RNA-directed DNA methylation
- 格式:pdf
- 大小:593.20 KB
- 文档页数:37

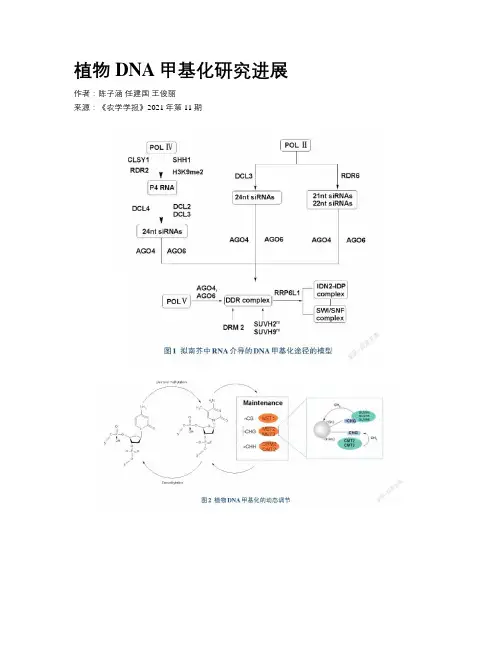
植物DNA甲基化研究进展作者:陈子涵任建国王俊丽来源:《农学学报》2021年第11期摘要:DNA甲基化是一種重要的表观遗传修饰,能够有效调控基因组稳定性。
为了了解DNA甲基化对植物生长发育的影响,本文归纳了近年来植物DNA甲基化的模式,总结了植物DNA甲基化的生物学功能,概括了DNA甲基化的研究方法,最后总结了植物DNA甲基化研究中存在的问题,并指明了研究方向,为后续植物基因组研究提供理论依据。
关键词:植物;DNA甲基化;表观遗传;修饰;生长发育;逆境胁迫;基因组;稳定性中图分类号:S184文献标志码:A论文编号:cjas2020-0152Research Advances on Plant DNA MethylationChen Zihan, Ren Jianguo, Wang Junli(School of Public Health, the key Laboratory of Enviromental Pollution Monitoring and Disease Control,Ministry of Education, Guizhou Medical University, Guiyang 550025, Guizhou, China)Abstract: DNA methylation is an important epigenetic modification that can effectively regulate genome stability. In order to understand the impact of DNA methylation on plant growth and development, this article summarizes plant DNA methylation patterns, concludes the physiological functions of plant DNA methylation, and reviews the research methods of DNA methylation. At last, this article sums up the problems in the study of plant DNA methylation and points out the research directions in the future, providing a theoretical basis for subsequent plant genome research.Keywords: Plants; DNA methylation; Epigenetic; Modification; Growth and Development; Adversity Stress; genome; stability0引言DNA甲基化(DNA methylation)是目前表观遗传学研究较为清晰的机制之一,广泛存在于生物界中,是真核细胞中最为常见的一种基因组修饰方式,它在调节基因组功能的同时不改变DNA的碱基序列。



RNA 干扰技术及其应用摘要:RNA 干扰(RNAinterference,RNAi)是由双链 RNA 分子在 mRNA 水平关闭相应序列基因表达或使其沉默的过程。
RNA 干扰技术作为新兴的基因阻断技术具有明显优势,已经广泛地被应用于基因功能研究、抗肿瘤和抗病毒等热门领域。
关键词:Rna 干扰技术生物学功能作用机理应用前言:基因沉默是指生物体中特定基因由于种种原因不表达或者是表达减少的现象。
基因沉默现象首先在转基因植物中发现,接着在线虫、真菌、水螅、果蝇以及哺乳动物中陆续发现。
基因沉默主要发生在两种情况,一种是转录水平上的基因沉默,另一种是转录后基因沉默。
RNA干扰是近几年发展起来的转录后基因阻断技术,RNAi在2002年被Science评为全球十大科技突破之一,作为一种在细胞水平的基因敲除工具,RNAi正在功能基因组学领域掀起一场革命。
一.简介RNA 干扰是通过双链 RNA介导,特异性地降解靶 mRNA,使同源的靶基因发生沉默,即序列特异性转录后基因沉默,从而诱使细胞表现出特定基因缺失的表型。
RNAi 现象普遍存在于从真菌到植物、从无脊椎动物到哺乳动物的各种生物中[1-3]如真菌的静息作用、植物的转录后基因沉默、动物的 RNAi,虽然在不同物种中的名称不同,但其分子机制极其相似.由于 RNAi 潜在的重要意义和近几年来的研究进展,Science 在 2001 年将其列为十大科学成就之一,2002 年又将其列为当年十大科学成就之首[4]。
RNAi 作为基因沉默的一个工具,已被广泛应用于研究基因功能、基因治疗、病毒防治等方面。
二.RNA 干扰生物学功能RNA 干扰是植物、线虫、真菌、昆虫、原生动物以及高等动物在进化中形成的一种内在基因表达的调控机制,在生物体的生长发育及防御系统的构成等方面均具有十分重要的作用[5]。
(一) 防止病毒感染所有复杂生物的基因组在长期的进化过程中均面临着病毒等外来核酸序列的侵入,如人类基因组约有 54%的序列含有这种入侵留下的遗迹。


转录水平siRNA介导的基因沉默左刚毛建平*(军事医学科学院放射和辐射医学研究所北京100850)摘要siRNA(small interference RNA)在转录后水平有效地沉默基因表达的现象称为RNAi(RNA interference,RNA干涉),传统认为它能在转录后水平上有效地沉默基因的表达(post transcriptional gene silence,PTGS)。
然而,最近实验研究表明,siRNA是一些甲基化转移酶活化的起始信号,在siRNA的作用下,甲基化转移酶使DNA区域包括胞嘧啶 鸟嘌呤核苷酸连续区(称为CG岛),CNG(N:A/T/C/G),CHH(H:A/C/T)中的胞嘧啶核苷C和组蛋白H3亚基第9个赖氨酸K(lysine residue K9in histone H3,H3K9)发生甲基化,基因表达为此而受到抑制,即siRNA能在转录水平调控基因的表达(transcriptional gene silence,TGS).同时,siRNA在异染色质的形成中也起着重要的作用,RNAi突变会激活原异染色质区域中沉默基因的表达。
关键词siRNA表观遗传RdRM异染色质收稿日期:2005 08 22修回日期:2005 09 21*通讯作者,电子信箱:maojp@表观遗传学(epigenetics)主要研究非基因序列改变所致基因表达水平的变化,包括DNA甲基化和组蛋白甲基化、乙酰化、磷酸化和泛素化等修饰导致的染色质重塑[1]。
DNA甲基化及其相应组蛋白甲基化引起的异染色质的形成是表观遗传学研究的首要内容。
DNA甲基化一方面在细胞发育过程中对基因表达调节起着重要作用;另一方面,它与肿瘤等疾病的发生发展也有着极为密切的关系,特别是抑癌基因CG岛的甲基化问题在近年来愈来愈受到重视。
RNAi是由双链RNA(dsRNA)诱导的在植物、动物和许多真菌中均存在的序列特异性基因沉默现象。
自Fire等[2]在线虫(C.elegans)中发现该现象以来,RNAi已成为基因功能研究的主要工具之一。


研究植物基因功能的策略和方法研究植物基因功能主要有两种策略:正向遗传学(forward genetics)和反向遗传学(reverse genetics)策略。
正向遗传学即通过生物个体或细胞基因组的自发突变或人工诱变,寻找相关表型或性状改变,然后通过图位克隆并结合一些基因差异表达筛选技术(如差减杂交、差异显示PCR、差异显示分析等)从这些特定性状变化的个体或细胞中找到对应的突变基因,并揭示其功能,例如遗传病基因的克隆。
反向遗传学的原理正好相反,人们首先是改变某个特定的基因或蛋白质,然后再去寻找与之有关的表型变化,例如基因剔除技术或转基因研究。
简单地说,正向遗传学是从表型变化研究基因变化,而反向遗传学则是从基因变化研究表型变化。
研究植物体内基因功能的方法主要有以下几种:(1)基因功能丧失或减少,即筛选目的基因功能部分丧失或全部丧失的突变体,比较其与野生型的表型差异来确定该基因功能;(2)基因功能增加或获得,即筛选目的基因高水平表达的植株,比较其与相应对照植株(野生型植株,功能丧失突变体或模式植物植株)差异,观察其表型性状变化来鉴定基因功能;(3)基因异位表达(Ectopic expression),通过定向调控靶基因的时空表达模式来研究基因功能;(4)微阵列(Microarray)是一种在全基因组水平对基因表达进行高通量检测的技术;(5)酵母双杂交技术(Yeast two-hybrid system)用于分析基因产物即蛋白质之间的互作。
1 基因功能丧失或减少以前,通常通过筛选自然突变体来获得基因功能部分或全部丧失的突变体,但概率较低;现在一般通过各种人工方法来获得合适突变体。
人工产生基因功能丧失的方法有插入突变、反义抑制(antisense suppression)、共抑制(cosuppression)、双链RNA干扰(double-stranded RNA interference, dsRNAi)。

分子遗传学复习题1.名词解释:DNA甲基化(DNA methylation):是指由DNA甲基化转移酶介导,催化甲基基团从S-腺苷甲硫氨酸向胞嘧啶的C-5位点转移的过程。
ENCODE计划(The Encyclopedia of DNA Elements Project):即“DNA元件百科全书计划”,简称ENCODE计划,是在完成人类基因组全序列测定后的2003年9月由美国国立人类基因组研究所(National Human Genome Research Institute,NHGRI)组织的又一个重大的国际合作计划,其目的是解码基因组的蓝图,鉴定人类基因组中已知的和还不知功能的多个物种的保守序列等在内的所有功能元件。
ENCODE计划的实施分为3个阶段:试点阶段(a pilot phase)、技术发展阶段(a technology development phase)和生产阶段(a producttion phase)。
gRNA (guide RNA):既指导”RNA(gRNA,guide RNA),能通过正常的碱基配对途径,或通过G—U配对方式与mRNA上的互补序列配对,指导编辑的进行。
GT--AG规律(GT-AG rule):真核生物所有编码蛋白质的结构基因,其RNA前体在内含子和外显子交界处有两个较短的保守序列,内含子的左端均为GT,右端均为AG,此规律称GT-AG规律。
miRNA:即小RNA,长度为22nt左右,5′端为磷酸基团、3′端为羟基。
miRNA广泛存在于真核生物中,不具有开放阅读框架,不编码蛋白质,其基因的转录产物是发夹状结构,在RNaseⅢ酶切后以双链形式存在,是近几年在真核生物中发现的一类具有调控功能的非编码RNA,它们主要参与基因转录后水平的调控。
RNA编辑(RNA editing) :是指通过碱基修饰、核苷酸插入或删除以及核苷酸替换等方式改变RNA的碱基序列的转录后修饰方式。

