The primers are attached to the flow cell's surface and act as anchor points for the DNA fragments.The next step involves the incorporation of fluorescently labeled nucleotides into the growing DNA strands. Each nucleotide is labeled with a different color, allowing for the detection and identification of the incorporated nucleotide during sequencing. The nucleotides are added one at a time in a process called sequencing by synthesis.As the nucleotides are added, a camera captures images of the flow cell, recording the fluorescence signal from each cluster of DNA fragments. The camera takes multiple images to ensure accurate detection of the fluorescent signals.These fluorescence signals are then analyzed by sophisticated software to determine the order of nucleotides in each DNA fragment. The software aligns the reads from the overlapping nucleotides, reconstructing the full DNA sequence.Once the reads are generated, they undergo quality control measures to ensure their accuracy. Sequencing errors can occur due to various factors such as machine errors, DNA damage, or low-quality samples. Quality control measures include checking for base call accuracy, read length distribution, error rates, and other metrics to ensure the reliability of the sequencing data.The reads produced by Illumina sequencing are typically short, ranging from 50 to 300 base pairs. However, the sheer number of reads generated is immense, with current sequencing platforms capable of producing billions of reads per run. This high-throughput nature of Illumina sequencing enables researchers toobtain a comprehensive view of the DNA being analyzed.Illumina reads have revolutionized the field of genomics by enabling researchers to analyze not only individual genomes but also entire populations of organisms. The massive amount of data generated from Illumina sequencing allows for the identification of genetic variations, such as single-nucleotide polymorphisms (SNPs) and structural variants. These genetic variations are crucial in understanding the genetic basis of diseases, the evolution of species, and the effectiveness of drugs.One of the applications of Illumina reads is in cancer research. By sequencing the DNA of cancer cells, researchers can identify the specific mutations that drive tumor growth. This information is vital in developing targeted therapies and personalized treatment plans for cancer patients.Another application is in agriculture, where Illumina sequencing is used to analyze the genetic makeup of crops and livestock. This allows for the identification of desirable traits, such as disease resistance or increased yield, and the development of improved breeding strategies.In addition to medical and agricultural research, Illumina reads are also used in forensics, evolutionary biology, and microbiome studies. The ability to sequence DNA quickly and accurately has opened up