缺氧条件下的microrna表达谱
- 格式:pdf
- 大小:555.11 KB
- 文档页数:9

㊃综述㊃微小RNA-210在缺氧致心血管系统疾病中的研究进展马明仁㊀蔡晓庆㊀张鹏㊀焦丕奇㊀赵瑞㊀王菲㊀马凌730050兰州,中国人民解放军联勤保障部队第九四〇医院心血管内科注:马明仁和蔡晓庆为共同第一作者通信作者:王菲,电子信箱:22785439@;马凌,电子信箱:1764535920@DOI:10.3969/j.issn.1007-5410.2023.05.016㊀㊀ʌ摘要ɔ㊀微小RNA-210(miR-210)是缺氧高度相关的微小RNA之一,其通过抑制细胞凋亡㊁促进血管生成㊁调控红系分化以及参与炎症反应发挥关键调节作用㊂miR-210启动子区域具有缺氧诱导因子1和核转录因子κB结合位点,该结构使其具有作为缺氧致心血管系统疾病生物标记物的临床应用前景㊂基于此,本文系统总结miR-210当前研究取得的成果以及存在的局限和挑战,以期为缺氧致心血管系统疾病的防治提供新思路㊂ʌ关键词ɔ㊀微小RNA-210;㊀缺氧;㊀心血管系统;㊀凋亡;㊀血管生成;㊀红系分化;㊀炎症基金项目:甘肃省自然科学基金资助项目(21JR1RA181㊁GSWSKY2020-01);联勤保障部队第940医院院内项目(2021yxky080)Research progress of microRNA-210in hypoxia induced cardiovascular diseases㊀Ma Mingren,CaiXiaoqing,Zhang Peng,Jiao Piqi,Zhao Rui,Wang Fei,Ma LingDepartment of Cardiology,the940th Hospital of Joint Logistics Support Force of Chinese PLA,Lanzhou730050,ChinaMa Mingren and Cai Xiaoqing are co-first authorsCorresponding author:Wang Fei,Email:22785439@;Ma Ling,Email:1764535920@ʌAbstractɔ㊀MicroRNA-210(miR-210)is one of the microRNAs highly related to hypoxia,whichplays a key regulatory role by inhibiting apoptosis,promoting angiogenesis,regulating erythroid differentiation,and participating in inflammatory responses.The miR-210promoter region has the bindingsite for hypoxia-inducing factor1and nuclear transcription factorκB,which makes it promising for clinical application as a biomarker for cardiovascular diseases caused by hypoxia.Based on this,this paper systematically summarizes the achievements of current research on miR-210as well as the existing limitationsand challenges,in order to provide new ideas for the prevention and treatment of cardiovascular diseasescaused by hypoxia.ʌKey wordsɔ㊀MicroRNA-210;㊀Hypoxia;㊀Cardiovascular system;㊀Apoptosis;㊀Angiogenesis;Erythroid differentiation;㊀InflammationFund program:Natural Science Foundation of Gansu Province(21JR1RA181,GSWSKY2020-01);Key Project of940th Hospital of Joint Logistics Support Force of Chinese PLA(2021yxky080)㊀㊀迄今为止心血管疾病(cardiovascular diseases,CVD)仍是导致全球人类死亡的主要原因之一,在我国CVD的患病率呈逐年攀升趋势[1]㊂心血管系统作为体内运输㊁传递氧的重要通道对缺氧十分敏感,缺氧已成为CVD独立危险因素,通常会导致不可逆的组织损伤[2]㊂当前临床应用广泛的CVD指标以反映心肌损伤㊁心脏功能及心血管炎症为主,而缺氧致CVD诊疗缺乏稳定性㊁特异性及敏感性高的标志物㊂微小RNA(microRNA,miRNA)是长度为20~26个核苷酸大小的单链小分子,通过结合靶mRNA的3 非编码区来调节转录后水平上蛋白质编码基因表达[3]㊂大量基础与临床研究佐证了miRNA参与心血管系统的病理生理进程[4],其中miR-210可谓缺氧相关疾病研究领域的明星分子,在缺氧致CVD中miR-210显著上调,通过调节缺氧细胞增殖㊁分化㊁迁移以及线粒体代谢发挥心血管系统保护作用,然而持续高表达的miR-210在特定环境下也会对机体产生损害[5]㊂基于此,本文聚焦miR-210在缺氧致CVD中的关键调控作用,以细胞凋亡㊁血管生成㊁红系分化以及炎症反应为切入点总结该研究领域当前最新进展,以期为缺氧致CVD的防治提供新策略㊂1㊀miR-210概述人类miR-210位于第11号染色体短臂末端(11p15.5),是由20~24个核苷酸组成的高度保守miRNA,包括miR-210-3p和miR-210-5p两个转录本,miR-210-3p整合到RNA诱导的沉默复合体中的引导链,而miR-210-5p降解失活的随从链[6],通过与靶mRNA序列互补抑制蛋白质翻译,在转录水平发挥调控作用㊂miR-210启动子区域含有一个大约40bp的功能性缺氧反应原件能被缺氧诱导因子1α(hypoxia inducible factor-1α,HIF-1α)识别,从而在缺氧条件下诱导miR-210转录,调节细胞缺氧应答反应使机体适应缺氧环境㊂miR-210是主要的缺氧诱导性miRNA,已证实它在缺氧条件下的多种细胞类型中显著上调,参与机体生物学进程以及疾病的发生发展[7]㊂miR-210结构稳定㊁耐高温及耐降解的特性使其具备作为缺氧致CVD稳定可靠生物标记物的潜质㊂2㊀miR-210抑制细胞凋亡细胞凋亡的发生由与细胞表面受体相关的外在途径和通过线粒体和内质网发生作用的内在途径介导,内质网应激在缺氧心肌细胞凋亡中发挥关键作用㊂Marwarha等[8]最新研究表明,miR-210减弱了缺氧诱导的内在凋亡途径激活,并首次揭示在AC-16心肌细胞中抑制丝氨酸/苏氨酸激酶糖原合酶激酶3β活性是miR-210减弱缺氧诱导心肌细胞凋亡的基础㊂在受到缺氧/复氧应激的AC-16心肌细胞中,该研究团队还证实了miR-210减弱缺氧驱动的内在凋亡途径,同时显著增强复氧诱导的csapase-8介导的外在凋亡途径[9]㊂心肌随年龄增长再生能力逐渐降低乃至消失的生理特性使研究者对干细胞治疗CVD产生了浓厚兴趣,缺氧诱导能增强间充质干细胞(mesenchymal stem cells,MSCs)及其分泌的外泌体中miR-210和中性鞘磷脂酶2的活性,增加对心脏的获益[10]㊂Cheng等[11]揭示,缺氧刺激MSCs来源的外泌体miR-210通过靶向AIFM3调节PI3K/AKT和p53信号通路,从而减少心肌梗死后的心肌细胞凋亡,内源性外泌体miR-210在此过程中发挥关键调控作用㊂铁-硫簇支架蛋白1/2 (iron-sulfur cluster scaffold protein,ISCU1/2)作为线粒体代谢的关键组成部分在肺动脉高压(pulmonary arterial hypertension,PAH)发病机制中起重要作用,且ISCU1/2受到miR-210调控㊂缺氧是人肺动脉平滑肌细胞(human pulmonary artery smooth muscle cell,HPASMC)增殖和PAH的重要刺激因素,Gou等[12]发现,miR-210是HPASMC缺氧诱导的主要miRNA,在慢性缺氧诱导的PAH小鼠肺组织中miR-210通过抑制E2F3表达减少缺氧诱导的HPASMC凋亡㊂血管平滑肌细胞(vascular smooth muscle cell,VSMC)凋亡在血管重构和动脉粥样硬化(atherosclerosis,AS)斑块不稳定中起重要作用,缺氧诱导的VSMC模型中发现miR-210通过靶向MEF2C抑制细胞凋亡[13]㊂缺氧诱导的细胞凋亡是多因素参与的复杂过程,miR-210在其中发挥的关键调控作用已得到大量研究证实,然而据当前研究可知miR-210下游靶点较多且调控网络复杂,以miR-210为节点展开miRNA-miRNA交互网络研究对阐明缺氧条件下miR-210在细胞凋亡中发挥的调控作用机制至关重要㊂3㊀miR-210促进血管生成缺氧条件下心血管系统会出现代偿性的血管新生以改善血流供应,这对维持机体内环境稳态尤为重要㊂HIF-1α是缺氧条件下细胞内稳态恢复的关键调节因子,研究证实HIF-1α/miR-210/miR-424/sFLT1轴通过血管内皮生长因子(vascular endothelial growth factor,VEGF)信号通路调节缺氧条件下人脐静脉内皮细胞和人真皮微血管内皮细胞的血管生成[14]㊂Mirzaei等[15]探讨饥饿素对缺氧相关心脏血管新生的影响发现,在Wistar大鼠缺氧模型中HIF-1α㊁VEGF和miR-210参与缺氧诱导的血管生成,缺氧增加心肌血管生成和心脏miR-210㊁VEGF和HIF-1α表达,而饥饿素可通过反向调节miR-210信号通路抑制这些缺氧诱导的变化㊂MSC 衍生的细胞外囊泡(mesenchymal stem cell-derived small extracellular vesicles,MSC-sEVs)作为细胞间通讯和维持细胞内环境稳态的重要介质已被证实在缺氧条件下参与血管生成,缺氧预处理MSC-sEVs中的miR-210-3p通过HIF-1α诱导表达上调,过表达的miR-210-3p负调节EFNA3表达并增强PI3K/AKT通路的磷酸化而促进血管生成[16]㊂侧支循环的早期建立可有效降低急性心肌梗死(acute myocardial infarction,AMI)后心肌细胞死亡的风险并促进心肌重构诱导心肌血管再生,在AMI小鼠模型中发现miR-210控制线粒体代谢,通过靶向特定基因诱导血管生成并抑制心肌细胞凋亡而发挥保护心脏的功能[17]㊂Fan等[18]在Sprague-Dawley 大鼠AMI模型中也证实,AMI+miR-210激动剂组的miR-210㊁肝细胞生长因子(hepatocyte growth factor,HGF)表达水平显著高于其他亚组,miR-210通过靶向调节HGF促进梗死心肌血管生成改善左心室重构㊂miR-210通过调控HIF㊁VEGF及HGF等在缺氧条件下促进血管生成,对缺氧致CVD的发生㊁发展及预后产生重要影响,事实上驱动miR-210在缺氧条件下促进血管生成可能涉及多种细胞和分子机制,目前的研究证据以缺氧经典通路为主而忽略了相关信号通路和因子在其中发挥的协同调控作用㊂4㊀miR-210调控红系分化红系分化作为产生红细胞的重要通路由多种转录因子及信号通路综合调控协同进行㊂脯氨酸羟化酶结构域蛋白(prolyl hydroxylase domain protein,PHD):HIF途径是氧依赖调控红细胞数量的关键途径,PHD位点特异性地使HIF-1α脯氨酸羟基化,然而在缺氧条件下这种修饰会被减弱,从而使稳定的HIF-1α激活包括促红细胞生成素在内的下游靶基因㊂Sarakul等[19]在K562细胞和β-地中海贫血/HbE红系祖细胞研究氧分压对红细胞生成的影响发现,缺氧诱导的K562细胞和β-地中海贫血/HbE红系祖细胞中miR-210表达上调,抑制miR-210表达导致细胞中珠蛋白基因表达减少和红细胞成熟延迟,表明缺氧条件下高表达的miR-210有利于红系分化㊂低压缺氧是高海拔地区独有的地理特征,不同海拔人群外周血细胞和血浆中miR-210-3p水平的研究证实,高原低氧环境下外周血miR-210-3p表达升高,血浆miR-210-3p浓度与血细胞中miR-210-3p水平呈正相关,且血细胞和血浆中的miR-210-3p水平与红细胞计数㊁血红蛋白和血细胞比容值高度相关[20]㊂GATA结合蛋白1(GATA binding protein1,GATA-1)是血细胞生成中核心造血转录因子之一,大多数已知的红系分化相关调控因子启动子区域均发现GATA结合位点㊂Hu等[21]在缺氧诱导的K562细胞中发现miR-210-3p以GATA-1依赖性方式上调,并发现SMAD2可能是miR-210-3p下游靶基因,有趣的是双荧光素酶报告基因分析发现miR-210-3p并没有直接与SMAD2的3 UTR结合,推测miR-210-3p通过抑制TGF信号通路的活性间接抑制SMAD2表达从而促进红系分化㊂缺氧条件下miR-210-3p对红系分化的正向调节作用可降低CVD风险,然而红系过度增殖㊁分化也会增加血液黏滞度,影响血液循环导致心㊁肺㊁脑等脏器功能受损,引发诸如高原红细胞增多症㊁AMI㊁急性冠状动脉综合征(acute coronary syndromes, ACS)等CVD的发生㊂低氧条件下miR-210调控红系分化的研究尚处起步阶段且临床研究证据有限,理清缺氧条件下miR-210调控红系分化的作用机制或许会为高原特殊环境下CVD的诊疗提供机会㊂5㊀miR-210参与炎症反应炎症是机体应对内外刺激的重要反应,由多种细胞因子介导产生,缺氧条件下促炎细胞因子增加㊁炎症反应增强[22]㊂HIF在缺氧时可激活一系列炎症因子导致炎性损伤,在所有炎症反应中核转录因子κB(nuclear factor-κB, NF-κB)转录调控蛋白家族发挥关键作用㊂van Uden等[23]和Fitzpatrick等[24]研究表明,组织与细胞缺氧时HIF-1和NF-κB通路启动了复杂且相互关联的炎症信号级联放大效应,且在心血管损伤㊁癌症等疾病的发生发展中发挥着双刃剑的作用㊂NF-κB亚单位p50(NF-κB1)是一种与炎症和DNA修复相关的蛋白,该蛋白定位于miR-210茎环上游的200bp核心启动子区域,在缺氧条件下NF-κB诱导miR-210表达发生变化[25]㊂miR-210通过调控TGF-β信号通路抑制胆固醇脂水解酶,进而促进AS形成㊂引起AS的系统性炎症㊁氧化应激和内皮功能障碍与缺氧密切相关,且缺氧影响脂质代谢促进AS斑块进展㊂Qiao等[26]探讨AS中miR-210-3p及其靶基因在巨噬细胞脂质沉积和炎症反应中的作用机制,发现miR-210-3p通过抑制IGF2/IGF2R来抑制CD36和NF-κB的表达,从而减轻氧化低密度脂蛋白诱导的巨噬细胞脂质积累和炎症反应㊂miR-210通过抑制三羧酸循环途径中的关键蛋白质来阻止能量生成,减少氧气消耗引起乳酸堆积改变线粒体膜电位最终破坏线粒体结构,也可通过调控细胞色素氧化酶COX10和琥珀酸脱氢酶的D亚基抑制线粒体呼吸作用,炎症激活将巨噬细胞的线粒体功能从氧化磷酸化转变为活性氧生成,这或许会促进AS中坏死核心的形成㊂Karshovska等[27]证实HIF-1α诱导的miR-210降低炎症巨噬细胞的线粒体呼吸,增加活性氧产生和坏死性细胞死亡, HIF-1α通过调节miR-210和miR-383促进巨噬细胞坏死㊂ATG7是一类泛素化激活酶,有证据表明ATG7的缺失导致白细胞介素1β表达及NLRP3通路激活,在LPS诱导的人冠状动脉内皮细胞炎症模型中miR-210通过靶向ATG7抑制细胞自噬及炎症发挥细胞保护作用[28]㊂ACS发生的直接原因是斑块破裂,而炎症细胞及其产物的形成和释放是不稳定斑块破裂的关键因素,ACS患者血清miR-210与炎症水平相关,miR-210可能通过促进炎症反应也可能作为炎症反应的下游信号参与ACS的发生发展[29]㊂炎症反应伴随大多数CVD发生发展,miR-210作为缺氧标志性miRNA在缺氧致CVD与炎症反应中所起的中枢纽带作用已被证实,然而缺氧条件下miR-210在炎症反应中扮演的双重角色以及如何转变多种调控方式的机制有待阐明㊂6㊀miR-210应用转化miR-210作为与缺氧高度相关的miRNAs之一,在肿瘤㊁心血管系统(表1)及深静脉血栓等伴随缺氧机制的疾病中表达显著上调㊂miR-210在缺氧致CVD中涉及细胞凋亡㊁血管生成㊁红系分化以及炎症反应方面已有大量基础研究佐证,但基础研究到临床转化尚需大量临床研究数据提供支撑㊂Karakas等[41]对1112例冠心病患者进行了为期4年的随访,其中包括430例ACS患者和682例稳定性心绞痛患者,确定了血浆中8种miRNAs(miR-19a㊁miR-19b㊁miR-132㊁miR-140-3p㊁miR-142-5p㊁miR-150㊁miR-186和miR-210)有助于ACS的诊断,这是迄今为止评估循环miRNAs在CVD中预后价值的最大规模临床研究,除此之外miR-210涉及临床研究甚少,迫切需要多中心㊁更大规模队列研究以推进临床转化应用㊂7㊀展望miR-210在缺氧致CVD中发挥关键调控作用无可非议,但是将miR-210应用临床仍存在不足和挑战,如:(1)缺氧条件下miR-210在多种细胞类型中表达发生变化[7],以此为靶点研发诊断试剂和治疗药物可能会存在特异性差㊁副作用强等诸多问题;(2)miR-210在高原缺氧环境久居人群中表达量普遍升高[20],在该类人群中miR-210的表达变化是否具有诊疗价值目前无相关研究报道;(3)临床治疗老年慢性阻塞性肺疾病合并Ⅱ型呼吸衰竭患者往往采取保持适度缺氧以刺激呼吸中枢兴奋,缺氧诱导miR-210在此类患者中持续高表达对心脏发挥保护作用还是产生损害仍然未知;(4)EVs可大幅增强miR-210对RNase降解的抵抗力,缺氧条件下EVs中的miR-210发挥保护心血管系统功能,然而EVs的提取㊁鉴定㊁储存㊁给药方式及剂量仍未标准化,其临床应用还有待探究[42];(5)铜死亡㊁胞葬㊁细胞焦亡及mRNA m6A甲基化修饰等是当前生命科学领域研究热点,但miR-210在缺氧条件下是否参与其中未见文献报道,随着单细胞测序㊁代谢组学等前沿技术的发展应用,它们之间的相关性表1㊀miR-210参与缺氧致CVD 的生理病理过程miR-210AIFM3PI3K /AKT 减少心肌细胞凋亡,保护心功能[11]miR-210E2F3miR-210/E2F3抑制肺血管平滑肌细胞凋亡[12]miR-210MEF2C miR-210/MEF2C 抑制平滑肌细胞凋亡[13]miR-210-3pNfkb1PI3K /AKT促进心肌细胞凋亡[30]miR-210CXCR4SMAD /mTOR加重缺氧诱导的H9C2细胞损伤[31]miR-210PDK1P13K /Akt /mTOR 诱导内皮细胞凋亡[32]miR-210E2F3miR-210/E2F3抑制OGD /R 诱导的心肌细胞凋亡[13]miR-210BNIP3miR-210/BNIP3在H 2O 2诱导的心肌细胞凋亡中发挥保护作用[33]血管生成miR-210-3pEFNA3PI3K /AKT 增强PI3K /AKT 磷酸化,促进血管生成[16]miR-210APCVEGF促进心肌细胞增殖,增加新生血管生成,改善心脏功能[34]miR-210Efna3miR-210/Efna3促进血管生成,改善心脏功能[35]miR-210MKP-1miR-210/MKP-1促进HPASMC 增殖及血管新生[36]红系分化miR-210α㊁β和γ珠蛋白miR-210/α㊁β和γ珠蛋白有助于缺氧诱导的红系分化[19]miR-210-3pSMAD2miR-210/SMAD2通过抑制TGF 信号通路的活性间接抑制SMAD2表达从而促进红系分化[21]miR-210PLCβ1miR-210/PLCβ1促进红系分化[37]miR-210BCL11A miR-210/BCL11A 参与红系分化[38]炎症反应miR-210-3pIGF2NF-κB减轻脂质蓄积和炎症反应[26]miR-210HIF-1αmiR-210/HIF-1α促进巨噬细胞坏死[27]miR-210ATG7miR-210/ATG7抑制细胞自噬及炎症发挥细胞保护作用[28]miR-210RIPK2NF-κB 细菌诱导的炎症反应中发挥调节作用[39]miR-210CEH TGF-β调节炎症反应,促进AS 形成[40]㊀㊀注:miR-210:微小RNA-210;CVD:心血管疾病;nSMase2:中性鞘磷脂酶2;HIF-1α:缺氧诱导因子1α;AIFM3:线粒体凋亡诱导因子2;PI3K /AKT:磷脂酰肌醇3激酶/丝氨酸苏氨酸蛋白激酶;E2F3:E2F 转录因子3;MEF2C:肌细胞增强因子2C;Nfkb1:核转录因子κB1;CXCR4:CXC 趋化因子受体4;SMAD /mTOR:母亲DPP 同源物/雷帕霉素靶蛋白;PDK1:磷酸肌醇依赖性蛋白激酶1;BNIP3:腺病毒E1B 相互作用蛋白3;EFNA3:肝配蛋白A3;APC:活化蛋白;VEGF:血管内皮生长因子;Efna3:酪氨酸蛋白激酶A3受体;MKP-1:丝裂原激活蛋白激酶磷酸酶-1;HPASMC:人肺动脉平滑肌细胞;SMAD2:母亲DPP 同源物2;TGF:转化生长因子;PLCβ1:磷脂酶Cβ1;BCL11A:淋巴瘤基因11A;IGF2:胰岛素样生长因子2;NF-κB:核转录因子κB;ATG7:自噬相关基因7;RIPK2:丝氨酸苏氨酸激酶2;CEH:胆固醇脂水解酶;TGF-β:转化生长因子β;AS:动脉粥样硬化或许是高原CVD 研究的又一个热点;(6)miR-210是缺氧高度相关的miRNAs 之一,HIF-1α已被证实在缺氧条件下激活,且两者之间存在结合位点,但它们在缺氧致CVD 中发挥的调控机制仍未阐明,如何构建以HIF-1α/miR-210为中心的调控网络对于理解缺氧致CVD 的发生至关重要㊂利益冲突:无参㊀考㊀文㊀献[1]吴超群,李希,路甲鹏,等.