siRNA介导的染色质基因沉默摘要探讨了siRNA在染色质调控中的作用,指出siRNA可使靶DNA发生甲基化并参与异染色质的形成以抑制基因表达。
关键词异染色质;siRNA;RNAiTheGeneSilenceingonsiRNAMediatedby chromationREN LiZHANG Mei-pingSUN Chun-yuJIANG Shi-cuiWANG Yi*(Jilin Agriculture University,ChangchunJilin 130118)AbstractThe effect of siRNA on chromatin regulation was studied. It was pointed out that RNA interfering could make targeted DNA methylation and participates formation of heterochromatin to inhibit gene expression.Key wordsheterochromatin;siRNA;RNA interferenceRNA干扰(RNA interference,RNAi)是近期发现的一种新的基因调控机制,研究发现同源dsRNA引发的基因沉默在生物体中是一种普遍现象[1]。
随着研究的深入,染色质水平的调控已成为研究热点,生物的染色质可以分为转录活性区的常染色质以及低转录水平的异染色质。
真核异染色质通常包括大量重复序列和转座子等,外源基因在异染色质区域的插入会导致此基因的沉默而不会被转录。
染色质水平的基因沉默主要依赖组蛋白H3的去乙酰化以及其第9位赖氨酸的甲基化,使得甲基化的第9位赖氨酸能够结合异染色质蛋白,从而终止转录。
RNAi中的重要因子siRNA不仅可在mRNA水平上抑制靶基因表达,还可在染色质水平上沉默基因[2],而后者近来更为人们所关注。
分子遗传学分子遗传学复习重点名词解释:RNA编辑:mRNA因核苷酸的插入、缺失或替换而改变了源自DNA模板的遗传信息,翻译出不同于基因编码的氨基酸序列,称为RNA编辑(RNA editing)C值及C值悖论:生物体的单倍体基因组所含DNA的总量称为C 值。
生物基因组的大小同生物在进化上所处地位的高低及复杂性之间无严格的对应关系,这种现象通常称为C值悖理假基因:核苷酸序列与相应正常功能基因基本相同,但没有编码蛋白质能力的基因或不产生有功能产物的基因RNA干涉(RNA interference,RNAi)是正常生物体内一些小的双链RNA,可有效地阻断靶基因表达的现象。
当向细胞中导入与内源性mRNA同源的双链RNA (double stranded RNA,dsRNA)小分子时,可导致该mRNA降解,从而高效、特异的阻断体内特定基因的表达,导致基因沉默。
转座子:是存在于染色体DNA上可自主复制和转位的基本单位。
程序性细胞死亡(PCD):多细胞生物体的一些细胞当不再为生物体所需或是已受到损伤时,会激活受遗传控制的自杀机构而自我毁灭。
抗原:一类能诱导机体发生免疫应答并能与相应的应答产物(如抗体)发生特异性免疫反应的大分子物质。
又称免疫原半抗原:缺乏免疫原性而有免疫反应性的物质。
抗体:在抗原物质的刺激下,由浆细胞产生的一类能与相应抗原在体内外发生特异性结合的免疫球蛋白DNA甲基化:在DNA甲基转移酶的催化下,利用S-腺苷蛋氨酸提供的甲基,将胞嘧啶第5位碳原子甲基化,从而使胞嘧啶转化为5甲基胞嘧啶。
遗传图谱:又称遗传连锁图,是指基因或DNA标记在染色体上的相对位置与遗传距离。
物理图谱:是指各遗传标记之间或DNA序列两点之间,以物理距离来表示其在DNA分子上的位置而构成的位置图,以实际的碱基对(bp)或千碱基对(Kb)或百万碱基对(Mb)长度来度量其物理距离。
Kazak序列:许多真核生物mRNA的5'端起始密码子附近有一段短的保守序列,可促进核糖体小亚基识别起始密码子,该序列为(GCC)RCCA TGG.miRNA:即小RNA,长度为22nt左右,5'端为磷酸基团,3'端为羟基。
基因组学1. 简述基因组的概念和其对生命科学的影响。
基因组:指一个物种的全套染色体和基因。
广义的基因组:核基因组,线粒体基因组,叶绿体基因组等。
基因组计划对生命科学的影响:①研究策略的高通量,彻底认识生命规律:基因组研究高通量,研究手段和研究策略的更新,加强了生命科学研究的分工与协作,从不同层次深入研究生命现象。
②促进了相关学科的发展:分子生物学遗传学生物信息学生物化学细胞生物学生理学表观遗传学等③物种的起源与进化:Ⅰ.重要基因的发掘、分离和利用:遗传疾病相关基因,控制衰老的基因,工业价值的细菌基因,重要农艺性状基因等。
Ⅱ.充分认识生命现象:基因的表达、调控,基因间的相互作用,不同物种基因组的比较研究,揭示基因组序列的共性,探讨物种的起源和进化。
④伦理学法律问题:伦理问题,知识产权问题,法律问题,社会保险问题。
2. Ac/Ds转座因子Ac因子有4563bp,它的大部分序列编码了一个由5个外显子组成的转座酶基因,成熟的mRNA有3500bp。
该因子本身的两边为11bp的反向重复末端(IR),发生错位酶切的靶序列长度8bp。
Ds因子较Ac因子短,它是由Ac因子转座酶基因发生缺失而形成的。
不同的Ds因子的长度差异由Ac因子发生不同缺失所致。
Ac/Ds因子转座引起的插入突变方式:玉米Bz基因是使糊粉层表现古铜色的基因,当Ac/Ds转座插入到Bz基因座后,糊粉层无色。
当Ac/Ds因子在籽粒发育过程,部分细胞发生转座,使Bz靶基因发生回复突变,从而形成斑点。
Ac/Ds两因子系统遗传特点:1)Ac具有活化周期效应,有活性的Ac+因子被甲基化修饰后会形成无活性的ac-因子,反之无活性的ac-因子去甲基化成有活性的Ac+因子。
2)Ac与Ds因子有时表现连锁遗传但更多表现独立遗传。
3)Ac对Ds的控制具有负剂量效应。
4)Ac/Ds可引发靶基因表现为插入钝化、活性改变、表达水平改变和缺失突变等。
5)Ds的结构不同,插入同一靶基因的位点可能不同,形成的易变基因的表型也不同。
DNA甲基化动态:发生、保持和去甲基化一、甲基化DNA 甲基化是最早发现的基因表观修饰方式之一,真核生物中的甲基化仅发生于胞嘧啶,即在DNA 甲基化转移酶(DNMTs )的作用下使CpG 二核苷酸5’ -端的胞嘧啶转变为5’-甲基胞嘧啶。
DNA 甲基化通常抑制基因表达,去甲基化则诱导了基因的重新活化和表达。
中文名甲基化检测外文名detection of methylation目录1. 1 甲基化检测2.2检测程序3.3送样要求甲基化检测编辑DNA 甲基化是最早发现的基因表观修饰方式之一,真核生物中的甲基化仅发生于胞嘧啶,即在DNA 甲基化转移酶(DNMTs )的作用下使CpG 二核苷酸5’ -端的胞嘧啶转变为5’-甲基胞嘧啶。
DNA 甲基化通常抑制基因表达,去甲基化则诱导了基因的重新活化和表达。
这种DNA 修饰方式在不改变基因序列前提下实现对基因表达的调控。
脊椎动物DNA 的甲基化状态与生长发育调控密切相关,比如在肿瘤发生时,抑癌基因CpG 岛以外的CpG 序列非甲基化程度增加,CpG 岛中的CpG 则呈高度甲基化状态,导致抑癌基因表达的下降。
检测程序编辑1.甲基化特异性的PCR ( Methylation-specific PCR ,MSP )用亚硫酸氢盐处理基因组DNA,所有未发生甲基化的胞嘧啶被转化为尿嘧啶,而甲基化的胞嘧啶不变;随后设计针对甲基化和非甲基化序列的引物进行PCR 。
通过电泳检测MSP扩增产物,如果用针对处理后甲基化DNA 链的引物能得到扩增片段,则说明该位点存在甲基化;反之,说明被检测的位点不存在甲基化。
2.亚硫酸氢盐测序法( Bisulfite sequencing PCR ,BSP )用亚硫酸氢盐处理基因组DNA,则未发生甲基化的胞嘧啶被转化为尿嘧啶,而甲基化的胞嘧啶不变。
随后设计BSP 引物进行PCR ,在扩增过程中尿嘧啶全部转化为胸腺嘧啶,最后对PCR 产物进行测序就可以判断CpG 位点是否发生甲基化称为BSP- 直接测序方法。
RNAi技术1. RNAi定义RNA干扰(RNA interference,RNAi)[1]是由双链RNA(double stranded RNA,dsRNA)分子在mRNA水平关闭相应序列基因表达或使其沉默的过程。
dsRNA可以抑制不同类型细胞的靶向基因表达,用特异性的抗体几乎检测不到靶向基因所表达的蛋白质。
因此,RNAi 技术又被形象地称为基因敲除(knock out)或基因沉默(gene silencing)。
RNAi是一种典型的转录后基因调控方法,又称转录后基因沉默(post transcriptional gene silencing,PTGS)。
RNAi现象广泛存在于生物界中。
它在真菌中被称为quelling,在拟南芥、烟草等植物中被称为PTGS或共抑制(cosuppression),在无脊椎动物如果蝇、水螅、线虫以及脊椎动物斑马鱼、小鼠等动物界中被称为RNAi。
PTGS可能是生物界,包括植物和动物等普遍存在的一种古老、保守而又极其重要的遗传行为。
RNA 干涉是生物基因组抵抗转座子或病毒之类的外来遗传元件入侵的一种保护性机制,具有以下4 个重要特征:①RNAi 属于转录后水平的基因沉默(PTGS) 机制,迄今为止的研究认为RNAi 对染色体DNA 序列的复制和转录过程不产生任何影响;②具有高度的序列特异性,最近将人工合成的siRNAs (短小RNA,是RNi 过程中直接起作用的分子)转入宿主细胞的实验表明,即使siRNAs 中只有一个碱基与靶序列错配,也会使干涉效应大大减弱;③具有抑制基因表达的高效性, 可以引起该基因缺失突变的表型;而且远远少于内源mRNA 数量的dsRNA 就可以实现完全的抑制效应, 即其发生过程中存在着正反馈的信号放大效应;④RNAi 的效应可以在不同细胞间长距离传递和维持,而且在某些生物中(比如线虫) 具有遗传性。
2. RNAi的发现与探索1995年康乃尔大学的Guo[2]博士在实验中想通过反义RNA阻断美丽线虫(C.elegans)的par-1基因表达,同时她还用正义RNA做了一个对照,试图观察到对照试验组基因表达增强的现象,但结果却发现了反义和正义RNA都阻断了该基因的表达。
分子遗传学复习题1.名词解释:DNA甲基化(DNA methylation):是指由DNA甲基化转移酶介导,催化甲基基团从S-腺苷甲硫氨酸向胞嘧啶的C-5位点转移的过程。
ENCODE计划(The Encyclopedia of DNA Elements Project):即“DNA元件百科全书计划”,简称ENCODE计划,是在完成人类基因组全序列测定后的2003年9月由美国国立人类基因组研究所(National Human Genome Research Institute,NHGRI)组织的又一个重大的国际合作计划,其目的是解码基因组的蓝图,鉴定人类基因组中已知的和还不知功能的多个物种的保守序列等在内的所有功能元件。
ENCODE计划的实施分为3个阶段:试点阶段( a pilot phase)、技术发展阶段(a technology development phase)和生产阶段(a producttion phase)。
gRNA (guide RNA):既指导”RNA(gRNA,guide RNA),能通过正常的碱基配对途径,或通过G—U 配对方式与mRNA上的互补序列配对,指导编辑的进行。