new avenues of research, leading to groundbreaking discoveries and advancements in various fields.However, Illumina reads are not without limitations. The shortread length makes it challenging to assemble the complete genomes of complex organisms or accurately analyze repetitive regions of the genome. This is where complementary technologies, such as long-read sequencing or optical mapping, come into play. In conclusion, Illumina reads are the foundation of modern genomics research. The ability to accurately sequence DNA on a massive scale has transformed our understanding of genetics and its implications in various fields. The power of Illumina sequencing lies in its ability to generate vast amounts of data quickly and cost-effectively, opening up new possibilities for personalized medicine, agriculture, and scientific discovery.。

![illumina Hiseq [Miseq] 测序原理](https://uimg.taocdn.com/a1122bdab8f67c1cfbd6b82e.webp)
Hiseq/Miseq测序原理简介主要内容一.Solexa技术简介二.测序原理①Follow Cell②可逆阻断技术③边合成边测序④桥式PCR⑤碱基读取三.工作流程四.测序类型五.仪器构造六.测序结果展示一.Solexa测序技术前世今生—Illumina平台•Solexa高通量测序技术是由英国剑桥大学派生的Solexa公司建立起来的。
该方法以单分子阵列技术为基础,是对合成测序技术的发展与延伸。
二.测序原理Solexa是一种基于边合成边测序技术(Sequencing-By-Synthesis,SBS)的新型测序技术。
通过单分子阵列实现在小型芯片(Flow Cell)上进行桥式PCR反应。
通过可逆阻断技术实现每次只合成一个碱基,再利用相应的激光激发荧光集团,捕获激发光,从而读取碱基信息。
Flow Cell70mm×25mm可逆阻断技术边合成边测序(SBS )➢加入测序引物➢Cycles1a)合成第一个碱基b)清除未反应的碱基和试剂c)激发荧光信号并读取d)去除荧光基团和阻断基团➢Cycle2……Cycle nATG C C CA TTGGGG AA TA A T G T T C C G G G G A A A A TT T T C C C 5’-3’-可逆阻断技术桥式PCR——簇生成之“起始延伸”桥式PCR——簇生成之“扩增”桥式PCR——簇生成之“测序”碱基读取三.工作流程四.测序类型a)单端测序(single-read sequencing,SR)b)双端测序(Paired-end sequencing,PE)c)混样测序(Index sequencing)Follow Cell接头差异Single Read Paired EndPeriodate Linearization 高碘酸盐切割位点Uracil Specific Excision Reagent (USER)尿嘧啶切割位点Formamidopyrimidine Glycosylase (fpg)糖基化酶切割位点a)双端测序➢Paired-End sequencing(PE/双端测序)−检测基因片段两端的基因信息。

Illumina测序平台介绍简介博奥生物基于Genome AnalyzerIIx,采用最新的TruSeq试剂,推出了小RNA测序(small RNA sequencing)、转录组测序(tanscriptome sequencing)、基于测序的基因表达研究(gene expression profiling based on sequencing)、全外显子组测序(exome sequencing)、目标区域深度测序(targeted sequence deep sequencing)、DNA甲基化测序(DNA methylation sequencing)和染色质免疫共沉淀测序(chromatin immunoprecipitation sequencing)高通量测序服务。
博奥生物具有近10年的高通量技术服务经验,实验室通过标准化认证并具备严格的质量控制管理系统,服务内容涵盖基因组、转录组、表观遗传组、蛋白质组各个层次和环节,同时拥有高通量测序平台、多种芯片检测平台、验证平台、高性能计算平台及专业的实验技术和生物信息团队,致力于为客户提供高质量的技术服务。
平台设备cBot Paired-endmodualGenome Analyzer IIx技术原理Illumina测序技术结合了微阵列技术和专有的可逆终止子技术进行大规模平行的边合成边测序,通过接头序列将基因组DNA或cDNA的随机片断附着到光学透明的流动槽(flow cell)表面,然后通过桥式扩增,形成数以亿计的DNA 簇(clusters),每个cluster约有1000~6000拷贝的相同DNA模板,然后用4种末端被封闭的不同荧光标记的碱基进行边合成边测序。
对读测序时,可在单端测序完成后进行原位反应,生成对读测序所需的模板,然后应用第二个测序引物对同一cluster进行测序,生成相应的数据。
Illumina测序技术一定程度上确保了高精确度和单碱基逐个测序,可有效减少同聚物和重复序列的测序错误。