中国居民心血管疾病危险因素分布报告[J].中国循环杂志,2021,36(1):4-13.DOI:10.3969/j.issn.1000-3614.2021.01.002.㊀Wu CQ,Li X,Lu JP,et al.Report on Geographical Disparity of Cardiovascular Risk Factors in China[J].Chin Circu J,2021,36(1):4-13.DOI:10.3969/j.issn.1000-3614.2021.01.002.[2]Mallet RT,Burtscher J,Richalet JP,et al.Impact of HighAltitude on Cardiovascular Health:Current Perspectives [J ].Vasc Health Risk Manag,2021,17:317-335.DOI:10.2147/VHRM.S294121.[3]Ouyang Z,Wei K.miRNA in cardiac development andregeneration[J].Cell Regen,2021,10(1):14.DOI:10.1186/s13619-021-00077-5.[4]储时春,成威.MicroRNA-590在心血管疾病中的研究进展[J].中国心血管杂志,2020,25(3):278-281.DOI:10.3969/j.issn.1007-5410.2020.03.016.㊀Chu SC,Cheng W.Research progress of microRNA-590in cardiovascular disease[J].Chin J Cardiovasc Med,2020,25(3):278-281.DOI:10.3969/j.issn.1007-5410.2020.03.016.[5]Guan Y,Song X,Sun W,et al.Effect of Hypoxia-Induced MicroRNA-210Expression on Cardiovascular Disease and the Underlying Mechanism [J ].Oxid Med Cell Longev,2019:4727283.DOI:10.1155/2019/4727283.[6]Lin KH,Ng SC,Paul CR,et al.MicroRNA-210repression facilitates advanced glycation end-product (AGE )-induced cardiac mitochondrial dysfunction and apoptosis via JNK activation[J].J Cell Biochem,2021,122(12):1873-1885.DOI:10.1002/jcb.30146.[7]Zaccagnini G,Greco S,Voellenkle C,et al.miR-210hypoxamiR in Angiogenesis and Diabetes [J].Antioxid Redox Signal,2022,36(10-12):685-706.DOI:10.1089/ars.2021.0200.[8]Marwarha G,Røsand Ø,Slagsvold KH,et al.GSK3βInhibition Is the Molecular Pivot That Underlies the Mir-210-Induced Attenuation of Intrinsic Apoptosis Cascade during Hypoxia[J].Int J Mol Sci,2022,23(16):9375.DOI:10.3390/ijms23169375.[9]Marwarha G,Røsand Ø,Scrimgeour N,et al.miR-210Regulates Apoptotic Cell Death during Cellular Hypoxia and Reoxygenation in a Diametrically Opposite Manner [J ].Biomedicines,2021,10(1):42.DOI:10.3390/biomedicines10010042.[10]Zhu J,Lu K,Zhang N,et al.Myocardial reparative functions of exosomes from mesenchymal stem cells are enhanced by hypoxia treatment of the cells via transferring microRNA-210in an nSMase2-dependent way[J].Artif Cells Nanomed Biotechnol,2018,46(8):1659-1670.DOI:10.1080/21691401.2017.1388249.[11]Cheng H,Chang S,Xu R,et al.Hypoxia-challenged MSC-derived exosomes deliver miR-210to attenuate post-infarctioncardiac apoptosis[J].Stem Cell Res Ther,2020,11(1):224.DOI:10.1186/s13287-020-01737-0.[12]Gou D,Ramchandran R,Peng X,et al.miR-210has anantiapoptotic effect in pulmonary artery smooth muscle cellsduring hypoxia[J].Am J Physiol Lung Cell Mol Physiol,2012,303(8):L682-691.DOI:10.1152/ajplung.00344.2011. [13]Bao Q,Jia H,A R,et al.MiR-210inhibits hypoxia-inducedapoptosis of smooth muscle cells via targeting MEF2C[J].Int JClin Exp Pathol,2019,12(5):1846-1858.[14]Zhao H,Wang X,Fang B.HIF1A promotes miR-210/miR-424transcription to modulate the angiogenesis in HUVECs andHDMECs via sFLT1under hypoxic stress[J].Mol CellBiochem,2022,477(8):2107-2119.DOI:10.1007/s11010-022-04428-x.[15]Mirzaei Bavil F,Karimi-Sales E,Alihemmati A,et al.Effect ofghrelin on hypoxia-related cardiac angiogenesis:involvement ofmiR-210signalling pathway[J].Arch Physiol Biochem,2022,128(1):270-275.DOI:10.1080/13813455.2019.1675712.[16]Zhuang Y,Cheng M,Li M,et al.Small extracellular vesiclesderived from hypoxic mesenchymal stem cells promotevascularized bone regeneration through the miR-210-3p/EFNA3/PI3K pathway[J].Acta Biomater,2022,150:413-426.DOI:10.1016/j.actbio.2022.07.015.[17]Song R,Dasgupta C,Mulder C,et al.MicroRNA-210ControlsMitochondrial Metabolism and Protects Heart Function inMyocardial Infarction[J].Circulation,2022,145(15):1140-1153.DOI:10.1161/CIRCULATIONAHA.121.056929. [18]Fan ZG,Qu XL,Chu P,et al.MicroRNA-210promotesangiogenesis in acute myocardial infarction[J].Mol Med Rep,2018,17(4):5658-5665.DOI:10.3892/mmr.2018.8620.[19]Sarakul O,Vattanaviboon P,Tanaka Y,et al.Enhancederythroid cell differentiation in hypoxic condition is in partcontributed by miR-210[J].Blood Cells Mol Dis,2013,51(2):98-103.DOI:10.1016/j.bcmd.2013.03.005. [20]Yan Y,Wang C,Zhou W,et al.Elevation of Circulating miR-210-3p in High-Altitude Hypoxic Environment[J].FrontPhysiol,2016,7:84.DOI:10.3389/fphys.2016.00084. [21]Hu C,Yan Y,Fu C,et al.Effects of miR2103p on the erythroiddifferentiation of K562cells under hypoxia[J].Mol Med Rep,2021,24(2):563.DOI:10.3892/mmr.2021.12202. [22]王一斐,张萍.心房颤动与炎症反应之交互关系[J].中国心血管杂志,2023,28(1):62-66.DOI:10.3969/j.issn.1007-5410.2023.01.013.㊀Wang YF,Zhang P.Interaction between atrial fibrillation andinflammation[J].Chin J Cardiovasc Med,2023,28(1):62-66.DOI:10.3969/j.issn.1007-5410.2023.01.013. [23]van Uden P,Kenneth NS,Rocha S.Regulation of hypoxia-inducible factor-1alpha by NF-kappaB[J].Biochem J,2008,412(3):477-484.DOI:10.1042/BJ20080476. [24]Fitzpatrick SF,Tambuwala MM,Bruning U,et al.An intactcanonical NF-κB pathway is required for inflammatory geneexpression in response to hypoxia[J].J Immunol,2011,186(2):1091-1096.DOI:10.4049/jimmunol.1002256. [25]Zhang Y,Yang M,Yang S,et al.Role of noncoding RNAs anduntranslated regions in cancer:A review[J].Medicine(Baltimore),2022,101(33):e30045.DOI:10.1097/MD.0000000000030045.[26]Qiao XR,Wang L,Liu M,et al.MiR-210-3p attenuates lipidaccumulation and inflammation in atherosclerosis by repressingIGF2[J].Biosci Biotechnol Biochem,2020,84(2):321-329.DOI:10.1080/09168451.2019.1685370.[27]Karshovska E,Wei Y,Subramanian P,et al.HIF-1α(Hypoxia-Inducible Factor-1α)Promotes MacrophageNecroptosis by Regulating miR-210and miR-383[J].Arterioscler Thromb Vasc Biol,2020,40(3):583-596.DOI:10.1161/ATVBAHA.119.313290.[28]李明.MicroRNA-210在LPS诱导的人冠状动脉内皮细胞炎症模型中对自噬和炎症的作用及机制研究[D].吉林大学,2020.㊀Li M.The role and regulation mechanism of microRNA-210onautophagy and inflammation in human coronary artery endothelialcell induced by LPS[D].Jilin University,2020. [29]许士海,王进,王丽,等.急性冠脉综合征患者血清microRNA-210的临床意义[J].中国处方药,2021,19(12):153-155.DOI:10.3969/j.issn.1671-945X.2021.12.067.㊀Xu SH,Wang J,Wang L,et al.Clinical significance of serummicroRNA-210levels in patients with Acute Coronary Syndromes[J].Journal of China Prescription Drug,2021,19(12):153-155.DOI:10.3969/j.issn.1671-945X.2021.12.067. [30]Zhong L,Jia J,Ye G.Rian/miR-210-3p/Nfkb1Feedback LoopPromotes Hypoxia-Induced Cell Apoptosis in MyocardialInfarction Through Deactivating the PI3K/Akt Signaling Pathway[J].J Cardiovasc Pharmacol,2020,76(2):207-215.DOI:10.1097/FJC.0000000000000824.[31]Feng M,Li Z,Wang D,et al.MicroRNA-210aggravateshypoxia-induced injury in cardiomyocyte H9c2cells by targetingCXCR4[J].Biomed Pharmacother,2018,102:981-987.DOI:10.1016/j.biopha.2018.03.151.[32]Li Y,Yang C,Zhang L,et al.MicroRNA-210inducesendothelial cell apoptosis by directly targeting PDK1in the settingof atherosclerosis[J].Cell Mol Biol Lett,2017,22:3.DOI:10.1186/s11658-017-0033-5.[33]Diao H,Liu B,Shi Y,et al.MicroRNA-210alleviates oxidativestress-associated cardiomyocyte apoptosis by regulating BNIP3[J].Biosci Biotechnol Biochem,2017,81(9):1712-1720.DOI:10.1080/09168451.2017.1343118.[34]Arif M,Pandey R,Alam P,et al.MicroRNA-210-mediatedproliferation,survival,and angiogenesis promote cardiac repairpost myocardial infarction in rodents[J].J Mol Med(Berl),2017,95(12):1369-1385.DOI:10.1007/s00109-017-1591-8.[35]Wang N,Chen C,Yang D,et al.Mesenchymal stem cells-derived extracellular vesicles,via miR-210,improve infarctedcardiac function by promotion of angiogenesis[J].BiochimBiophys Acta Mol Basis Dis,2017,1863(8):2085-2092.DOI:10.1016/j.bbadis.2017.02.023.[36]Jin Y,Pang T,Nelin LD,et al.MKP-1is a target of miR-210and mediate the negative regulation of miR-210inhibitor onhypoxic hPASMC proliferation[J].Cell Biol Int,2015,39(1):113-120.DOI:10.1002/cbin.10339.[37]Bavelloni A,Poli A,Fiume R,et al.PLC-beta1regulates theexpression of miR-210during mithramycin-mediated erythroiddifferentiation in K562cells[J].Oncotarget,2014,5(12):4222-4231.DOI:10.18632/oncotarget.1972. [38]Gasparello J,Fabbri E,Bianchi N,et al.BCL11A mRNATargeting by miR-210:A Possible Network Regulatingγ-GlobinGene Expression[J].Int J Mol Sci,2017,18(12):2530.DOI:10.3390/ijms18122530.[39]Su H,Chang R,Zheng W,et al.microRNA-210andmicroRNA-3570Negatively Regulate NF-κB-MediatedInflammatory Responses by Targeting RIPK2in Teleost Fish[J].Front Immunol,2021,12:617753.DOI:10.3389/fimmu.2021.617753.[40]吴洁.miR-210对动脉粥样硬化斑块形成的调控及机制研究[D].第三军医大学,2017.㊀Wu J.miR-210regulate the formation of atherosclerotic plaqueand its mechanism research[D].The Third Military MedicalUniversity,2017.[41]Karakas M,Schulte C,Appelbaum S,et al.CirculatingmicroRNAs strongly predict cardiovascular death in patients withcoronary artery disease-results from the large AtheroGene study[J].Eur Heart J,2017,38(7):516-523.DOI:10.1093/eurheartj/ehw250.[42]岳霏霏,宋煜,王晓蓓,等.心肌梗死后心脏修复中细胞外囊泡的应用及作用[J].中国组织工程研究,2023,27(10):1610-1617.㊀Yue FF,Song L,Wang XP,et al.Application and effect ofextracellular vesicles in cardiac repair after myocardial infarction[J].Chinese Journal of Tissue Engineering Research,2023,27(10):1610-1617.(收稿日期:2022-11-07)(本文编辑:谭潇)。