GT--AG规律(GT-AG rule):真核生物所有编码蛋白质的结构基因,其RNA前体在内含子和外显子交界处有两个较短的保守序列,内含子的左端均为GT,右端均为AG,此规律称GT-AG 规律。
miRNA:即小RNA,长度为22nt左右,5′端为磷酸基团、3′端为羟基。
miRNA广泛存在于真核生物中,不具有开放阅读框架,不编码蛋白质,其基因的转录产物是发夹状结构,在RNaseⅢ酶切后以双链形式存在,是近几年在真核生物中发现的一类具有调控功能的非编码RNA,它们主要参与基因转录后水平的调控。
RNA编辑(RNA editing) :是指通过碱基修饰、核苷酸插入或删除以及核苷酸替换等方式改变RNA的碱基序列的转录后修饰方式。
3RNA-directed DNA methylationMarjori Matzke,Tatsuo Kanno,Bruno Huettel,Estelle Jaligot, M.Florian Mette,David P.Kreil,Lucia Daxinger,Philipp Rovina,Werner Aufsatz and Antonius J.M.Matzke3.1Introduction3.1.1RNA interferenceRNA interference(RNAi)has provided a new paradigm for understanding gene regulation in many eukaryotic organisms.Although plant scientists laid much of the groundwork for the discovery of RNAi(Matzke&Matzke,2004), it was the identification of double-stranded RNA as the trigger for gene silencing in Caenorhabditis elegans by Fire et al.(1998)that provided the means to downregulate gene expression reproducibly in plants,animals and many fungi.The canonical RNAi pathway is now well established:double-stranded RNA is processed by a ribonuclease III–type enzyme called Dicer into short interfering RNAs(siRNAs)of around21–24nt in length.The siRNAs associate with the RNA-induced silencing complex(RISC)and guide cleavage of complementary mRNAs(Novina&Sharp,2004).A second type of small RNA,the microRNA(miRNA),is also produced via Dicer cleavage of longer duplex RNAs.The miRNAs associate with an RISC-like complex and,depending on the degree of complementarity to the target mRNA,elicit either mRNA cleavage or translational repression.In addition to Dicer,other core proteins of the RNAi machinery include Argonaute,an RISC component that binds small RNAs and in some cases executes mRNA cleavage,and RNA-dependent RNA polymerase(RDR),which can synthesize double-stranded RNA from single-stranded RNA templates to initiate or amplify the RNAi reaction(Bartel,2004;He&Hannon,2004).In addition to making substantial contributions to elucidating the classical RNAi pathway,plant scientists have been at the forefront of research on RNA-mediated epigenetic modifications.Epigenetics refers to mitotically and/or meiotically heritable changes in gene expression that do not involve a change in DNA sequence.Epigenetic modifications include DNA cytosine(C)methy-lation and various posttranslational modifications of histones,including acet-ylation and methylation.The connection between RNAi and epigenetic regulation is exemplified by the phenomenon of RNA-directed DNA methy-lation(RdDM),which is the subject of this chapter.70PLANT EPIGENETICS3.1.2Discovery and characteristics of RNA-directed DNA methylation Originally detected in viroid-infected tobacco plants,RdDM was the first RNA-guided epigenetic modification of the genome to be reported(Wasse-negger et al.,1994).Viroids are minute plant pathogens consisting exclusively of a non-protein-coding,circular rod-shaped RNA several hundred base pairs in length(Tabler&Tsagris,2004).In the experiments that revealed RdDM,viroid cDNAs integrated as transgenes into tobacco chromosomes became methylated de novo during RNA–RNA replication of the cognate viroid.Further analysis demonstrated that Cs in all sequence contexts are modified and that methylation is largely confined to the region of RNA–DNA sequence homology(Pe´lissier et al.,1999).DNA regions as short as30bp can be targeted for methylation in the RdDM pathway(Pe´lissier& Wassenegger,2000).The identification of RdDM occurred in the framework of homology-dependent gene-silencing phenomena,which were postulated at the time to be due variously to DNA–DNA,DNA–RNA or RNA–RNA interactions (Jorgensen,1992;Matzke&Matzke,1993).Indeed,one of the initial papers describing‘cosuppression’,a process in which a resident gene is silenced in the presence of a homologous transgene,proposed that transgene RNA might interact with DNA of the resident gene to block transcription(van der Krol et al.,1990).Even though cosuppression turned out to be a posttran-scriptional gene-silencing phenomenon that is now considered the plant equivalent of RNAi(De Carvalho et al.,1992;Matzke&Matzke,2004; van Blokland et al.,1994),this example illustrates that RNA–DNA inter-actions were considered as a possible trigger of silencing during the early stages of gene-silencing research.The viroid experiments supplied experi-mental documentation of this possibility and revealed the characteristic features of RdDM.Shortly thereafter,cosuppression(also known as post-transcriptional gene silencing(PTGS))of a reporter transgene in tobacco was shown to be accompanied by de novo methylation of the corresponding genomic DNA sequences(Ingelbrecht et al.,1994).Thus,a transgene subject to posttranscriptional regulation could also be targeted for DNA methylation, suggesting that cytoplasmic and