The Best Next-Gen Sequencing Workflow Just Got BettercBot is a revolutionary automated clonal amplification system at the core of the Illumina sequencing workflow (Figure 1, upper panel). cBot replaces a lab full of equipment with a single compact device, deliver-ing unsurpassed efficiency and ease of use for the highest quality sequencing results.With cBot, hands-on time is reduced to less than 10 minutes, com-pared to more than six hours of hands-on effort for emulsion PCR methods. The process of creating sequencing templates is complete in about four hours, compared to more than 24 hours for emulsion PCR-based protocols (Figure 1, lower panel).Breakthrough System for Cluster GenerationThe Illumina sequencing workflow is based on three simple steps: libraries are prepared from virtually any nucleic acid sample, amplified to produce clonal clusters, and sequenced using massively parallel synthesis. The cBot clonal amplification system has innovative features that eliminate user intervention, reduce potential failure points, and increase sequencing productivity.TruSeq Cluster Generation reagents are packaged in ready-to-use96-well plates, completely removing reagent preparation errors, potential sources of contamination, and decreasing storage require-ments. cBot features a single unique, plate-piercing manifold for intervention-free runs. Cluster generation occurs within the sealed, eight-channel Illumina flow cell, bypassing the frequent handling and contamination issues inherent to emulsion PCR-based protocols. cBot is capable of processing > 96 samples within a single flow cell, resulting in substantial cost savings without incremental effort and wasted reagents. Innovative instrument features ensure seamless operation for your sequencing workflow (Figure 2).Better Results with Less EffortcBot software enhancements and user interaction features ensure high productivity:• Integrated 8-inch touch screen provides simplified operation in a small, lab-friendly footprint• On-screen, step-by-step instructions with embedded multimedia help enable user operation with no prior training • Real-time progress indicators provide at-a-glance monitoring • Remote monitoring allows a single user to manage multiple systems from any web browser or phone• Status emails are sent when the run is complete or when intervention is requiredcBot Cluster Generation ProcessPrior to sequencing, single-molecule DNA templates are bridge amplified to form clonal clusters inside the flow cell. (Figure 3).cBotFully automated clonal cluster generation for Illumina sequencing.Illumina cBot Highlights• Fast, Efficient Workflow:Amplify > 96 samples in ~4–5 hours with < 10 minutes ofhands-on time• Easiest to Use:Pre-packaged 96-well TruSeq™ reagents, and simple touch screen interface simplifies operation• Innovative System Design:Real-time fluidic monitoring, integrated system sensors and remote monitoring ensure robust instrument operation• Highest Quality Results:Improved chemistry generates higher density clusters and sequencing accuracy LibraryPreparation SequencingCluster GenerationEight-channel flow