低氧训练调控microRNA表达的研究现状氧气是生命活动必不可少的重要物质,细胞利用氧气进行呼吸代谢,产生能量和二氧化碳。
但是,在特定的环境中,如高海拔地区和深海,氧气的浓度较低,这就需要细胞对低氧环境进行适应和应对。
低氧训练是一种体育训练方式,通过在低氧环境中进行训练,能够提高人体的肺活量、氧气摄取量和耐力,同时增加心肌的收缩能力,预防心脏病和中风等疾病。
然而,低氧训练对细胞的分子机制和代谢调控仍存在许多未解之谜。
最新的研究表明,低氧训练能够调控microRNA表达,从而影响基因表达和细胞功能。
microRNA是一类长度为21-25个核苷酸的小分子RNA,在细胞中具有重要的调控作用。
它们能够与靶基因的mRNA相结合,从而抑制该基因的转录和翻译,影响其表达。
最近的研究发现,在低氧环境下,许多microRNA的表达发生了变化,进而影响基因表达和细胞代谢。
例如,在人类肺癌细胞中,低氧环境下miR-210的表达显著上调,进而促进细胞的存活和增殖,增强其对低氧环境的适应能力。
类似地,低氧环境下,miR-29b的表达水平下调,进而影响胶原蛋白的合成和细胞支架的重构。
低氧训练能够模拟低氧环境,促进体内的氧气交换和代谢调节。
一些研究表明,低氧训练能够调控多个microRNA的表达,进而影响基因转录和蛋白质合成。
例如,在人类肌肉细胞中,低氧训练能够上调miR-21和miR-146a的表达,进而促进血管内皮细胞的增殖和血管生成。
类似地,在小鼠脊髓中,低氧训练能够下调miR-124a的表达,进而促进神经前体细胞的增殖和分化,促进神经系统的修复和重建。
然而,目前对低氧训练调控microRNA表达的相关研究还很少,有待进一步验证和确认。
此外,不同类型的细胞和不同低氧训练方式对microRNA表达的调控效应也不同。
因此,需要进一步深入研究低氧训练对microRNA的调控作用,促进其在临床上的应用和推广。
总之,低氧训练调控microRNA表达的研究是生命科学领域的一个热点,有望揭示低氧环境下细胞的分子机制和代谢调控,并为开发低氧训练的新技术和新方法提供理论支持。

低氧训练调控microRNA表达的研究现状
随着全球气候变化及高海拔地区的不断开发,人们越来越需要了解高原低氧环境对身体的影响及应对策略。
低氧环境下,人体为了适应供氧不足的状态,会发生一系列的生理改变,其中包括调节基因表达水平。
microRNA(miRNA)是一类具有调节基因表达功能的非编码小RNA分子,其通过靶向清除mRNA前体或抑制其翻译而发挥作用。
近年来,越来越多的研究表明,低氧环境可以通过改变miRNA表达来调节一系列的基因表达。
二、气道疾病
高海拔地区居民容易出现气道炎症、哮喘和氧气饱和度下降等问题。
miRNA在气道疾病的发生发展中发挥着重要作用,而低氧环境会影响miRNA的表达。
例如miRNA-146a和miRNA-155在哮喘的免疫反应中发挥着重要作用,但其在低氧环境下的表达不同,需要进一步深入研究。
三、心血管代谢反应
低氧环境下,人体代谢转变,可以通过miRNA的调控来实现。
例如miRNA-23a在心肌小细胞中有重要的调控作用,可以促进心肌细胞的增殖和生长,维持心血管系统的稳定。
然而,目前对低氧环境下miRNA参与心血管代谢反应的机制还不十分清楚,需要进一步探索。
在低氧训练中,研究miRNA的表达调控可以为制定正确的训练方案提供重要的指导意义。
通过研究不同低氧训练模式下miRNA的表达调控,可以深入理解训练对基因表达的影响,并提高训练效果。
同时,研究miRNA的表达调控还可以为特定疾病的预防和治疗提供新的方法和思路。
需要更多科学家加入相关研究,并不断推动其在理论和实践中的应用和发展。

第 45卷第1期2024 年1月Vol.45 No.1January 2024中山大学学报(医学科学版)JOURNAL OF SUN YAT⁃SEN UNIVERSITY(MEDICAL SCIENCES)MiRNA调控脑缺血/再灌注诱导的自噬信号通路研究进展刘筱蔼1,罗友根2(1. 广东食品药品职业学院,广东广州 510520; 2. 江苏医药职业学院盐城市偏瘫康复工程技术研究中心,江苏盐城 224005)摘要:脑卒中时缺血缺氧致脑组织功能损伤,且缺血后脑组织再恢复血液供应时,大量自由基、钙超载等引起脑缺血/再灌注损伤,进一步加重病情。
自噬是一种维持细胞内环境稳态的自我保护机制,但过度自噬引起脑组织损伤。
MiRNA为小的内源性非编码RNA分子,通过与其靶基因mRNA的3’ -UTR中的互补序列结合,导致翻译抑制或mRNA降解,从基因水平上调控多种生理活动。
MiRNA不仅直接作用于自噬相关蛋白,还可通过多种信号通路,参与缺血/再灌注诱导的自噬调控。
但关于miRNA调控脑缺血/再灌注诱导的自噬信号通路尚缺乏系统性归纳与分析。
本文综述了miRNA-124、miRNA-298、miRNA-202-5p、miRNA-142以及miRNA-26b等miRNA通过不同信号通路调控脑缺血/再灌注中的细胞自噬,为脑卒中的自噬研究提供了系统的理论思路。
关键词:脑缺血/再灌注损伤;微小RNA;自噬;信号通路;自噬相关蛋白;调控中图分类号:R363.2 文献标志码:A 文章编号:1672-3554(2024)01-0021-07DOI:10.13471/ki.j.sun.yat-sen.univ(med.sci).20240004.003MiRNA Regulating Autophagy Signaling Pathway Induced byCerebral Ischemia/ReperfusionLIU Xiaoai1, LUO Yougen2(1. Institute of Nursing, Guangdong Food and Drug Vocational College, Guangzhou 510520, China;2. Yancheng Hemiplegia Rehabilitation Engineering Technology Research Center, Jiangsu Vocational College ofMedicine, Yancheng 224005, China)Correspondence to: LUO Yougen; E-mail:***************Abstract:Ischemia and hypoxia cause functional damage to brain tissues during stroke, and when blood supply is re⁃stored to brain tissues after ischemia, a large number of free radicals and calcium overload cause cerebral ischemia-reper⁃fusion injury, which further aggravates the condition. Autophagy is a self-protection mechanism that maintains the homeo⁃stasis of the intracellular environment, but excessive autophagy causes brain tissue damage. MiRNA is a small endogenous non-coding RNA molecule that regulate various physiological activities at the gene level by binding to complementary se⁃quences in the 3 '- UTR of its target gene mRNA, leading to translation inhibition or mRNA degradation. MiRNA not only directly acts on autophagy related proteins, but also participates in autophagy regulation induced by ischemia/reperfusion through various signaling pathways. However, there is still a lack of systematic induction and analysis of miRNA regulation of autophagy signaling pathways induced by cerebral ischemia/reperfusion. This article reviews the regulation of cellular au⁃·综述·收稿日期:2023-10-16 录用日期:2023-12-11基金项目:国家自然科学基金(31660271),江苏省产学研合作项目(BY20221030),江苏省双创计划(JSSCBS20211166);广东省医学科学技术研究基金(A2020164);广东食品药品职业学院自然科学研究项目(2016YZ009),江苏医药职业学院博士科研启动项目(20200010)作者简介:刘筱蔼,第一作者,副教授,研究方向:神经损伤与修复,E-mail:***************.cn;罗友根,通信作者,教授,E-mail:***************第45卷中山大学学报(医学科学版)tophagy during cerebral ischemia/ reperfusion by miRNA-124, miRNA-298, miRNA-202-5p, miRNA-142, miRNA-26b and so on through different signaling pathways, providing a systematic and theoretical approach for the study of autoph⁃agy in stroke.Key words:cerebral ischemia reperfusion injury;MicroRNA;autophagy;signal pathway;autophagy related pro⁃teins; regulation[J SUN Yat⁃sen Univ(Med Sci),2024,45(1):21-27]脑卒中具有高致残率和高死亡率的特点,早期预防具有十分重要的意义。
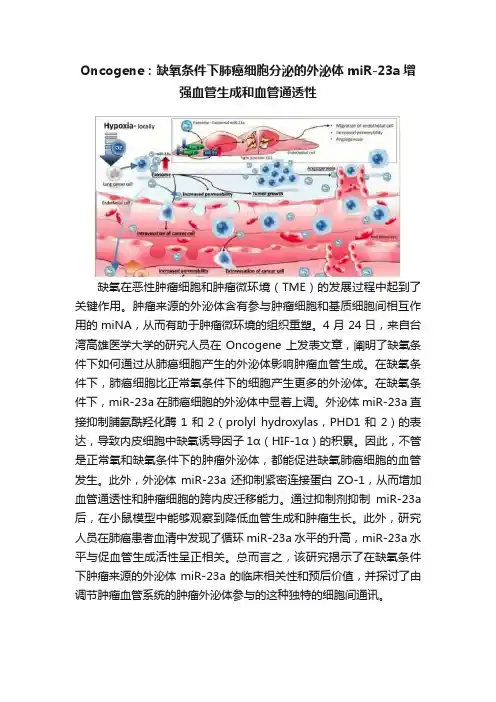
Oncogene:缺氧条件下肺癌细胞分泌的外泌体miR-23a增强血管生成和血管通透性缺氧在恶性肿瘤细胞和肿瘤微环境(TME)的发展过程中起到了关键作用。
肿瘤来源的外泌体含有参与肿瘤细胞和基质细胞间相互作用的miNA,从而有助于肿瘤微环境的组织重塑。
4月24日,来自台湾高雄医学大学的研究人员在Oncogene上发表文章,阐明了缺氧条件下如何通过从肺癌细胞产生的外泌体影响肿瘤血管生成。
在缺氧条件下,肺癌细胞比正常氧条件下的细胞产生更多的外泌体。
在缺氧条件下,miR-23a在肺癌细胞的外泌体中显着上调。
外泌体miR-23a直接抑制脯氨酰羟化酶1和2(prolyl hydroxylas,PHD1和2)的表达,导致内皮细胞中缺氧诱导因子1α(HIF-1α)的积累。
因此,不管是正常氧和缺氧条件下的肿瘤外泌体,都能促进缺氧肺癌细胞的血管发生。
此外,外泌体miR-23a还抑制紧密连接蛋白ZO-1,从而增加血管通透性和肿瘤细胞的跨内皮迁移能力。
通过抑制剂抑制miR-23a 后,在小鼠模型中能够观察到降低血管生成和肿瘤生长。
此外,研究人员在肺癌患者血清中发现了循环miR-23a水平的升高,miR-23a水平与促血管生成活性呈正相关。
总而言之,该研究揭示了在缺氧条件下肿瘤来源的外泌体miR-23a的临床相关性和预后价值,并探讨了由调节肿瘤血管系统的肿瘤外泌体参与的这种独特的细胞间通讯。
图1:肺癌细胞分泌的外泌体。
图2:缺氧条件下的肺癌细胞分泌的外泌体促进血管生成、内皮细胞通透性,增强肿瘤细胞的内皮迁移。
图3:肺癌病人体内的循环miR-23a情况。
参考文献:Hsu YL, Hung JY, Chang WA, Lin YS, Pan YC, Tsai PH, Wu CY, Kuo PL. Hypoxic lung cancer-secreted exosomal miR-23a increased angiogenesis and vascular permeability by targeting prolyl hydroxylase and tight junction protein ZO-1. Oncogene. 2017 Apr 24. PubMed PMID: 28436951.。

低压低氧暴露条件下肺动脉高压大鼠循环microRNA 表达及其意义徐娇阳;司马玲;是文辉;付勇;刘江伟;周瑾;余伍忠;桂俊豪【摘要】目的:研究低压低氧暴露条件下肺动脉高压(HPH)模型大鼠循环microRNA(miRNA)表达的变化。
方法利用基因芯片技术定量检测、分析 HPH 组及对照组大鼠血清中潜在的循环miRNA的表达及其差异。
基于病例‐对照设计,采用荧光定量PCR技术对差异化表达miRNA进行验证。
受试者工作特征曲线(ROC)分析测试4种差异化表达miRNA对HPH组及对照组的鉴别效能。
结果与对照组比较,HPH组大鼠13种miRNA表达上调,10种miRNA表达下调,其中,荧光定量PCR初步证实miR‐451、miR‐505及let‐7d表达上调,而miR‐214表达下调。
ROC分析结果显示,miR‐451、miR‐505及let‐7 d用于鉴别HPH组大鼠和对照组大鼠的曲线下面积(AUC)分别为0.979、0.938和0.993。
结论低压低氧暴露条件下 HPH组大鼠与对照组大鼠循环miRNA的表达存在显著差异,提示血清miRNA差异化表达可能与HPH病理变化相关。
%Objective To investigate the expression of circulating microRNA (miRNA) of rats with hypobaric hypoxia‐induced pulmonary hypertension (HPH) .Methods Commercial rat miRNA microarray was employed to detect and analyze the circulating miRNA profile in the serum samples of Sprague‐Dawley rats with hypobaric hypoxia‐induced HPH andcontrols .Furthermore ,differentially expressed candidate circulating miRNAs be tween HPH and control groups were validated by Real‐time quantitative PCR based on the case‐control study ,and receiver operating characteristic curve (ROC ) analysis was used to test the performance offour differentially expressed circulating miRNAs in discriminating HPH and control groups .Results Compared with those in the control group ,13 upregulated miRNAs and 10 downregulated miRNAs were identified in hypobaric hypoxia‐induced HPH rats by using miRNA microarray . And differentially expressed miR‐451, miR‐505 , let‐7d and miR‐214 were validated by using RT‐PCR .ROC analysis showed that the area under the curve of miR‐451 ,miR‐505 and let‐7d was 0 .979 ,0 .938 and 0 .993 in discriminating HPH and control groups ,respectively .Conclusion The aberrant e xpression of circulating miR‐451 ,miR‐505 and let‐7d in serum may be correlated with the pathogenesis of HPH .【期刊名称】《西安交通大学学报(医学版)》【年(卷),期】2016(037)004【总页数】4页(P556-559)【关键词】低氧性肺动脉高压;循环miRNA;基因芯片;荧光定量PCR【作者】徐娇阳;司马玲;是文辉;付勇;刘江伟;周瑾;余伍忠;桂俊豪【作者单位】开封市中心医院检验科,河南开封 475000; 兰州军区乌鲁木齐总医院,新疆乌鲁木齐 830000;兰州军区乌鲁木齐总医院,新疆乌鲁木齐 830000;兰州军区乌鲁木齐总医院,新疆乌鲁木齐 830000;兰州军区乌鲁木齐总医院,新疆乌鲁木齐 830000;兰州军区乌鲁木齐总医院,新疆乌鲁木齐 830000;兰州军区乌鲁木齐总医院,新疆乌鲁木齐 830000;兰州军区乌鲁木齐总医院,新疆乌鲁木齐830000;兰州军区乌鲁木齐总医院,新疆乌鲁木齐 830000【正文语种】中文【中图分类】R446.1◇基础研究◇低压低氧暴露能够诱发机体的一系列生理病理变化,其中,慢性缺氧诱发的低氧性肺动脉高压(hypoxic pulmonary hypertension, HPH)是高原心脏病、高原肺水肿等高原特有的多发疾病发生发展的中心环节,低压低氧暴露条件下HPH相关生物标志物的发掘对于疾病的早期预警和治疗具有重要意义。