nuclear events are induced by a common silencing mechanism.A further connection between PTGS and DNA methylation was observed in transgenic pea plants infected with an RNA virus that replicates exclusively in the cytoplasm.Virus replication initiated PTGS of a transgene encoding the viral replicase gene,which was accompanied by de novo methylation of replicase transgene sequences integrated into nuclear DNA(Jones et al., 1998).The pattern of methylation was consistent with RdDM,occurring at Cs in both symmetrical(CG and CNG,where N is A,T or C)and asymmet-rical(CNN)sequence contexts within the region of RNA–DNA sequenceRNA-DIRECTED DNA METHYLATION71 homology.These findings suggested that a sequence-specific signal,most likely RNA,produced during virus replication and/or PTGS diffused from the cytoplasm into the nucleus to induce methylation of homologous DNA sequences.In an extension of these experiments,replicating potato virus X RNA vectors modified to contain sequences homologous to the35S promoter were able to trigger methylation and transcriptional silencing of35S pro-moter–driven nuclear transgenes(Jones et al.,1999).The experiments with viroids and viruses hinted that RdDM is initiated by a double-stranded RNA because these RNA pathogens replicate via a double-stranded RNA intermediate.An unequivocal demonstration that double-stranded RNA is required for RdDM came from studies on a non-pathogenic,transgenic system.These experiments grew from work that suggested a role for RNA in mediating methylation of transgene promoters (Mette et al.,1999;Park et al.,1996).To further investigate this possibility, RNAs that contained promoter sequences were tested for their ability to induce methylation and transcriptional silencing of unlinked homologous promoters in transgenic tobacco plants.Cre-lox-mediated recombination was used to convert a transcribed direct repeat of target promoter sequences into a tran-scribed inverted repeat in planta.Methylation and silencing of a homologous target promoter was observed only after generation of the inverted repeat and initiation of double-stranded RNA synthesis(Mette et al.,2000).The double-stranded RNA that induced RdDM was shown to be processed to short RNAs 21–24nt in length,similar to those involved in RNAi,reinforcing a link between the two silencing processes.Double-stranded RNAs containing promoter sequences have subsequently been used to silence and methylate different transgene promoters in Arabidopsis thaliana(Aufsatz et al.,2002a, 2002b;Kanno et al.,2004)and several endogenous promoters in petunia (Sijen et al.,2001)and Arabidopsis(Melquist&Bender,2003).Therefore, RNA-mediated transcriptional gene silencing(TGS)and promoter methyla-tion appears to be a general process in plants.3.2RNAi-mediated pathways in the nucleusViewed as a plant-specific phenomenon for several years after its discovery, RdDM is now regarded as one of several RNAi-mediated pathways in the nucleus.In addition to RdDM,the other nuclear pathways include:(1)RNAi-mediated heterochromatin formation that has been observed infission yeast,plants,Drosophila melanogaster and vertebrates;(2)elimination of intergenic DNA that has been packaged into heterochro-matin via the RNAi pathway during nuclear differentiation in Tetrahy-mena thermophila and other ciliated protozoa;72PLANT EPIGENETICS(3)meiotic silencing of DNA that is unpaired during meiosis in Neurosporacrassa and C.elegans(reviewed by Matzke&Birchler,2005).Here we focus on RdDM and on RNAi-mediated heterochromatin formation, particularly as it relates to DNA methylation.3.2.1RNAi-mediated heterochromatin formationThe involvement of the RNAi pathway in heterochromatin formation was first discovered in fission yeast(Schizosaccharomyces pombe).Mutants defective in the core RNAi proteins–Dicer,Argonaute and RDR–were shown to be unable to assemble functional heterochromatin at the centromeres(Volpe et al., 2002,2003).In wild-type cells,transient double-stranded RNA molecules originating from forward and reverse transcription of centromere outer repeats are processed by Dicer to siRNAs(Reinhart&Bartel,2002).The siRNAs are thought to guide histone H3lysine9(H3K9)methylation that is catalyzed by cryptic loci regulator4(Clr4),the S.pombe ortholog of the histone methyl-transferase Su(var)3–9(from Drosophila suppressor of variegation3–9).Swi6, the S.pombe ortholog of Drosophila heterochromatin protein1(HP1)can bind via its chromodomain to histone H3methylated on K9,which promotes lateral spreading of heterochromatin from the siRNA-targeted nucleation site.The formation of heterochromatin leads to binding of cohesin,which fosters sister chromatid cohesion and proper chromosome segregation.Chromatin-associ-ated RDR primes RNA synthesis from the reverse transcript of the outer centromere repeat,which