cell reduces risk of contamination and eliminates the needfor extra equipment Manifold clamps for leak-free connections and superior thermal contactTouch screen monitor simplifies operation and provides real-timeImmobilization of Single-Molecule DNA TemplatesHundreds of millions of templates are hybridized to a lawn of oligo-nucleotides immobilized on the flow cell surface. The templates are copied from the hybridized primers by 3’ extension using a high-fidelity DNA polymerase to prevent misincorporation errors. The original templates are denatured, leaving the copies immobilized on the flow cell surface.Isothermal Bridge AmplificationImmobilized DNA template copies are amplified by isothermal bridge amplification. The templates loop over to hybridize to adjacent lawn oligonucleotides. DNA polymerase copies the templates from the hybridized oligonucleotides, forming dsDNA bridges, which are dena-tured to form two ssDNA strands. These two strands loop over and hybridize to adjacent oligonucleotides and are extended again to form two new dsDNA loops. The process is repeated on each template by cycles of isothermal denaturation and amplification to create millions of individual, dense clonal clusters containing ~2,000 molecules. Linearization, Blocking, and Primer HybridizationEach cluster of dsDNA bridges is denatured, and the reverse strand is removed by specific base cleavage, leaving the forward DNA strand. The 3’-ends of the DNA strands and flow cell-bound oligonucleotides are blocked to prevent interference with the sequencing reaction. The sequencing primer is hybridized to the complementary sequence on the Illumina adapter on unbound ends of the templates in the clusters. The flow cell now contains >200 million clusters with ~1,000 mol-ecules/cluster, and is ready for sequencing.SummaryIllumina sequencing with cBot automated cluster generation sets the new standard for simplified next- generation sequencing. Ready-to-use reagents, smart instrumentation improvements, and new cluster generation chemistry offers significant advantages over emulsion PCR-based workflows and promotes even higher data density and sequencing accuracy. By streamlining the critical clonal amplification step in the next-generation sequencing workflow, Illumina continues to accelerate your landmark discoveries and publications.Ordering InformationDescriptioncBotCatalog No.HiSeq System Genome AnalyzercBot Instrument Includes cBot, flow cell adapter plate,one year warranty, user manualSY-301-2002cBot Flow Cell Manifold (Optional)SY-301-2014TruSeq Single-Read Cluster Generation Kits include flow cell,reagent plate, manifold, user instructionsGD-401-3001GD-300-2001TruSeq Paired-End Cluster Generation Kits include flow cell,reagent plate, manifold, PE reagents, user instructionsPE-401-3001PE-300-2001Illumina, Inc. •9885TowneCentreDrive,SanDiego,CA92121USA•1.800.809.4566toll-free•1.858.202.4566tel•************************• For