低氧训练调控microRNA表达的研究现状随着越来越多的人们对低氧训练的关注,越来越多的实验研究表明低氧训练正逐渐成为重要的一种训练方法。
低氧训练可以有效地提高人体体能和心肺功能、调控代谢和脑功能等,因此越来越多的运动员和普通人们开始重视这种训练方式。
但是在低氧训练中,microRNA(miRNA)也扮演了重要的角色,因此本文将详细介绍低氧训练调控miRNA表达的研究现状。
MiRNA是一种小分子RNA,通常在许多细胞过程中被发现。
它们主要通过抑制靶基因的翻译来发挥作用,并且已经被证明在多种疾病中发挥了关键作用。
研究表明,在低氧训练中,运动导致的氧分压降低和mitochondrial dysfunction会导致有关miRNA的表达变化,这些miRNA的表达变化进一步影响着人体的代谢和适应性调节。
具体来说,已经有一些研究证明,低氧训练可以刺激miRNA-21、miRNA-210、miRNA-199a、miRNA-214和miRNA-38等miRNA的表达。
这些miRNA具有广泛的生物功能,可以调节细胞增殖、损伤修复、代谢和脑功能等。
其中,miRNA-21和miRNA-210可能是最重要的miRNA之一,它们已经被证明可以通过靶向特定的信号通路和转录因子来调节细胞增殖和凋亡、氧化应激和炎症等过程。
此外,miRNA-199a和miRNA-214也扮演了重要的角色,它们可以增强肌肉代谢和线粒体功能,并通过抑制氧化应激和干细胞分化来增强人体对低氧环境的适应性。
除了上述miRNA之外,还有一些miRNA的表达受到低氧训练的负调节,例如miRNA-486,miRNA-1和miRNA-133a。
这些miRNA分别控制肌肉细胞的分化、肌肉损伤修复和巨噬细胞的代谢等重要过程。
因此,低氧训练会削弱这些miRNA的表达,从而影响上述过程的正常进行。
总之,低氧训练通过调节miRNA的表达影响着肌肉代谢、神经功能和干细胞分化等生物过程。

㊃论 著㊃D O I :10.3969/j.i s s n .1672-9455.2023.24.009新生大鼠缺氧缺血性脑病中m i R N A 表达谱的改变及分析*袁 昊,李 婷,张腾伟,肖 娟,杨 刘,黎 巧,阮 滔,朱 昉,肖咏梅,彭湘莲ә湖南省妇幼保健院新生儿一科,湖南长沙410000摘 要:目的 通过基因芯片检测新生大鼠缺氧缺血性脑病(H I E )模型中差异性表达m i R N A ,构建H I E中m i R N A 及其靶基因调控网络㊂方法 将7d 龄新生S D 大鼠12只随机分为H I E 组及假手术对照组,每组6只㊂采用R i c e 法建立新生大鼠H I E 模型,造模24h 取新生大鼠海马组织,基因芯片检测H I E 组和假手术对照组海马组织中差异性表达的m i R N A ,采用生物信息软件分析相关m i R N A 调控的靶基因及信号通路㊂结果筛选出差异性表达的m i R N A 共22个,其中表达上调9个(mm u -m i R -215-5p ,mm u -m i R -1249-3p,mm u -m i R -3072-5p ,mm u -m i R -324-3p ,mm u -m i R -690,mm u -m i R -874-5p,11_10767,11_10888,13_12928_s t a r )㊁表达下调共13个(mm u -m i R -141-3p ,mm u -m i R -182-5p ,mm u -m i R -183-5p ,mm u -m i R -190a -3p ,mm u -m i R -200a -3p,mm u -m i R -200b -3p ,mm u -m i R -200c -3p ,mm u -m i R -429-3p ,mm u -m i R -455-3p ,mm u -m i R -760-5p,mm u -m i R -96-5p,6_5971_s t a r ,9_8784)㊂G O 功能分析显示,差异性表达的m i R N A 调控基因在生物过程中主要参与调节生物生长发育㊁神经元形成与分化过程,K E G G 通路分析显示主要有丝裂原活化蛋白激酶信号通路被激活㊂结论 差异性表达的m i R N A 可能参与调控H I E 发病的机制,将为H I E 的机制研究㊁临床诊断及药物设计提供理论依据和新的思路㊂关键词:缺氧缺血性脑病; m i R N A ; 基因芯片中图法分类号:R 722.19文献标志码:A 文章编号:1672-9455(2023)24-3619-04C h a n g e a n d a n a l y s i s o f m i R N A e x p r e s s i o n s pe c t r u m i n n e o n a t a l r a t s w i t h h y p o x i c -i s c h e m i c e n c e p h a l o p a t h y*Y U A N H a o ,L I T i n g ,Z HA N G T e n gw e i ,X I A O J u a n ,Y A N G L i u ,L I Q i a o ,R U A N T a o ,Z HU F a n g ,X I A O Y o n g m e i ,P E N G X i a n gl i a n әF i r s t D e p a r t m e n t o f N e o n a t o l o g y ,H u n a n P r o v i n c i a l M a t e r n a l a n d C h i l d H e a t h C a r e H o s pi t a l ,C h a n gs h a ,H u n a n 410000,C h i n a A b s t r a c t :O b je c t i v e T o d e t e c t t h e d if f e r e n t i a l e x p r e s s i o n o f m i R N A i n t h e n e o n a t a l r a t m o d e l o f h y p o x i c -i s c h e m i c e n c e p h a l o p a t h y (H I E )b yg e n e m i c r o a r r a y f o r c o n s t r u c t i n g th e mi R N A a n d i t s t a r g e t g e n e r e g u l a t o r yn e t w o r k i n H I E .M e t h o d s T w e l v e n e o n a t a l S D r a t s a g e d 7d w e r e r a n d o m l y d i v i d e d i n t o H I E g r o u p an d s h a m o p e r a t i o n c o n t r o l g r o u p ,w i t h 6r a t s i n e a c h g r o u p .T h e H I E m o d e l w a s e s t a b l i s h e d b y th e R i c e m e t h o d ,a n d t h e h i p p o c a m p u s t i s s u e o f n e o n a t a l r a t s w a s t a k e n a t 24h a f t e r m o d e l i n g .T h e m i R N A d i f f e r e n t i a l l y e x pr e s s -i n g i n t h e h i p p o c a m p u s t i s s u e o f t h e H I E g r o u p a n d c o n t r o l g r o u p w a s d e t e c t e d b y t h e g e n e m i c r o a r r a y,a n d t h e t a r g e t g e n e s a n d s i g n a l i n g p a t h w a y s r e g u l a t e d b y r e l a t e d m i R N A w e r e a n a l y z e d b yt h e b i o i n f o r m a t i c s o f t -w a r e .R e s u l t s A t o t a l o f 22d i f f e r e n t i a l l y e x pr e s s e d m i R N A s w e r e s c r e e n e d o u t ,i n w h i c h t h e 9m i R N A s e x -p r e s s i o n s w e r e u p -r e g u l a t e d (mm u -m i R -215-5p ,mm u -m i R -1249-3p ,mm u -m i R -3072-5p ,mm u -m i R -324-3p,mm u -m i R -690,mm u -m i R -874-5p ,11_10767,11_10888,13_12928_s t a r ),a n d 13m i R N A s w e r e d o w n r e gu l a t e d (mm u -m i R -141-3p ,mm u -m i R -182-5p ,mm u -m i R -183-5p ,mm u -m i R -190a -3p ,mm u -m i R -200a -3p,mm u -m i R -200b -3p ,mm u -m i R -200c -3p ,mm u -m i R -429-3p ,mm u -m i R -455-3p ,mm u -m i R -760-5p ,m m u -m i R -96-5p ,6_5971_s t a r ,9_8784).T h e G O f u n c t i o n a l a n a l y s i s s h o w e d t h a t d i f f e r e n t i a l l y m i R N A r e g u l a t o r y g e n e s w e r e m a i n l yi n -v o l v e d i n r e g u l a t i n g t h e b i o l o g i c a l g r o w t h ,d e v e l o pm e n t ,n e u r o n a l f o r m a t i o n a n d d i f f e r e n t i a t i o n p r o c e s s e s .T h e K E G G p a t h w a y s h o w e d t h a t t h e m i t o g e n a c t i v a t e d p r o t e i n k i n a s e s i g n a l i n g p a t h w a y w a s m a i n l y ac t i v a t ed .C o n c l u s i o n D i f fe r e n t i a l l y e x p r e s s e d m i R N A m a y p a r t i c i p a t e i n t h e r e g u l a t i o n of t h e p a t h og e n e s i s o f H I E ,whi c h w i l l p r o v i d e a t h e o r e t i c a l b a s i s a n d n e w i d e a s f o r t h e m e c h a n i s m r e s e a r c h ,c l i n i c a l d i a g n o s i s a n d d r u g d e -s i gn o f H I E .K e y wo r d s :h y p o x i c -i s c h e m i c e n c e p h a l o p a t h y ; m i R N A ; g e n e m i c r o a r r a y ㊃9163㊃检验医学与临床2023年12月第20卷第24期 L a b M e d C l i n ,D e c e m b e r 2023,V o l .20,N o .24*基金项目:湖南省妇幼保健院科研基金课题(201926)㊂ 作者简介:袁昊,女,主治医师,主要从事新生儿神经系统方面的研究㊂ ә通信作者,E -m a i l :340273789@q q.c o m ㊂新生儿缺氧缺血性脑病(H I E)是指足月和近足月新生儿由于围生期缺氧导致的急性脑损伤㊂我国报告H I E发生率为活产婴儿的3ɢ~6ɢ,其中15%~20%患儿在新生儿期死亡,存活者中有25%~ 30%可遗留严重的神经功能障碍,包括脑瘫㊁癫痫㊁视听障碍㊁认知障碍㊁行为异常等,成为影响我国儿童生活质量的重要疾病之一[1-2]㊂因此,积极研究H I E的发生机制㊁早期诊断及针对性干预治疗,在临床上具有重大意义㊂m i c r o R N A(m i R N A)是一种内源性非编码R N A(由18~23个核苷酸组成),其功能广泛,参与细胞周期㊁增殖和分化㊁信号传导和凋亡等过程[3]㊂有研究证明,许多m i R N A在神经系统损伤修复过程中发挥重要的调控作用[4],将成为H I E具有潜力的治疗靶点及有效的生物预测指标㊂本研究利用基因芯片技术,分析出新生大鼠H I E中差异表达的m i R N A,筛选特异性的m i R N A,探讨其可能的发生机制,为H I E的机制研究㊁临床诊断及药物设计提供理论依据和新的治疗思路㊂1材料与方法1.1实验动物及分组7d龄S D大鼠12只,雌雄不限,体质量12~16g,购自广东省实验动物检测所㊂随机分为H I E组及假手术对照组,每组6只㊂1.2主要材料及仪器基因芯片筛选m i R N A由新开源晶锐(广州)生物医药科技有限公司完成㊂1.3方法1.3.1造模采用R i c e法制作H I E模型:7d龄S D 新生大鼠乙醚吸入麻醉后,仰卧位固定于手术台,消毒切开颈部皮肤,分离左侧颈总动脉并在远㊁近心端进行双结扎,缝合切口,术后放入37ħ恒温箱中30 m i n,以1.5L/m i n不断输入含8%氧气和92%氮气的低氧气源2h㊂假手术对照组:7d龄S D新生大鼠只游离左侧颈总动脉穿线但不结扎,缝合伤口后不进行缺氧处理㊂两组手术时间均低于10m i n,环境温度26~28ħ㊂造模后观察新生大鼠行为情况㊂1.3.2取新生大鼠海马组织造模后24h,取海马组织㊂给予新生大鼠乙醚吸入麻醉,在低温环境下快速断头取脑,冰上分离海马组织㊂将左右两侧海马放入冻存管中,液氮速冻后放入-80ħ低温冰箱备用㊂1.3.3基因芯片筛选差异性m i R N A 取新生大鼠海马组织,采用T r i z o l法进行样品的总R N A抽提纯化并质检㊂将提取的总R N A送检,进行微阵列杂交㊁芯片扫描,利用生物信息软件筛选显著差异的m i R-N A㊂本实验采用i l l u m i n a H i s e q测序平台的单端50 b p测序模式对样本进行高通量测序㊂原始数据需经过引物与a d a p t o r序列去除,并经过对测序片段碱基的质量检验和长度筛选,最终选择质量可靠的测序片段㊂将每个样本的r e a d s比对到已有的m i R N A数据库(m i R B a s e)和新m i R N A预测的结果上,从而计算m i R N A表达量㊂利用D E S e q软件进行差异表达分析,筛选差异表达的m i R N A,计算每个样本的表达量和组内均值,并且计算组间差异F o l d C h a n g e,再计算l o g2(F o l d C h a n g e)用于后续筛选差异基因㊂1.3.4目的基因G O功能集分析及富集分析将全部基因作为背景列表,目的基因列表作为从背景列表中筛选出来的候选列表,利用F i s h e r精确检验计算G O功能集在目的基因列表中是否显著富集的P值,再对P值经B e n j a m i n i&H o c h b e r g多重检验纠正后得到F D R㊂针对这些基因进行K E G G数据库中通路的功能注释和归类,以及D i s G e N E T疾病数据库中疾病类型的功能注释和归类㊂1.4统计学处理利用S e q t k(1.0-r82-d i r t y)对预处理数据进行质量控制分析㊂序列比对使用B o w t i e (1.0.0)软件分析,采用D E S e q软件进行差异表达分析㊂以P<0.05为差异有统计学意义㊂2结果2.1行为学评估 H I E组术后1只新生大鼠表现为持续精神萎靡,其余5只均出现先兴奋后抑制行为:首先躁动不安;术后30m i n,出现呼吸增快㊁站立不稳㊁全身震颤症状;术后1h,出现精神萎靡㊁嗜睡㊁易激惹㊁夹尾左旋症状㊂假手术对照组活动正常㊂2.2 m i R N A检测结果分析2.2.1测序数据质控针对每个样本预处理后所有序列和去重复后的唯一序列,分别采用b o w t i e软件与该物种的参考基因组㊁R f a m序列数据库㊁R e p B a s e序列数据库㊁m i R B a s e数据库进行比对,平均质控率大于89.00%,平均对比率约为47.96%,所获得数据能满足进一步分析要求㊂2.2.2差异表达的m i R N A 通过比较两组m i R N A 表达量,利用D E S e q软件对两组进行差异表达分析,筛选出共22个表达存在差异的m i R N A,H I E组表达上调的m i R N A共9个(m m u-m i R-215-5p,m m u-m i R-1249-3p,m m u-m i R-3072-5p,m m u-m i R-324-3p,m m u-m i R-690,m m u-m i R-874-5p,11_10767,11_10888,13_ 12928_s t a r),表达下调的m i R N A共13个(m m u-m i R-141-3p,m m u-m i R-182-5p,mm u-m i R-183-5p,mm u-m i R-190a-3p,mm u-m i R-200a-3p,mm u-m i R-200b-3p,mm u-m i R-200c-3p,mm u-m i R-429-3p,mm u-m i R-455-3p,mm u-m i R-760-5p,mm u-m i R-96-5p,6_5971_ s t a r,9_8784),见图1和表1㊂当满足P<0.05并且|l o g2(F o l d C h a n g e)|ȡ0.58时,认为该基因表达在组间差异有统计学意义㊂2.2.3差异性m i R N A的靶基因预测及富集分析使用m i R a n d a对差异表达的m i R N A进行靶标预测,共得到1495个靶基因㊂针对目的基因集,采用T o p G O 软件进行G