is continuously transcribed,ensuring a constant supply of double-stranded RNA even when transcription of the forward strand is repressed by heterochromatin formation(reviewed by Maison&Almouzni, 2004).A similar process occurs at the S.pombe silent mating type locus,which contains a copy of the centromeric outer repeat(Hall et al.,2002).In addition, synthetic hairpin RNAs can induce heterochromatin formation at the sites of genes that are normally euchromatic(Schramke&Allshire,2003).Another natural target of RNAi-mediated heterochromatin in S.pombe is retrotranspo-son long terminal repeats(LTRs)that are transcribed bidirectionally.Spread-ing of heterochromatin from LTRs into adjacent genes can contribute to cellular differentiation by silencing stage-specific gene expression(Schramke &Allshire,2003).A nuclear complex called RNA-induced initiation of transcriptional gene silencing(RITS)has been isolated,which directly links siRNAs to heterochromatin in S.pombe.The RITS complex contains siRNAs from known heterochromatic regions,such as centromeres,as well as Chp1,a chromodomain protein that binds centromeres,the S.pombe ortholog of Ago1, and Tas3,a serine-rich protein that is found only in S.pombe(Verdel et al., 2004).The RITS complex is tethered to silenced loci that contain H3K9RNA-DIRECTED DNA METHYLATION73 methylation.This tethering faciliates siRNA production and maintenance of heterochromatin(Noma et al.,2004).The RNAi-mediated heterochromatin pathway is conserved in animals. Heterochromatin in Drosophila,assessed by H3K9methylation and localiza-tion of the heterochromatin proteins HP1and HP2,is disrupted in mutants defective in piwi and aubergine,which are both members of the PAZ/piwi domain family that contains Argonaute,and in spindle-E(homeless),a DEAD-box RNA helicase important for RNAi(Pal-Bhadra et al.,2004).In vertebrate cells,formation of centromeric heterochromatin depends on functional Dicer activity.This was shown by generating a conditional loss-of-function Dicer mutant in chicken–human hybrid cells that contain human chromosome21. Aberrant accumulation of transcripts from human centromeric a-satellite repeats and delocalization of two heterochromatin proteins,Rad21cohesinand the mitotic checkpoint protein BubR1,were observed in the Dicer-deficient cells,which also displayed mitotic defects and frequently died in interphase(Fukagawa et al.,2004).The most comprehensive study on RNAi-mediated heterochromatin in plants has been carried out on the heterochromatic knob on chromosome4of A.thaliana.This work is considered in the context of RdDM(see Section3.2.2AQ1).3.2.2RdDM and RNAi-mediated heterochromatin assembly:one pathwayor two?While RdDM and RNAi-mediated heterochromatin are likely to be inter-related,it is not yet clear whether they are the outcomes of a single pathwayor two separate pathways.Both RdDM and RNAi-mediated heterochromatin formation are initiated by double-stranded RNAs that are substrates of Dicer cleavage,but they might diverge after this step to yield distinct primary epigenetic marks that differ in terms of chemistry,mitotic heritability and reversibility.Historically,RdDM has been studied from the angle of C methylation, which we assume is the primary epigenetic mark associated with this process.This assumption is based on the pattern of methylation,which is largely confined to the region of RNA–DNA sequence homology and which can encompass sequences as short as30bp.This contrasts to histone modificationsthat occur in a nucleosomal context that comprises147bp of DNA.Often referred to as the‘fifth base’in eukaryotic DNA(Hergersberg,1991),5-methylcytosine represents a covalent modification of the DNA molecule itself.By contrast,the primary epigenetic mark that is evaluated for RNAi-mediated heterochromatin formation involves covalent modification of histone proteins,most typically methylation of lysine9of histone H3,which is a hallmark of silent heterochromatin.C methylation is carried out by enzymes known as DNA methyltransferases.The cytosine-5-methyltransferases transfer a methyl group from S-adenosyl-l -methionine to carbon 5of C residues (Finnegan &Kovac,2000).When present in symmetrical CG and CNG nucleotide groups,methylation can be copied faithfully during DNA replication and passed on to daughter cells (see Figure 3.1).In addition to passive loss of methylation,which results from deficiencies in maintenance methylation,there is increasing evidence that CG methylation can be actively removed from non-replicating DNA through the activity of DNA glycosylases in plants (Choi et al .,2002;Gong et al .,2002;Kinoshita et al .,2004)and in vertebrates (reviewed by Kress et al .,2001).Thus,CG and CNG methylation are mitotically heritable but also potentially revers-ible epigenetic modifications.It is not yet clear