research use only© 2011 Illumina, Inc. All rights reserved.Illumina, illuminaDx, BeadArray, BeadXpress, cBot, CSPro, DASL, Eco, Genetic Energy, GAIIx, Genome Analyzer, GenomeStudio, GoldenGate, HiScan, HiSeq, Infinium, iSelect, MiSeq, Nextera, Sentrix, Solexa, TruSeq, VeraCode, the pumpkin orange color, and the Genetic Energy streaming bases design are trademarks or registered trademarks of Illumina, Inc. All other brands and names contained herein are the property of their respective owners. Pub. No. 770-2009-032 Current as of 27 April 2011at the address below.Laser radiationDo not stare into the visible-light beam of the barcode scanner. The barcode scanner is a Class 2 laser product.SY-301-2002Instrument ConfigurationCE Marked and ETL Listed instrument, Installation, setup, and accessoriesInstrument Control ComputerMini-ITX Board with Celeron M Processor 1 GB RAM, 80 GB Hard Drive Windows Embedded OSIntegrated 8” Touch Screen Monitor Operating Environment Temperature: 22°C ± 3°CHumidity: Non-Condensing 20%–80%Altitude: Less than 2,000 m (6,500 ft)Air Quality: Pollution Degree Rating of II For Indoor Use Only LaserClass 2 Laser: 630 –650 nm DimensionsW×D×H: 38 cm × 62 cm × 40 cm Weight: 34 kg Crated Weight: 36 kg Power Requirements100−240V AC 50/60 Hz, 4A, 400 Watts。

illumina sequencing原理
Illumina sequencing(Illumina 测序)是一种广泛使用的高通量DNA 测序技术,其原理基于边合成边测序(Sequencing-by-Synthesis,SBS)的方法。
Illumina 测序的基本步骤如下:
1. 文库制备:将待测DNA 片段连接到测序载体上,形成文库。
2. 桥接和扩增:将文库加载到测序芯片上,通过桥式PCR 进行扩增,每个DNA 片段都被扩增成许多相同的拷贝。
3. 测序:在测序过程中,DNA 聚合酶会将荧光标记的核苷酸逐个添加到延伸的DNA 链上。
每次添加一个核苷酸,都会释放出相应的荧光信号,通过光学检测系统记录。
4. 图像采集和分析:光学检测系统会捕捉每个核苷酸添加时产生的荧光信号,并将其转换为数字图像。
这些图像被用于分析和识别每个核苷酸的身份。
5. 数据处理和质量控制:测序得到的原始数据需要经过处理和质量控制步骤,包括去除低质量的序列、比对到参考基因组等,以获
得最终的测序结果。
Illumina 测序技术具有高通量、高准确性和低成本的特点,使其成为基因组学研究、生物医学研究和临床诊断等领域的重要工具。

本指南提供有关Illumina ®NextSeq ®500和NextSeq ®550系统安装、维修以及操作的重要安全信息。
另外还包含产品合规性和监管声明。
对系统执行任何程序之前,请先阅读本文档。
安全注意事项和标记此部分指出与本仪器安装、维修和操作相关的潜在危险。
请不要以会使您面临以下危险的方式操作或使用仪器。
只要遵照《NextSeq 500系统指南》(文档号15046563)或《NextSeq 550系统指南》(文档号15069765)中介绍的标准操作程序进行操作,便可避免所述的所有危险。
一般安全警告确保所有人员都受过有关仪器正确操作和任何潜在安全注意事项的培训。
在标有此标签的区域使用时,请遵照所有操作指示,以最大限度降低人员或仪器面临的风险。
激光安全警告NextSeq 系统是嵌入了3B 类二极管的1类激光产品。
1类辐射水平并不视为有危险性。
照射到操作员的所有激光辐射均符合适用于1类激光产品的IEC 60825-1可达极限。
电气安全警告请不要取下仪器上的外部面板。
本仪器内部并没有用户可维修的部件。
取下任意面板对仪器进行操作可能会造成线电压和直流电压触电。
本仪器由100–240伏交流电以50–60赫兹的频率供电。
危险电压源位于后面板和左侧面板后,但如果取下其他面板,可能会碰触到它们。
即使仪器关闭时,上面仍然会有一些电压。
请在所有面板都原封不动的情况下操作仪器,以免触电。
电源规格线路电压100–240伏交流电,50/60赫兹电源额定功率最高600瓦电气连接将仪器连接到至少能够提供以下电流等级的接地电路:}15安培(如电源为100-110伏)}10安培(如电源为220-240伏)有关详细信息,请参见《NextSeq 系统场地准备指南》(文档号15045113)。
保护接地仪器通过外壳进行保护接地连接。
电源线的安全接地可将保护接地返回到安全参照点。
使用此设备时电源线的保护接地连接必须处于良好的工作状态。

illumina测序特点Illumina测序是一种常用的高通量测序技术,具有以下几个特点。
1. 高通量:Illumina测序平台可以同时进行数千万到上亿个DNA片段的并行测序,大大提高了测序的通量,使得高通量测序成为可能。
高通量测序可以应用于大规模基因组、转录组和表观基因组的测序,以及人群的全基因组测序等。
2. 短读长:Illumina测序技术产出的测序读长通常在25-300个碱基左右,其中以100个碱基左右的reads最常见。