O功能分析㊂图2展示预测靶基因在生物过程㊁细胞组成㊁分子功能的前10个最显著的G O㊂结果显示差异m i R N A调控基因在生物过程中主要参与调节生物生长发育㊁神经元形成与分化过程,在细胞组成中主要与突触发生㊁突触可塑性有关,在分子功能中调节蛋白结合㊁酶结合及谷氨酸受体活性等㊂㊃0263㊃检验医学与临床2023年12月第20卷第24期 L a b M e d C l i n,D e c e m b e r2023,V o l.20,N o.24图1 H I E 组与假手术对照组对比的火山图表1 H I E 组与假手术对照组差异性m i R N A 筛选差异性m i R N Al o g 2(Fo l d C h a n g e )P表达m m u -m i R -215-5p2.440.00上调m m u -m i R -1249-3p 0.770.04上调m m u -m i R -3072-5p 0.830.04上调m m u -m i R -324-3p 0.970.02上调m m u -m i R -6900.790.04上调m m u -m i R -874-5p1.000.02上调11_107671.490.00上调11_108880.710.03上调13_12928_s t a r 0.930.01上调m m u -m i R -141-3p -0.700.02下调m m u -m i R -182-5p -0.780.01下调m m u -m i R -183-5p -0.710.02下调m m u -m i R -190a -3p -0.770.00下调m m u -m i R -200a -3p -0.800.01下调m m u -m i R -200b -3p-0.780.01下调m m u -m i R -200c -3p -0.690.03下调m m u -m i R -429-3p -0.680.04下调m m u -m i R -455-3p -0.900.02下调m m u -m i R -760-5p -0.620.04下调m m u -m i R -96-5p-0.630.03下调6_5971_s t a r -0.890.01下调9_8784-0.710.02下调注:l o g 2(F o l d C h a n g e )为H I E 组相对于假手术对照组评价表达量的差异倍数的l o g 2转化值㊂ 注:横坐标代表-l g(P 值),纵坐标代表显著富集的G O 名称㊂图2 显著富集G O 柱状图针对这些基因进行K E G G 数据库中通路的功能注释和归类㊂图3显示富集到的主要通路有丝裂原活化蛋白激酶(MA P K )信号通路㊁神经节鞘磷脂生物合成㊁细胞内吞作用等㊂其中以MA P K 信号通路最为显著,H I E 组表达上调的11-10767及mm u -m i R -874-5p 调控的8个靶基因(B r a f ,M a p t ,P p p5c ,M a p k 8i p 2,F g f 6,F l t 1,N g f r ,T gf b 1)共同参与此信号通路㊂注:横坐标代表显著富集的K E G G 通路名称,纵坐标代表-l g (P 值)㊂纵坐标越显著表示该通路越富集显著,红色柱表示显著的通路(P <0.05),蓝色柱表示不显著的通路㊂图3 显著富集K E G G 通路柱状图3 讨 论新生儿H I E 病理生理改变主要包括脑血流低灌注或过度灌注㊁脑细胞能力代谢障碍㊁自由基损伤及神经元坏死或过度凋亡等㊂临床治疗策略主要通过增加氧分压㊁维持脑血流灌注等多方法联合治疗,但不能有效阻断神经损伤㊂如何进行早期诊断及高效治疗,最大程度上减少后遗症发生,是近年来国内外研究热点㊂m i R N A 被认为参与多种疾病的发病机制,能够通过转录后调控靶基因的表达来调节不同的细胞过程[5],可用于疾病分子生物诊断㊁靶向治疗和评估㊂目前,已有大量研究证明,m i R N A 在H I E 损伤修复过程中发挥重要的调控作用,将有助于新生儿H I E 早期诊断㊁评估预后及靶向治疗[6-7]㊂本研究通过构建H I E 新生大鼠模型,在H I E 发生24h 进行基因测序,共筛选出22个表达差异的m i R N A ,其中9个表达上调,13个表达下调㊂通过靶基因预测,将目的基因进行G O 功能富集分析,显示调控基因主要涉及调节生物生长发育㊁神经元形成与分化的过程,促进突触发生㊂因此,笔者推测在H I E 发生脑损伤早期,m i R N A 大多富集在与神经元功能相关的生物通路中,修复受损神经元㊂在H I E 表达上调的m i R N A 中,mm u -m i R -215-5p 差异倍数最为明显,大量研究认为,其参与调控肿瘤发生㊁发展㊂在非小细胞肺癌,m i R -215-5p 通过调控MA P K /E R K 信号通路靶向调节Z E B 2表达,参与细胞凋亡及死亡[8]㊂在恶性胸膜间皮瘤中,m i R -215-5p 通过激活基因p53功能,参与D N A 损伤修复㊁细胞周期调控㊁细胞凋亡及抑制血管生成等过程,其低表达与该疾病不良预后有关[9]㊂在其他肿瘤疾病如乳腺癌㊁结肠直肠癌㊁髓系白血病等中均有异常表㊃1263㊃检验医学与临床2023年12月第20卷第24期 L a b M e d C l i n ,D e c e m b e r 2023,V o l .20,N o .24达[10]㊂本研究中发现mm u-m i R-215-5p表达明显上调,考虑可能参与缺氧缺血损伤后神经元凋亡及死亡的调控机制,需进一步研究验证,其有望成为评估H I E的生物标志物㊂在H I E表达下调的m i R N A中,m i R-200家族最为明显㊂m i R-200家族成员包括m i R-200a㊁m i R-200b㊁m i R-200c㊁m i R-141㊁m i R-429,其参与多种疾病的发病机制,包括神经退行性病变㊁纤维化㊁肿瘤等[11]㊂在缺氧缺血发生后,少突胶质前体细胞迅速反应㊁增殖㊁迁移到损伤部位,分化为成熟的少突胶质细胞,包绕轴突形成髓鞘,促进神经修复㊂在脑卒中的研究中发现,m i R N A-200过表达可抑制血清反应因子表达,从而抑制少突胶质前体细胞分化[12]㊂在缺氧研究中发现,抑制m i R N A-200表达可诱导内皮细胞血管生成,改善组织缺氧[13]㊂由此可见,m i R N A-200参与神经系统损伤修复的病理过程㊂本研究发现m i R N A-200表达下调,G O功能富集提示主要与神经元形成与分化有关,推测新生大鼠神经系统损伤后,机体修复神经元及促进血管生成,与上述研究结论一致㊂故本研究认为,m i R-200家族有望成为H I E治疗靶点之一㊂在H I E中K E G G通路分析结果显示MA P K信号通路激活㊂MA P K家族主要包括细胞外调节蛋白激酶(E R K)㊁c-J u n氨基末端激酶(J N K)和p38丝裂原活化蛋白激酶(p38MA P K),通过调节细胞内相关基因转录,参与细胞增殖㊁分化㊁癌变㊁转移㊁凋亡等生理过程,介导生长发育㊁炎症反应等[14]㊂研究表明, MA P K参与调节脑缺血再灌注损伤的发生㊁发展过程,但存在脑保护及脑损伤双重作用[15-16]㊂本研究发现H I E组表达上调的11-10767及mm u-m i R-874-5p 调控的8个靶基因(B r a f,M a p t,P p p5c,M a p k8i p2, F g f6,F l t1,N g f r,T g f b1)共同参与此信号通路,可为治疗靶点提供参考方向㊂综上所述,本研究初步筛选出H I E中差异m i R-N A表达谱,构建靶基因调控网络,提示H I E发生机制复杂多样,需进一步对差异表达的m i R N A进行验证㊁分子机制及信号通路调控的研究㊂随着不断发现及深入研究m i R N A及其靶位点,m i R N A在H I E中发挥的作用将逐步被揭开,分子机制也将逐渐阐明㊂这将为H I E找到新的生物标志物和治疗靶点提供思路及理论基础㊂参考文献[1]邵肖梅,叶鸿瑁,丘小汕.实用新生儿学[M].5版.北京:人民卫生出版社,2019.[2]WA N G Q,L V H,L U B,e t a l.N e o n a t a l h y p o x i c-i s c h e m i ce n c e p h a l p a t h y:e m e r g i n g t h e r a p e u t i c s t r a t e g i e s b a s e d o np a t h o p h y s i o l o g i c p h a s e o f t h e i n j u r y[J].M a t e r n F e t a l N e o n a t a l M e d,2019,32(21):3685-3692.[3]K L O O S T E R MA N W P,P L A S T E R K R H.T h e d i v e r s ef u n c t i o n o f m i c r o R N A s i n a n i m a l d e v e l o p m e n t a n d d i s-e a s e[J].D e v C e l l,2006,11(4):441-450.[4]P E E P L E S E S,S A HA R N E,S N Y D E R W,e t a l.T e m p o-r a l b r a i n m i c r o R N A e x p r e s s i o n c h a n g e s i n a m o u s e m o d e l o f n e o n a t a l h y p o x i c-i s c h e m i c i n j u r y[J].P e d i a t r R e s, 2022,91(1):92-100.[5]M E N D E L L J T,O L S O N E N.M i c r o R N A s i n s t r e s s s i g-n a l i n g a n d h u m a n d i s e a s e[J].C e l l,2012,148:1172-1187.[6]P E E P L E S E S,M i c r o R N A t h e r a p e u t i c t a r g e t s i n n e o n a t a lh y p o x i c-i s c h e m i c b r a i n i n j u r y:a n a r r a t i v e r e v i e w[J].P e d i-a t r R e s,2023,93(4):780-788.[7]P O N N U S AMY V,Y I P P K.T h e r o l e o f m i c r o R N A i nn e w b o r n b r a i n d e v e l o p m e n t a n d h y p o x i c i s c h a e m i c e n-c e p h a l o p a t h y[J].N e u r o p h a r m a c o l o g y,2019,149(1):55-65.[8]H O U Y,Z H E N J,X U X,e t a l.m i R-215f u n c t i o n s a s a t u m o r s u p p r e s s o r a n d d i r e c t l y t a r g e t s Z E B2i n h u m a n n o n-s m a l l c e l l l u n g c a n c e r[J].O n c o l L e t t,2015,10:1985-1992.[9]S I N G H A,B HA T T A C HA R Y Y A N,S R I V A S T A V A A,e t a l.M i c r o R N A-215-5p t r e a t m e n t s u p p r e s s e s m e s o t h e l i-o m a p r o g e s s i o n v i a t h e M D M2-p53-s i g n a l i n g a x i s[J].M o l T h e r,2019,4:1665-1680.[10]L I W,L I N G D I L,X I Q I A N G D,e t a l.M i c r o R N A-215-5pi n h i b i t s t h e p r o l i f e r a t i o n a n d m i g r a t i o n o f W i l m's t u m o rc e l l s b y t a r g e t i n g C R K[J].T e c h n o l C a n c e r R e s T r e a t,2021,20:15330338211036523.[11]S U N D A R A R A J A N V,B U R K U C,B A J D A K-R U S I N E KK.R e v i s i t i n g t h e m i R-200F a m i l y:a c l a n o f f i v e s i b l i n g s w i t h e s s e n t i a l r o l e s i n d e v e l o p m e n t a n d d i s e a s e[J].B i o-m o l e c u l e s,2022,12(6):781.[12]B U L L E R B,C HO P P M,U E N O Y,e t a l.R e g u l a t i o n o fs e r u m r e s p o n s e f a c t o r b y m i R N A-200a n d m i R N A-9 m o d u l a t e s o l i g o d e n d r o c y t e p r o g e n i t o r c e l l d i f f e r e n t i a t i o n [J].G l i a,2012,60(12):1906-1914.[13]C HA N Y C,K HA N N A S,R O Y S,e t a l.M i R-200b t a r-g e t s E t s-1a n d i s d o w n-r e g u l a t e d b y h y p o x i a t o i n d u c e a n-g i o g e n i c r e s p o n s e o f e n d o t h e l i a l c e l l s[J].J B i o l C h e m, 2011,286(3):2047-2056.[14]B O S T F,A O U A D I M,C A R O N L,e t a l.T h e r o l e o fMA P K s i n a d i p o c y t e d i f f e r e n t i a t i o n a n d o b e s i t y[J].B i o-c h i m i e,2005,87(1):51-56.[15]Z HA N G J,X I A J,Z HA N G Y,e t a l.HMG B1-T L R4s i g-n a l i n g p a r t i c i p a t e s i n r e n a l i s c h e m i a r e p e r f u s i o n i n j u r ya n d c o u l db e a t t e n u a t e d b y d e x a m e t h a s o n e-m e d i a t e d i n h i-b i t i o n o f t h e E R K/N F-κB p a t h w a y[J].A m J T r a n s l R e s, 2016,8(10):4054.[16]Z HA O J,L E M,L I J,e t a l.L I N C00938a l l e v i a t e s h y p o x i ai s c h e m i a e n c e p h a l o p a t h y i n d u c e d n e o n a t a l b r a i n i n j u r y b y r e g u l a t i n g o x i d a t i v e s t r e s s a n d i n h i b i t i n g J N K/p38 MA P K s i g n a l i n g p a t h w a y[J].E x p N e u r o l,2023,367: 1414449.(收稿日期:2023-06-23修回日期:2023-10-27)㊃2263㊃检验医学与临床2023年12月第20卷第24期 L a b M e d C l i n,D e c e m b e r2023,V o l.20,N o.24。

低氧训练调控microRNA表达的研究现状作者:鲁哲朱磊来源:《当代体育科技》2019年第21期摘 ;要:近年来,微RNA(miRNA,microRNA)被发现是一类与RNA家族基因表达调控密切相关的小分子。
miRNA是一类非编码的单链RNA分子,内源性基因编码长度为21-25个核苷酸,miRNA在许多生物过程中起着不可或缺的作用,包括免疫反应、发育、干细胞分化、脂肪细胞分化和脂质代谢。
低氧及低氧训练可以调控多种miRNA的表达,不仅在许多组织细胞中得到了证明,而且也参与了脂肪细胞分化、脂代谢等多种生物过程调控。
为了促进低氧和低氧训练调控miRNA表达相关方面的研究,本文通过综述miRNA的合成与功能、运动对miRNA的影响,探讨低氧和低氧训练与miRNA的关系,以期促进miRNA与心肌梗死,缺血低氧损伤、肿瘤、血管发生等之间的联系,为肥胖机体减重降脂以及脂代谢相关疾病的干预提供研究思路。
关键词:microRNA ;低氧 ;低氧训练 ;脂代谢中图分类号:G804 ; ; ; ; ; ; ; ; ; ; ; ; ; ; ; ; ; 文献标识码:A ; ; ; ; ; ; ; ; ; ; ; 文章编号:2095-2813(2019)07(c)-0017-05Abstract:In recent years, microRNA (miRNA), a class of small molecules closely related to gene expression regulation, has been found in the RNA family.Micrornas are a class of non-coding single-stranded RNA molecules, endogenous gene encoding length is 2-25 nucleotides, the miRNA plays an indispensable role in many biological processes, including immune response,development, differentiation of stem cells, fat cells and lipid metabolism, transcription factors.Hypoxia and hypoxia training can regulate the expression of a variety of mirnas, which has not only been proved in many tissue cells, but also participated in the regulation of adipocyte differentiation, lipid metabolism and other biological processes.In order to promote hypoxia and hypoxia training control the miRNA expression related research, this article through the synthesis of micrornas and function, the influence of exercise on the miRNA, hypoxia and hypoxia training and the relationship of micrornas, in order to promote the miRNA and myocardial infarction,ischemia hypoxia injury, the connection between the tumor and angiogenesis, lipid and body weight loss for obesity intervention provides research train of thought of the lipid metabolism related diseases.Key Words:MicroRNA;Hypoxia;Hypoxic training ;Fat metabolism最近的研究表明[1],低氧训练不仅可以减轻体重和体脂,而且可以调节人体功能,降低患病风险,并且与miRNA的表达也存在微妙的关系,因此,从高原训练演变而来的常压低氧训练逐渐引起了国内外体育工作者的关注。