how histone modifications can be inherited through rounds of DNA replication and cell divisions.Indeed,if the term ‘epigenetic’implies mitotic heritability,then some authors have questioned whether histone modifications fully qualify owing to uncertaintiesm m m Dnmt2 (?): vertebrates, plants,D. melanogaster , S. pombeRNAvs maintenance methylationRNADRM1, DRM2/Dnmt3a, b(MET1; CMT3?)de novo methylation AQ2Figure 3.1DNA methyltransferases catalyzing de novo and maintenance methylation.Plant enzymes are shown in bold.De novo methylation refers to methylation of a previously unmodified DNA sequence.One of the best-documented signals for de novo methylation is double-stranded RNA that can be processed by Dicer-like activity into short RNAs (dashed lines;not drawn to scale).DNA methyltransferases that are generally classified as de novo activities include mammalian Dnmt3a/b and the plant homologs,DRM1,DRM2.Methylation can be maintained in symmetrical CG (and in plants,CNG)nucleotide groups through subse-quent rounds of DNA replication (arrow)by the action of so-called maintenance methyltransferases,which recognize the methylated C in the parental strand and catalyze methylation of the opposite C in the newly synthesized strand.The CG maintenance activity in mammals is Dnmt1;the plant homolog is MET1.Plants have a special DNA methyltransferase,CMT3,that can maintain methylation in CNG trinucleotides.Recent work on RdDM indicates that MET1and CMT3can also participate in de novo methylation in the presence of RNA signals.Methylation in asymmetric CNNs cannot be maintained and requires the continuous presence of the triggering RNA.Members of the enigmatic Dnmt2class of DNA methyltransferases,whose function is not known,are present in vertebrates and plants and in organisms not normally thought to methylate their DNA,such as Drosophila melanogaster and fission yeast (Schizosaccharomyces pombe ).74PLANT EPIGENETICSabout their inheritance during mitosis (Bird,2002).Moreover,even though arginine methylation in histones can be altered by enzymes that convert methylated arginines to citrullines (Cuthbert et al .,2004;Wang et al .,2004),enzymes that can remove lysine methylation in histones have not yet been identified (Zhang,2004).Thus,despite an incomplete understanding of its mode of inheritance,H3K9methylation is thought to be a means for ensuring relatively stable,long-term silencing.RdDM and RNAi-mediated heterochromatin might be further distinguished by the manner in which short RNAs interact with the homologous target sequence (see Figure 3.2).The failure of RdDM to spread substantially beyond the region of RNA–DNA sequence homology hints that direct RNA–DNA base pairing provides a substrate for methylation.By contrast,models in which short RNAs base-pair to nascent RNAs transcribed from the target locus have been proposed for RNAi-mediated heterochromatin assembly (Grewal &Moazed,2003),which might somehow facilitate spreading beyond the siRNA-targeted nucleation site.DNA methylation can influence histone modifications and vice versa,but the order of events appears to vary depending on the system under investiga-tion (Lund &van Lohuizen,2004;Mutskov &Felsenfeld,2004;Tariq &Paszkowski,2004).For example,in N.crassa ,all DNA methylation is cata-lyzed by one DNA methyltransferase,DIM-2(Kouzminova &Selker,2001);this DNA methylation,however,is fully dependent on DIM-5,a histone H3K9methyltransferase (Tamaru &Selker,2001).In Neurospora ,therefore,histone methylation clearly precedes –and is a prerequisite for –DNA methylation.However,Neurospora might be exceptional.The structure of the DIM-2DNA methyltransferase differs from the four major families of DNAshort RNA short RNAnascent RNAdsDNA dsDNA RNA polymeraseRNA − RNARNA − DNAFigure 3.2Possible modes of interaction between short RNAs and the homologous target locus.Short RNAs can base-pair to a nascent RNA transcribed from the target locus (RNA–RNA)or to one strand of DNA (RNA–DNA).RNA-DIRECTED DNA METHYLATION 7576PLANT EPIGENETICS3.3.2.2)(Groll&Bestor,2005).Moreover, methyltransferase(see Section AQ3 unlike many other eukaryotes,Neurospora does not seem to use the RNAi machinery to guide epigenetic modifications(Chicas et al.,2004;Freitag et al., 2004).Instead,signals for de novo DNA methylation are generated by the unusual process of repeat-induced point mutation(RIP),which produces C:Gto T:A transition mutations in duplicated DNA regions by a poorly understood mechanism.