相比于其他测序技术如PacBio的长读长,Illumina的短读长具有读长廉价、高准确性以及较好的覆盖深度的优势。
短读长最适合于重测序、变异检测、转录本表达量定量和RNA剪接等短序列分析。
3. 高精度:Illumina测序技术具有高准确性,其单个碱基的错误率在0.1%以下。
这是因为Illumina采用了碱基按照碱基特异性保护的方法,通过不断侵蚀和反应,仅限制一个碱基的添加发生,保证了其高精度。
加之Illumina采用金属离子探针和碱基的标记来检测,进一步提高了准确性。
高精度的测序结果对于遗传变异和基因突变的检测非常关键。
4. 成本低廉:相比于其他测序技术,Illumina测序具有较低的测序成本。
通过应用高密度测序流片,Illumina平台能同时测序上千个样本,并通过分析软件实现高效的数据分析,从而大幅度降低了测序成本。
因此,Illumina测序成为许多研究所、大公司和医疗机构常用的测序方法。
5. 应用广泛:Illumina测序技术的广泛应用范围涵盖了基因组学、转录组学、表观基因组学等多个研究领域。
在基因组学研究中,Illumina测序被用于完成大规模全基因组测序、重测序以及SNP检测等。
在转录组学研究中,Illumina测序可以衡量基因表达水平、发现新的非编码RNA等。
在表观基因组学研究中,Illumina测序可以帮助寻找DNA甲基化区域、组蛋白修饰区域等。
总结起来,Illumina测序具有高通量、高精度和成本低廉的特点,广泛应用于基因组学、转录组学和表观基因组学等研究领域。
本指南提供Illumina ®iSeq™100定序系統有關安裝、維修和操作的重要安全資訊。
本手冊包含產品合規與法規聲明。
在系統上執行任何程序之前,請先閱讀此資訊。
系統的原產國和製造日期皆印製在儀器標籤上。
安全注意事項及標識本節旨在提出與安裝、維護和操作儀器有關的潛在危害。
請勿以任何會讓您曝露在這些危險的方式操作儀器或與儀器互動。
一般安全警告確認所有人員均接受訓練,了解如何正確操作儀器以及任何潛在的安全性考量。
在標有此標籤的區域工作時,請遵守所有操作指示,以將人員或儀器所受的風險降到最低。
電氣安全警告請勿取下任何儀器上的外部面板。
儀器內部並無可由使用者維修的零組件。
在移除任一面板的情況下操作本儀器,可能會接觸到線路電壓以及直流電壓而導致觸電。
本儀器電源要求為100至240伏特,50或60赫的交流電。
危險的電壓來源位於背板和側板後面,但如果其他面板移除仍然可能接觸到。
即使關閉儀器電源,儀器上仍會存在些許電壓。
請在所有面板均完好的情況下操作儀器,以免觸電。
電源規格100至240伏特、50或60赫交流電峰值耗電量80 瓦特必須進行電氣接地。
如果電壓波動超過10%,則需要電力線路調節器。
電源線取用請調整儀器的放置方式,以便從插座快速地拔除電源線。
保護性接地線本儀器有通過外殼連接至保護性接地線的線路。
電源線的安全接地使保護性接地線回到安全基準。
使用本裝置時,電源線的保護性接地線連接狀況務必處於良好的工作狀態。
保險絲電源輸入模組包含兩條位於高電壓輸入線路的輸入保險絲。
保險絲尺寸為5公釐×20公釐,額定為10安培,250VAC ,緩熔。
高溫表面安全警告請勿在移除任何面板的情況下操作儀器。
環境注意事項將實驗室溫度保持在15°C 至30°C 之間(22.5°C ±7.5°C )。
運行期間,不可讓周圍溫差大於±2°C 。
濕度將非冷凝相對濕度保持在20至80%之間。
IndexedSequencingOverviewGuide
Introduction3Single-IndexedSequencingOverview3Dual-IndexedSequencingOverview4Dual-IndexedWorkflowonaPaired-EndFlowCell4Dual-IndexedWorkflowonaSingle-ReadFlowCell6SequencingPrimersforHiSeqSystems8RevisionHistory9TechnicalAssistance11
Document#15057455v03February2017ILLUMINAPROPRIETARYForResearchUseOnly.Notforuseindiagnosticprocedures.ThisdocumentanditscontentsareproprietarytoIllumina,Inc.anditsaffiliates("Illumina"),andareintendedsolelyforthecontractualuseofitscustomerinconnectionwiththeuseoftheproduct(s)describedhereinandfornootherpurpose.Thisdocumentanditscontentsshallnotbeusedordistributedforanyotherpurposeand/orotherwisecommunicated,disclosed,orreproducedinanywaywhatsoeverwithoutthepriorwrittenconsentofIllumina.Illuminadoesnotconveyanylicenseunderitspatent,trademark,copyright,orcommon-lawrightsnorsimilarrightsofanythirdpartiesbythisdocument.
Theinstructionsinthisdocumentmustbestrictlyandexplicitlyfollowedbyqualifiedandproperlytrainedpersonnelinordertoensuretheproperandsafeuseoftheproduct(s)describedherein.Allofthecontentsofthisdocumentmustbefullyreadandunderstoodpriortousingsuchproduct(s).
FAILURETOCOMPLETELYREADANDEXPLICITLYFOLLOWALLOFTHEINSTRUCTIONSCONTAINEDHEREINMAYRESULTINDAMAGETOTHEPRODUCT(S),INJURYTOPERSONS,INCLUDINGTOUSERSOROTHERS,ANDDAMAGETOOTHERPROPERTY.
ILLUMINADOESNOTASSUMEANYLIABILITYARISINGOUTOFTHEIMPROPERUSEOFTHEPRODUCT(S)DESCRIBEDHEREIN(INCLUDINGPARTSTHEREOFORSOFTWARE).