?论著??基础研究?LncRNA 在缺氧诱导的胃癌细胞中表达谱的变化王亚芳1,刘理礼2,靳海峰1,张慧1,张宏博1,聂勇战1,吴开春1,樊代明1Study on expression profile of long non -coding RNA in gastric cancer cell lines underhypoxiaWang Yafang 1,Liu Lili 2,Jin Haifeng 1,Zhang Hui 1,Zhang Hongbo 1,Nie Yongzhan 1,Wu Kaichun 1,Fan Daiming 11State Key Laboratory of Cancer Biology &Xijing Hospital of Digestive Diseases ,the Forth Military Medical University ,Shaanxi Xi 'an710032,China ;2Department of Oncology ,Tangdu Hospital ,the Forth Military Medical University ,Shaanxi Xi 'a n 710038,China.【Abstract 】Objective :To analyze the expression profile variation of long non -coding RNA (lncRNA )in gastric cancer cell lines under normoxia and hypoxia.Methods :To use lncRNA microarray technology to inspect the differ-ence of lncRNA expression profile between the gastric cancer cell lines under normoxia and hypoxia ,screening out the lncRNA with diferential expression after pretreatment and homogenization of raw data and make hierarchical clustering and volcano scatter diagram analysis.Results :Compare with the gastric cancer cell lines under normoxia ,and 254lncRNA which have more than 1.5times variation and significant diference (P <0.05)by statistical analysis are re-garded as lncRNA with differential expression ,accounting for 3.6%of all lncRNA :while 136increase more than 1.5times and 118reduce more than 1.5times ;30increase more than 2times :25reduce more than 2times ;5increase more than 3times ;and 5reduce more than 3times.Conclusion :LncRNA expression profile of the gastric cancer cell lines under hypoxia changes significantly in comparison with the gastric cancer cell lines under noemoxia .The different lncRNAs may include a fair proportion of lncRNA exerts enhancer -like effect and lncRNA exerts inhibitory effect ,participating in the critical section of regulatory mechanism in the gastric cancer cells under hy-poxia.【Key words 】long non -coding RNA ;lncRNA microarray ;gastric cancer ;hypoxiaModern Oncology 2013,21(02):0225-0228【摘要】目的:观察长链非编码RNA (lncRNA )在常氧条件下的胃癌细胞及缺氧诱导的胃癌细胞中表达谱的变化。
缺氧环境下微RNA对肿瘤的调节作用王宇竞(综述);敖敏高娃;刘恩才(审校)【摘要】MiRNAs are recognized as a new class of master regulators under hypoxia that control gene expression and are responsible for many normal and pathological cellular processes. Studies have shown that hypoxia inducible factor 1(HIF1) regulates a panel of miRNAs,whereas some of miRNAs target HIF1. The interaction between miRNAs and HIF1 can account for many vital events relevant to tumorigenesis,such as angiogenesis,metabolism,apoptosis,cell cycleregulation,proliferation,metastasis,and resistance to antican-cer therapy. Recently, a few reports revealed that hypoxia regulates the expression of miRNAs, which are involved in tumor target gene regulation,thus influence a wide variety of human tumors on molecular level, such as metabolism,apoptosis,metastasis and treatment.%微RNAs( miRNAs)目前被认为是缺氧的新型调控基因,它能调节基因表达并且参与许多正常细胞和肿瘤细胞的生理过程。
缺氧相关microRNA在窒息死亡原因推断中的意义曾颜;马剑龙;陈龙【摘要】在缺氧条件下microRNA(miRNA)能与转录因子等相互作用,调节细胞代谢,血管再生,血细胞生成,细胞增殖、分化及凋亡等生理过程,这些过程可能在窒息导致的死亡中发挥重要作用.本文拟对缺氧条件下miRNA的调节功能及缺氧对miRNA生物合成的影响进行综述,以期为miRNA在法医学窒息死亡原因推断中的研究提供新思路.%U nder hypoxia condition, m icroR N A (m iR N A ) can interact w ith transcription factors for regu-lating the cell m etabolism , angiogenesis, erythropoiesis, cellular proliferation, differentiation and apoptosis. T he biological processes above m ay play an im portant role in m echanical asphyxia death. T his article re-view s the regulating function of m iR N A under hypoxia condition and the influence of hypoxia to biosynthesis of m iR N A , w hich m ay provide som e new ideas to the research of m iR N A on determ ining the cause of m echanical asphyxia death in the field of forensic m edicine.【期刊名称】《法医学杂志》【年(卷),期】2017(033)001【总页数】4页(P38-41)【关键词】法医病理学;窒息;综述;microRNA;缺氧;死亡原因【作者】曾颜;马剑龙;陈龙【作者单位】复旦大学上海医学院法医学系,上海200032;复旦大学上海医学院法医学系,上海200032;复旦大学上海医学院法医学系,上海200032【正文语种】中文【中图分类】DF795.1microRNA(miRNA)是生物体内合成的长度约为22个核苷酸的非编码RNA,通过与靶基因mRNA 3′-非翻译区(untranslated regions,UTR)互补结合,促进靶基因mRNA的降解或抑制其翻译,在转录后水平调控基因表达,广泛参与细胞包括缺氧在内的生理过程[1]。
缺氧环境下子宫内膜腺上皮细胞中微RNA表达谱及细胞凋亡变化王含必;窦帅杰;刘思邈;张婉玉;刘美芝;邓成艳【期刊名称】《协和医学杂志》【年(卷),期】2022(13)4【摘要】目的探究缺氧环境下子宫内膜腺上皮细胞(endometrial glandular epithelial cell,EEC)中微RNA(micro-RNA,miRNA)表达谱变化及其对凋亡的影响。
方法取对数生长期EEC,接种至六孔板(1×10^(5)个细胞/孔)并随机分为缺氧组和对照组。
缺氧组、对照组分别置于缺氧环境(O_(2)∶N_(2)∶CO_(2)体积比为1∶94∶5)和正常氧环境(O_(2)∶CO_(2)体积比为95∶5)中培养。
培养4 h后收集细胞,加入Trizol并提取总RNA。
采用高通量测序法测定两组EEC中miRNA表达谱变化,采用实时荧光定量PCR(realtime fluorescence quantitative polymerase chain reaction,RT-PCR)法检测miR-7704、miR-7974基因表达水平,采用流式细胞术检测EEC凋亡情况,采用蛋白印迹法(Western blot)检测p53和凋亡相关蛋白表达变化。
结果高通量测序对21种常见miRNA表达水平检测后发现,与对照组比较,缺氧组EEC中16种miRNA表达上调,5种miRNA表达下调;其中对照组表达丰度较高的miR-7704和miR-7974在缺氧组表达下降最为显著(P均<0.05)。
RT-PCR结果显示,与对照组比较,缺氧组EEC中miR-7704和miR-7974相对表达量分别降低20%、80%。
流式细胞术检测结果显示,缺氧组早期凋亡率及晚期凋亡率均高于对照组(P均<0.001)。
Western blot检测结果显示,与对照组比较,缺氧组EEC中p53表达升高,抗凋亡蛋白B细胞淋巴瘤-2表达降低(P均<0.05)。
高表达microRNA-22对缺氧心肌细胞的保护作用及机制凌琳;凌子成;顾少华【期刊名称】《中国动脉硬化杂志》【年(卷),期】2017(25)10【摘要】目的观察高表达microRNA-22对体外培养的缺氧心肌细胞的保护作用及机制。
方法用携带microRNA-22的腺病毒载体和空载体转染体外缺氧条件下培养的原代心肌细胞,检测高表达microRNA-22后心肌细胞生物学特性的改变。
采用MTT检测细胞活力,EdU检测细胞DNA合成能力,Caspase-3检测细胞凋亡情况。
结果表达microRNA-22的腺病毒载体成功转染原代心肌细胞,流式细胞仪检测绿色荧光蛋白阳性细胞比例>95%,高表达microRNA-22从蛋白水平显著下调PTEN表达。
与空载腺病毒组相比,高表达microRNA-22组细胞核增殖明显增加,细胞生长更快,细胞活力增加,细胞凋亡减少。
结论体外高表达microRNA-22能保护缺氧条件下的心肌细胞。
【总页数】5页(P997-1001)【关键词】microRNA-22;心肌细胞:细胞特性【作者】凌琳;凌子成;顾少华【作者单位】苏州大学附属第一医院心血管内科;扬州大学医学院;昆山市第三人民医院【正文语种】中文【中图分类】R541.4【相关文献】1.自噬和PI3K-Akt-mTOR信号转导通路在缺氧预处理对高糖心肌细胞缺氧/复氧损伤保护中的作用 [J], 孙超;杨桂珍;薛富善;李慧娴;刘亚洋;廖旭2.LXA4诱导HO-1高表达对心肌细胞缺氧/复氧损伤的保护作用 [J], 唐艳荣;吴升华3.缺氧后适应与心肌营养素-1对心肌细胞缺氧复氧的保护作用及机制探讨 [J], 周贻军;赵亚男;赵鑫;缪绯;王先宝;颜竞;张秀丽;刘映峰;杨平珍4.缺氧后处理对糖尿病性心肌细胞缺氧复氧时保护作用削弱的机制:与DJ-1表达的关系 [J], 周斌;夏中元;赵博;孙倩;薛锐;刘敏5.微小RNA-124-3p通过靶向抑制RIPK1的表达对缺氧/复氧心肌细胞损伤中的保护作用及其机制研究 [J], 汪鲁青; 姜花; 郑生因版权原因,仅展示原文概要,查看原文内容请购买。
microRNA在缺氧性肺高压中的功能研究
张小英;苟德明
【期刊名称】《老年医学与保健》
【年(卷),期】2014(20)3
【摘要】microRNA (miRNA)是一类高度保守、长度约20 ~ 25个核苷酸(nt)的非编码单链小RNA.miRNA通过与靶基因mRNA 3’非翻译区(3’-UTR)特异性结合,使靶mRNA降解或翻译受阻,从而在转录后水平调控基因的表达.大量文献表明,miRNA已经成为新的靶点用于疾病的早期诊断或治疗.最近研究表明,miRNA也参与了缺氧性肺高压的发生与发展.现就miRNA在缺氧性肺高压中的异常表达、作用机制,以及作为诊断生物标志物和治疗靶点的研究进展予以综述.【总页数】5页(P145-149)
【作者】张小英;苟德明
【作者单位】518060深圳市,深圳大学生命科学学院;518060深圳市,深圳大学生命科学学院
【正文语种】中文
【中图分类】R544.1
【相关文献】
1.Nω——硝甘——L——精氨酸甲酯增强大鼠慢性缺氧性肺高压的研究 [J], 黎文;赵斌
2.Caspase-1在大鼠缺氧相关性肺高压中的作用及茶多酚对其影响的研究 [J], 雷
敏;徐戈;黄琛;欧婷;王利兵
3.血浆microRNA-21水平与左室射血分数保留的心力衰竭相关性肺高压的关系[J], 吴美华;曾建斌;熊向阳
4.热休克蛋白70调控Bcl-2/Bax对缺氧性肺高压新生大鼠肺血管内皮细胞凋亡及肺动脉压力的作用 [J], 赵倩;曹静;李明霞
5.马西替坦联合维利西呱治疗缺氧性肺高压大鼠的作用机制 [J], 李晨;李鑫;刘春蕾;杨明会
因版权原因,仅展示原文概要,查看原文内容请购买。
缺氧对H9C2心肌细胞中miR-19a-3p表达的研究黄浩;陈文江;刘长召;王玲;沈艳芳【摘要】目的检测大鼠H9C2心肌细胞缺氧后细胞水平和培养液中循环微小RNA(miRNA,miR)-19a-3p的表达,并探讨其与可能靶基因低密度脂蛋白受体相关蛋白2(LRP2)的关系.方法对 H9C2心肌细胞进行缺氧(37 ℃,5% CO2,1% O2)0、4、8、12、16、20、24 h处理,采用荧光实时定量聚合酶链反应(qRT-PCR)检测其在不同时间点细胞水平和培养液中循环 miR-19a-3p的表达水平,使用ELISA、qRT-PCR、Western Blot 检测LRP2在培养液和细胞中mRNA和蛋白表达.结果H9C2细胞培养液中miR-19a-3p的表达在缺氧0、4、8、12、16 h差异无统计学意义(P>0.05),在缺氧20 h与24 h时,H9C2细胞培养液中miR-19a-3p的表达与缺氧0 h比较,明显高表达(P<0.05);在缺氧20 h与24 h时,H9C2细胞中miR-19a-3p的表达与缺氧0 h比较,明显下降(P<0.05);H9C2细胞培养液中LRP2的表达在缺氧16 h后表达明显升高,缺氧16 h[(675.2 ± 42.4) ng/mL,n=3]、缺氧20 h[(979.4 ± 204.2)ng/mL,n=3]和缺氧24 h[(1456.0 ± 363.6)ng/mL,n=3]较0 h [(245.0 ± 10.84)ng/mL,n=3]明显升高(P<0.05);H9C2细胞中LRP2 mRNA表达水平在缺氧16 h后表达明显升高,缺氧16 h[(0.000503 ± 0.000100)ng/mL,n=3]、缺氧20 h[(0.001303 ± 0.000090)ng/mL,n=3]和缺氧24 h[(0.001617 ±0.000110)ng/mL,n=3]较0 h[(0.0001394 ± 0.0000600)ng/mL,n=3]明显升高(P<0.05);20 h[(0.235000 ± 0.003427)ng/mL,n=3]与24 h[(0.535000 ±0.003427)ng/mL,n=3]LRP2蛋白表达与0 h[(0.14010 ± 0.01821)ng/mL,n=3]比较,差异有统计学意义(P<0.01).结论 miR-19a-3p可能参与大鼠H9C2心肌细胞的缺氧坏死过程,并且可能通过调控LRP2而发挥作用.%Objective To study the relationship of miRNA-19a-3p and LDL receptor related protein 2(LRP2)under hypoxia in rat H9C2 cardiomyocytes.Methods H9C2 cardiomyocytes were treated in hypoxia condition(37 ℃,5% CO2,1% O2)for 0,4,8,12,16,20,24 h.The miRNA-19a-3p level was detected by real time fluorescence quantitative PCR(qRT-PCR)in H9C2 cells and cell culture medium,and the probable target gene LRP2 level were test by ELISA,qRT-PCR and western blot in H9C2 cells and cell culture medium.Re-sults miRNA-19a-3p expression in cell culture medium was much higher under 20 h and 24 h(P<0.05), miRNA-19a-3p expression in H9C2 cells was much lower under 20 h and 24 h(P<0.05).LRP expression in cell culture medium was much higher under 16 h[(675.20 ± 42.40)ng/mL,n=3],20 h[(979.4 ± 204.2)ng/mL, n=3]and 24 h[(1 456.00 ± 363.60)ng/mL,n=3]than 0 h[(245.0 ± 10.84)ng/mL,n=3](P<0.05);the LRP2 mRNA level were much higher in 16 h[(0.000 503 ± 0.000 100)ng/mL,n=3],20 h[(0.001 303 ± 0.000090)ng/m L,n=3]and 24 h[(0.001 617 ± 0.000 110)ng/mL,n=3](P<0.05);than in the 0 h[(0.000 139 4 ± 0.000 060 0) ng/mL,n=3],the LRP2 protein level were much higher in 20 h[(0.235 000 ± 0.003 427),n= 3] and 24 h [(0.535 000 ± 0.003 427)ng/mL,n=3](P<0.05).Conclusion miR NA-19a-3p may partipate in H9C2 car-diomyocytes necrosis through regulating LRP2.【期刊名称】《检验医学与临床》【年(卷),期】2018(015)004【总页数】5页(P503-506,509)【关键词】循环微小RNA-19a-3p;H9C2心肌细胞;缺氧;低密度脂蛋白受体相关蛋白2【作者】黄浩;陈文江;刘长召;王玲;沈艳芳【作者单位】湖北省恩施土家族苗族自治州中心医院心血管病中心 445000;广东医科大学研究生学院,广东湛江524023;湖北省恩施土家族苗族自治州中心医院心血管病中心 445000;湖北省恩施土家族苗族自治州中心医院心血管病中心 445000;湖北省恩施土家族苗族自治州中心医院健康体检中心 445000【正文语种】中文【中图分类】R541.4急性心肌梗死(AMI)是冠状动脉闭塞,血流中断,造成部分心肌因严重持久性缺血而发生局部坏死,为心内科常见致死性疾病[1-2]。