‘RIPed’sequences are preferentially targeted for methylation, apparently because of their A:T richness and high density of TA dinucleotides (Tamaru&Selker,2003).In another example,silencing of the redundant X chromosome in female mammals appears to involve initially methylation of histone H3at Lys9andLys27,followed by DNA methylation(Okamoto et al.,2004).In Arabidopsis, there are conflicting reports about whether DNA methylation precedes (Malagnac et al.,2002;Soppe et al.,2002;Tariq et al.,2003)or follows (Gendrel et al.,2002;Jackson et al.,2002;Johnson et al.,2002)histone H3K9 methylation.Despite these uncertainties and differences among species,it is clear that RNAi-mediated H3K9methylation can be induced in the absence of detect-able DNA methylation,as is the case in S.pombe.Thus,the mechanisms of DNA methylation and H3K9methylation are not obligatorily coupled.It is conceivable that RdDM represents a distinct pathway that leads only second-arily to histone modifications.In this chapter,we discuss RdDM as a pathwayof RNA-guided de novo methylation in which histone modifications are imposed in later steps to maintain and/or reinforce C methylation.3.3Mechanism of RNA-directed DNA methylation:RNAand protein requirements3.3.1Systems used for genetic analyses of RdDMand transcriptional silencingGenetic investigations performed in our laboratory on RdDM and RNA-mediated transcriptional silencing have exploited well-defined,two-compon-ent transgene systems in A.thaliana.In these systems,methylation of a transgene promoter is induced experimentally by a double-stranded RNAthat is encoded by a second,unlinked transgene complex(Matzke et al., 2004).We have carried out genetic screens on two promoter systems.The nopaline synthase promoter(NOSpro)is a moderately active,constitutive plant promoter;the a’promoter(a’pro)is a strong seed-specific promoter. Both these promoters are several hundred base pairs in length and have the same overall GC content($45%)but the NOSpro contains about twice as many CG dinucleotides as the a’pro.Thus,one might anticipate a priori thatRNA-DIRECTED DNA METHYLATION77 NOSpro would be more sensitive to CG methylation than the a’pro.Indeed,as described below,the differences in sequence composition between the two promoters are a likely explanation for the different types of mutations that have been recovered for each system.Although transgenes provide well-defined and manipulatable systems for analysis,it is important to use the knowledge gained with them to understand RNA-mediated transcriptional regulation of endogenous genes.Parallel gen-etic analyses of several endogenous genes that are silenced(or likely to be silenced)by double-stranded RNA–such as SUPERMAN(SUP),PAI2and FWA–are proving particularly informative with regard to RdDM.Before discussing the results of genetic analyses,we briefly describe these three endogenous gene systems.SUP encodes a zinc finger transcription factor that is important for flower development(Ito et al.,2003).For reasons that are not understood,SUP becomes highly methylated and silenced in ddm1(decrease in DNA methyla-tion1)and methyltransferase1(met1)mutants that otherwise display genome-wide hypomethylation(Jacobsen et al.,2000).The silent loss-of-function ‘epialleles’of SUP,termed the clark kent(clk)alleles,are hypermethylated at Cs in all sequence contexts throughout the transcribed region and at the tran-scription start site(Jacobsen&Meyerowitz,1997).An approximately350-bp region near the5’end is rich in asymmetric CNNs and CNGs,but contains only one CG dinucleotide(Cao&Jacobsen,2002).To facilitate genetic analyses for the identification of loci important for methylation and silencing of SUP,a stable,non-reverting clk allele,clk-st,was created by introducing an additional SUP locus into clk-3plants(Lindroth et al.,2001).The extra SUP sequences at this locus are arranged as an inverted repeat(Cao&Jacobsen, 2002),which is probably transcribed to produce a double-stranded RNA that stabilizes silencing and methylation of the endogenous SUP gene.The tryptophan biosynthetic(phosphoribosylanthranilate isomerase,PAI) gene family in the Wassilewskija(WS)strain of Arabidopsis comprises four copies of the PAI gene that are densely methylated at CGs and non-CGs throughout their regions of sequence identity.Due to a naturally occurring duplication(perhaps generated by transposons),two copies–PAI1–PAI4–are arranged as an inverted repeat.PAI2and PAI3are unlinked singlet copies.In contrast,ecotype Columbia(Col)has three singlet,unmethylated PAI genes at the analogous loci(Melquist et al.,1999).In WS,only PAI1and PAI2encode functional enzymes but PAI1is the sole expressed copy because it is tran-scribed from an unrelated upstream promoter that is unmethylated.The PAI1–PAI4inverted repeat induces de novo methylation of unmethylated PAI sequences,apparently by encoding a double-stranded RNA that targets the singlet copies for methylation by RdDM(Melquist&Bender,2003,2004). Genetic screens have been carried out for suppressors of hypermethylation and silencing of the PAI2gene.78PLANT EPIGENETICSThe flowering Wageningen(FWA)gene(M.Koornneef,personal