©2017Illumina,Inc.Allrightsreserved.Illumina,BaseSpace,HiSeq,MiniSeq,MiSeq,Nextera,NextSeq,NovaSeq,TruSeq,thepumpkinorangecolor,andthestreamingbasesdesignaretrademarksofIllumina,Inc.and/oritsaffiliate(s)intheU.S.and/orothercountries.Allothernames,logos,andothertrademarksarethepropertyoftheirrespectiveowners.
Document#15057455v03ForResearchUseOnly.Notforuseindiagnosticprocedures.2
IndexedSequencingOverviewGuideIntroductionThisguideprovidesanoverviewofindexedsequencingforallIlluminasequencingsystems.Indexedsequencingisamethodthatallowsmultiplelibrariestobepooledandsequencedtogether.
Indexinglibrariesrequirestheadditionofauniqueidentifier,orindexsequence,toDNA samplesduringlibrarypreparation.BaseSpaceSequenceHub,LocalRunManager,orstandalonebcl2fastq2processthesetagstoidentifyeachuniquelytaggedlibraryfordownstreamanalysis.
SingleandDualIndexingThenumberofindexsequencesaddedtosamplesdiffersforsingle-indexedanddual-indexedsequencing.uSingle-indexedlibraries—Addsupto48unique6-baseIndex1(i7)sequencestogenerateupto48uniquelytaggedlibraries.
uDual-indexedlibraries—Addsupto24unique8-baseIndex1(i7)sequencesandupto16unique8-baseIndex2(i5)sequencestogenerateupto384uniquelytaggedlibraries.
Duringindexedsequencing,theindexissequencedinaseparateread,calledtheIndexRead,whereanewsequencingprimerisannealed.Whenlibrariesaredual-indexed,thesequencingrunincludes2additionalreads,calledtheIndex1ReadandIndex2Read.
Single-IndexedSequencingOverviewThesingle-indexedsequencingworkflowappliestoallIlluminasequencingplatforms,whereanIndexReadfollowsRead1.
Figure1Single-IndexedSequencing
1Read 1—Read 1followsthestandardRead1sequencingprotocolusingSBSreagents.TheRead1sequencingprimerisannealedtothetemplatestrandduringtheclustergenerationstep.
2IndexReadpreparation—TheRead 1productisremovedandtheIndex1(i7)sequencingprimerisannealedtothesametemplatestrand,producingtheIndex1(i7)Read.
3Index1(i7)Read—FollowingIndexReadpreparation,theIndex1(i7)Readisperformed.Thereadlengthdependsonthesystemandrunparameters.
Document#15057455v03ForResearchUseOnly.Notforuseindiagnosticprocedures.3
IndexedSequencingOverviewGuide4Read 2resynthesis—TheIndexReadproductisremovedandtheoriginaltemplatestrandisusedtoregeneratethecomplementarystrand.Then,theoriginaltemplatestrandisremovedtoallowhybridizationoftheRead2sequencingprimer.
5Read 2—Read 2followsthestandardpaired-endsequencingprotocolusingSBS reagents.
Dual-IndexedSequencingOverviewDual-indexedsequencingincludes2indexreadsafterRead1:theIndex1ReadandtheIndex2Read.SequencingkitsforHiSeq®systemsareavailablewithasingle-readorpaired-endflowcell.Forallothersystems,sequencingkitsincludeapaired-endflowcell.
Dual-IndexingWorkflowsThecontrolsoftwareperformsRead1,anyindexreads,andthenRead2basedontheparametersprovidedfortheruninthesamplesheetorduringrunsetup.
Forallindexingworkflows,theIndex1ReaddirectlyfollowsRead1.However,fordual-indexingonapaired-endflowcell,therestoftheworkflowdiffers:
uWorkflowA—TheIndex2ReadisperformedbeforeRead2resynthesis,sotheIndex2(i5)adapterissequencedontheforwardstrand.