M OLECULAR AND C ELLULAR B IOLOGY,Mar.2007,p.1859–1867Vol.27,No.5 0270-7306/07/$08.00ϩ0doi:10.1128/MCB.01395-06Copyright©2007,American Society for Microbiology.All Rights Reserved.A MicroRNA Signature of Hypoxia†ᰔRitu Kulshreshtha,1Manuela Ferracin,2Sylwia E.Wojcik,3Ramiro Garzon,3Hansjuerg Alder,3 Francisco J.Agosto-Perez,3Ramana Davuluri,3Chang-Gong Liu,3Carlo M.Croce,3Massimo Negrini,2George A.Calin,3and Mircea Ivan1*Molecular Oncology Research Institute,Tufts-New England Medical Center,Boston,Massachusetts021111;Department of Experimental and Diagnostic Medicine and Interdepartmental Center for Cancer Research,University of Ferrara, Ferrara44100,Italy2;and Comprehensive Cancer Center,Ohio State University,Columbus,Ohio432103Received28July2006/Returned for modification28August2006/Accepted10December2006Recent research has identified critical roles for microRNAs in a large number of cellular processes,includingtumorigenic transformation.While significant progress has been made towards understanding the mecha-nisms of gene regulation by microRNAs,much less is known about factors affecting the expression of thesenoncoding transcripts.Here,we demonstrate for thefirst time a functional link between hypoxia,a well-documented tumor microenvironment factor,and microRNA expression.Microarray-based expression profilesrevealed that a specific spectrum of microRNAs(including miR-23,-24,-26,-27,-103,-107,-181,-210,and-213)is induced in response to low oxygen,at least some via a hypoxia-inducible-factor-dependent mechanism.Selectmembers of this group(miR-26,-107,and-210)decrease proapoptotic signaling in a hypoxic environment,suggesting an impact of these transcripts on tumor formation.Interestingly,the vast majority of hypoxia-induced microRNAs are also overexpressed in a variety of human tumors.MicroRNAs represent approximately1%to2%of the eu-karyotic transcriptome and have been shown to play critical roles in the coordination of cell differentiation,proliferation, death,and metabolism(1,3,6,7,17,24)and more recently in tumorigenesis(2,5,6,10,13,20,21).Indeed,a significant percentage of microRNA-encoding genes are located at fragile sites,minimal loss-of-heterozygosity regions,minimal regions of amplification,or common breakpoint regions in cancers. Moreover,global microRNA expression changes have been described to occur in human cancers and in some cases shown to correlate with the clinico-pathological features of the tumor (2,13,29).However,no mechanism has been proposed to date for these profile alterations.Despite this wealth of data,relatively little is known about microRNA regulation and response to microenvironmental factors.One mechanism involves the activation of specific sig-nal transduction pathways that in turn promote the transcrip-tion of certain microRNAs.For example,it was reported that the miR-1genes are targets of serum response factor,a con-verging downstream effector for a variety of oncoproteins and growth factors(30).Another transcription factor,the c-myc oncogene product,was also found to activate the expression of a microRNA cluster(25).Hypoxia is an essential feature of the neoplastic microenvi-ronment.Tumors with widespread low oxygenation tend to exhibit increased invasion and resistance to conventional ther-apy(9).The molecular mechanisms responsible for the hypoxic survival of neoplastic cells are not fully characterized,and a better understanding of this process may lead to novel strate-gies for pharmacological intervention.Our data indicate that hypoxia leaves a specific mark on microRNA profiles in a variety of cell types,with a critical contribution of the hypoxia-inducible factor(HIF).Moreover, at least a subgroup of these hypoxia-regulated microRNAs (HRMs)seem to play a role in cell survival in a low-oxygen environment.Finally,by comparing hypoxia-associated microRNA spectra with published data from a large number of tumors(28),we propose that cancer-associated microRNA profiles exhibit a hypoxic signature.MATERIALS AND METHODSCell culture and growth conditions.Colon(HT29and HCT116)and breast cancer(MCF7and MDA-MB231)cell lines were obtained from Phil Hinds (Tufts-New England Medical Center,Boston,MA).HT29and HCT116cell lines were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10%fetal bovine serum and MCF7,and MDA-MB231cell lines were maintained in RPMI1640medium supplemented with10%fetal bovine serum.Hypoxic conditions were maintained in an InVivo200hypoxia workstation(Biotrace In-ternational,Ruskinn Life Sciences,United Kingdom)with oxygen maintained at 0.2%.Normoxic controls were propagated at37°C and5%CO2.Plasmids.The mutant versions of HIF-1␣and HIF-2␣with double proline-to-alanine substitutions have been described previously(14,15).The three-HRE–thymidine kinase(tk)–luciferase reporter is a HIF-responsive construct containing a tandem of hypoxia-responsive elements from the erythropoietin promoter.Both plasmids were provided by William Kaelin(Dana-Farber Cancer Institute,Boston,MA).MicroRNA microarray analysis.RNA was extracted by using TRIzol(Invitro-gen,Carlsbad,CA),according to the manufacturer’s protocol.RNA was labeled and hybridized on microRNA microarray chips as previously described(20). Briefly,5g of RNA from each sample was biotin labeled during reverse transcription using random hexamers.Hybridization was carried out on the second version of our microRNA chip(ArrayExpress accession number A-MEXP-258),which contains381probes for mature and precursor human microRNAs and393probes for mouse microRNAs,together with controls. There are four spot replicates for each probe on the chip.Hybridization signals were detected by biotin binding of a Streptavidin-Alexa647conjugate using a GenePix4000B scanner(Axon Instruments).Images were quantified using the*Corresponding author.Mailing address:750Washington Street, Box5609,Boston,MA02111.Phone:(617)636-7514.Fax:(617)636-6127.E-mail:mivan@.†Supplemental material for this article may be found at http://mcb/.ᰔPublished ahead of print on28December2006.1859 on January 10, 2016 by guest / Downloaded fromGenePix Pro 6.0apparatus (Axon Instruments).We used the microRNA no-menclature according to the microRNA Registry (miRBase http://microrna /sequences/)at the Sanger Institute and the Genome Browser ().Analysis of microarray data.Raw data were normalized and analyzed by GeneSpring GX software version 7.3(Agilent Technologies).The GeneSpring software generated a unique value for each microRNA,determining the average of the results from the four spots.Samples were normalized using the on-chip median normalization feature of GeneSpring software;the result for each cell line time course experiment was normalized to that at time zero.MicroRNAs showing an altered expression across the time course were identified using the filter of the n -fold-change tool.In detail,a GeneSpring GX filter with the n -fold-change tool was used to point out the microRNAs that,for at least two cell lines,were 1.5-fold upregulated at 24h compared to expression levels at time zero.The 1.5-fold-change threshold was chosen on the basis of its use in previ-ously published articles employing these particular types of microarrays.Analysis of variance (ANOVA)with the Benjamini and Hochberg correction for false-positive reduction was used to compare the average values obtained with microRNA probes in all cell lines at 24and 48h with values at time zero.Hierarchical cluster analysis was performed using average linkage and Pearson