commu-nication)encodes a homeodomain transcription factor that displays imprinted (maternal origin–specific)expression in endosperm.Normally FWA is kept silent during the plant vegetative phase by methylation in two transposon-derived direct repeats in the promoter region(Soppe et al.,2000;Lippman et al.,2004).Loss of methylation from the maternal allele in the central cell of the gametophyte through the activity of the DNA glycosylase protein DEMETER leads to maternal expression only in endosperm(Kinoshita et al. 2004).Short RNAs originating from the direct repeats suggest that transcrip-tional repression of FWA occurs by an RNAi-mediated pathway(Lippman et al.,2004).In contrast to SUP,which becomes hypermethylated in ddm1and met1mutants,FWA becomes hypomethylated in these mutants,producing gain-of-function epialleles that condition a late flowering phenotype(Kaku-tani,1997).3.3.2Steps in the RdDM pathwayThe RdDM pathway can be divided into three steps:(1)synthesis and processing of double-stranded RNA;(2)de novo methylation of Cs in all sequence contexts in the presence of RNAsignals;(3)maintenance of CG and CNG methylation in the absence of RNA signals(or reinforcement of methylation if RNA signals remain available). Forward and reverse genetic approaches are identifying the molecular machin-ery needed for each step.3.3.2.1Double-stranded RNA synthesis and processingThe issue of double-stranded RNA synthesis and processing is complicated in plants because of:(1)different ways to synthesize double-stranded RNA,including through theactivity of RDR,which is encoded by a multi-gene family in Arabidopsis;(2)the existence of multiple Dicer-like(DCL)enzymes and Argonaute(AGO)proteins that are differentially distributed into nuclear or cytoplas-mic compartments;(3)the production of two size classes of short RNAs that appear to befunctionally distinct:a longer class that is$24nt in length has been implicated in DNA and histone methylation and a shorter size class that is$21nt in length is involved in the mRNA degradation step of PTGS.RNA-DIRECTED DNA METHYLATION79 As discussed below,this distinction has generally held up but it is not yet known whether the24-nt class is exclusively capable of eliciting epigenetic modifications in all systems.Double-stranded RNA can be synthesized by:(1)bidirectional transcription of double-stranded DNA,producing sense andantisense RNAs that can anneal with each other;(2)transcription through an inverted DNA repeat,which forms an RNA that isself-complementary and can fold back on itself to form a hairpin RNA;(3)transcription of a single-stranded RNA template,which requires the ac-tivity of an RDR(see Figure3.3).Only the third way of producing double-stranded RNA is compromised by mutations in genes encoding RDRs.The A.thaliana genome contains at least three expressed genes that encode RDRs:RDR1,RDR2and RDR6(Xie et al., 2004).Forward genetic screens have demonstrated that RDR6(SGS2/SDE1) is required for PTGS triggered by sense transgenes(Dalmay et al.,2000; Mourrain et al.,2000).Mutations in RDR6reduce DNA methylation associ-ated with PTGS of sense transgenes but not inverted repeat transgenes(Be´clin et al.,2002;Dalmay et al.,2000;Mourrain et al.,2000).Reverse genetics approaches have revealed that RDR2is required for DNA methylation and H3K9methylation of several endogenous repeats and for accumulation of siRNAs originating from these repeats(Chan et al.,2004;Xie et al.,2004). Mutations of RDR1had no effect on the accumulation of these endogenous siRNAs and its function is unknown.The Arabidopsis genome encodes four DCL activities(Schauer et al., 2002).Three of these–DCL1,DCL2and DCL3–appear to be localized predominantly in the nucleus as assessed by the subcellular distribution of DCL–GFP fusion proteins(Papp et al.,2003;Xie et al.,2004).Forward genetic screens for mutants defective in RNA-mediated TGS of transgene promoters have not yet recovered mutants defective in any of the four DCL enzymes.This may be due to redundancy of these proteins,as has been observed for the two Dicers in Neurospora(Catalanotto et al.,2004).Another possibility is that unprocessed double-stranded RNA can participate to some extent in RdDM.Indeed,we have not yet found a way to eliminate siRNAs originating from the NOSpro or a’pro,which would allow us to confirm they are needed for RdDM in these systems.Neither RdDM of a target NOSpro nor accumulation of NOSpro siRNAs is impaired in dcl1partial loss of function mutants(Papp et al.,2003).Accumulation of siRNAs generated from hairpin RNAs that induce PTGS is also not reduced in dcl1partial loss of function mutants(Finnegan et al.,2003).Instead,the primary role of DCL1appears to be the processing of miRNA precursors(Park et al.,2002;Reinhart et al., 2002).Whether this is the only function of DCL1is unknown.。