correlation as a measure of similarity.Transfections.Plasmid transfections were performed using Lipofectamine 2000(Invitrogen)as per the manufacturer’s protocol.Fresh medium was added after 5h of transfection,and RNA and protein were harvested 24/48h post-transfection.Transfection of mature microRNAs or inhibitors (Ambion,Inc.)was performed using siPORT NeoFX transfection agent (Ambion,Inc.)accord-ing to the manufacturer’s protocol.Transfection complexes were added to cells at a final oligonucleotide concentration of 20nM,and medium was replaced 24h later.Luciferase assays using reporters based on HRM promoter fragments.We started by amplifying HRM promoter fragments predicted to encompass HIF sites,as shown in Table S1b in the supplemental material.Thus,the genomic region 4.9kb immediately upstream of miR-24-1was amplified using primers 5ЈATACTCGAGCTGCTAGGCCATGCGTGTCC3Ј(forward)and 5ЈATTAA GCTTCAAGAGAGAGTTCACCGCGC3Ј(reverse)(underlining indicates re-striction sites inserted).For miR-181c,a region of approximately 2.0kb imme-diately upstream was PCR amplified using 5ЈATAGGTACCCACTCCACAGC CTGAATG3Ј(forward)and 5ЈTATAAGCTTGGTGGGGTAGGTGGCAGG GAAC3Ј(reverse).For miR-26b,a region situated approximately 3kb upstream of the 5Јend (and encompassing a cluster of four predicted HIF sites)was PCR amplified using the following primers:5ЈATAGCTAGCGAGACAGATGTCC CGCTCCCAG3Ј(forward)and 5ЈATCGCTAGCACGCTCTTGAATGGGAC GG3Ј(reverse).Due to the size and CG contents,PCR amplification was done using the Herculase II fusion DNA polymerase (Stratagene).The resulting fragments were cloned in the pGL3basic vector (Promega)(miR-24-1and -181c)or pGL3-tk-luciferase vector (from David Fisher,DFCI)(miR-26b).MCF7or HT29cells were cotransfected using Lipofectamine 2000(Invitrogen)with the reporter plasmids and either of the HIF mutants or the empty vector.Luciferase assays were performed 24h posttransfection by following the manufacturer’s protocol (luciferase assay system;Promega).Luciferase activity (measured as relative light units)was scored using a Femtomaster FB12luminometer (Zylux Corporation)and normalized to that of an equal protein concentration in all samples.Apoptosis assays.Cells were plated in triplicate in a 96-well plate,transfected with microRNA precursors or inhibitors (Ambion,Inc.)as described above,and scored for caspase activation 72h posttransfection using an Apo-ONE homoge-neous caspase-3/7assay kit (Promega)according to manufacturer’s instructions.The protein concentration was determined using Bradford’s method,and caspase activities for all samples were normalized to that of an equal protein amount.The data are presented as means plus standard deviations from three independent experiments performed in triplicate.Northern hybridization.Total RNA isolation was performed using the TRIzol reagent (Invitrogen)according to the manufacturer’s instructions.RNA samples (20g each)were run on 15%acrylamide-denaturing (urea)Criterion precast gels (Bio-Rad Laboratories,Hercules,CA)and then transferred onto a Hy-bond-N ϩmembrane (Amersham Pharmacia Biotech).The hybridization with [␣-32P]ATP was performed at 42°C in 7%sodium dodecyl sulfate (SDS)–0.2M Na 2PO 4(pH 7.0)overnight.Membranes were washed at 42°C,twice in 2ϫSSPE (1ϫSSPE is 0.18M NaCl,10mM NaH 2PO 4,and 1mM EDTA [pH 7.7])–0.1%SDS and twice with 0.5ϫSSPE–0.1%SDS.Blots were stripped by boiling them in 0.1%aqueous SDS–0.1ϫSSC for 10min and were reprobed several times.As a loading control,we used 5S rRNA stained with ethidium bromide or probing for the U6snRNA.TaqMan RT-PCR.The method was optimized for microRNA,and reagents,primers,and probes were obtained from Applied Biosystems.Human 18S rRNA was used to normalize all RNA samples.Reverse transcriptase (RT)reactions and real-time PCR (PCR)were performed according to manufacturer protocols.RNA concentrations were determined with a NanoDrop apparatus (NanoDrop Technologies,Inc.),and 1nanogram per sample was used for the assays.All RT reaction mixtures,including no-template controls and RT-minus controls,were run in duplicate in an Applied Biosystems 9700thermocycler.Gene expression levels were quantified using the ABI Prism 7900HT sequence detection system (Applied Biosystems).Analysis was performed in duplicate,including with no-template controls.Relative expression was calculated using the comparative cycle threshold method.ChIP.HT29cells (60%confluent)were exposed to hypoxia (0.2%O 2)or normoxia (21%O 2)for 24h.Cross-linking was performed using 1%(vol/vol)formaldehyde for 10min,and the reaction was stopped with 125mM glycine for 5min.Cells were washed twice with ice-cold 1ϫphosphate-buffered saline and collected by scraping them in 1ml of 1ϫphosphate-buffered saline supple-mented with 1mM phenylmethylsulfonyl fluoride (PMSF),followed by centri-fugation and lysis in 400l of buffer (1%SDS,10mM EDTA,50mM Tris-Cl,pH 8.0,protease inhibitor cocktail,and 1mM PMSF).The resulting lysate was incubated on ice for 10min,followed by sonication for DNA shearing to frag-ments between 200bp and 1kb.The supernatant was recovered and diluted in chromatin immunoprecipitation (ChIP)dilution buffer (1%Triton X-100,2mM EDTA,150mM NaCl,20mM Tris-Cl,pH 8.0,protease inhibitor cocktail,1mM PMSF),followed by immunoprecipitation overnight at 4°C with a polyclonal anti-HIF-1␣antiserum (ab2185;Abcam)or whole rabbit antiserum (immuno-globulin G [IgG]control).Immunocomplexes were recovered by the addition of a 50%slurry of salmon sperm DNA-protein A-agarose (Upstate)to the samples and sequentially washed for 4min each in buffer I (20mM Tris,pH 8.0,200mM NaCl,0.5%Triton X-100,0.05%deoxycholate,0.5%NP-40,1mM PMSF),buffer II (20mM Tris,pH 8.0,500mM NaCl,0.5%Triton X-100,0.05%deoxycholate,0.5%NP-40,1mM PMSF),buffer III (10mM Tris,pH 8.0,250mM LiCl,1%Triton X-100,0.1%deoxycholate,0.5%NP-40,0.5mM EDTA,1mM PMSF),and buffer IV (10mM Tris,pH 8.0,5mM EDTA).The immuno-precipitated DNA was retrieved from the beads by incubation in elution buffer (10mM Tris,pH 8.0,5mM EDTA,1%SDS)at 65°C for 1h.Cross-linking was reversed using 200mM NaCl at 65°C overnight followed by proteinase K diges-tion at 47°C for 2h.DNA was then purified using a PCR purification kit (QIAGEN),and PCR was performed using primers spaced approximately 15to 250bp apart and encompassing predicted HIF binding sites in miR-210and -26a-2promoters (see below).5ЈGGAGCCTTGACGGTTTGACC 3Ј(forward)and 5ЈCGAGGACCAGGG TGACAGTG3Ј(reverse)were used to PCR amplify the miR-210promoter fragment (210-A)containing predicted HIF binding sites located at positions Ϫ1720and Ϫ1822.For the 210-B fragment containing a predicted HIF consen-sus at position Ϫ1166,the following pair was used:5ЈGGTCGTGAAGGAGC CTCTAAG3Ј(forward)and 5ЈGGACCATCCCACTCACTACAG3Ј(reverse).Finally,the miR-26a-2promoter fragment (26-A),encompassing a predicted HIF binding site at position Ϫ2860,was amplified using 5ЈCCAAGGACTATG CACATCACC3Ј(forward)and 5ЈGGAAAGGCAGTGAGATGGCAC3Ј(re-verse).The following primers were used to amplify a region in the promoter of miR-130b (negative control,hypoxia nonresponsive):5ЈGCGAAACCCCAGCT CTACTA3Ј(forward)and 5ЈACACTCTCACTCTGTCGCCC3Ј(reverse).For HIF site localization and sequences,see Table S1b in the supplemental material.The PCR products were resolved on a 1.5%agarose gel and visualized by ethidium bromide staining.RESULTSMicroRNA expression changes in hypoxia.We hypothesized that microRNAs could be involved in hypoxia response and began addressing this hypothesis using a microarray-based screening.We first analyzed a panel of four human cancer cell lines (MDA-MB231,MCF7,HT29,and HCT116),which were subjected to time course exposure to 0.2%oxygen,a concen-tration often present in the hypoxic regions of tumors.RNA was extracted at specific time points (0,8,24,and 48h),and microRNA expression profiling was done subsequently.Twenty-seven microRNA probes were identified to be at least 1.5-fold upregulated at 24h in at least two cell lines.The ANOVA,with1860KULSHRESHTHA ET AL.M OL .C ELL .B IOL .on January 10, 2016 by guest/Downloaded fromthe Benjamini and Hochberg correction for false-positive re-duction,was employed to compare the microRNA probes av-erage values in all cell lines at 24and 48h versus values at time zero,yielding in all cases significant P values (Ͻ0.05)(see Table 1for a summary of levels of induction at 24and 48h in all cell lines versus those at 0h and corresponding P values).A cluster analysis of time points for each cell line is shown in Fig.1.These probes correspond to 23distinct mature microRNAs,which will henceforth be termed HRMs.We went on to confirm the microarray data using Northern analysis (Fig.2a for miR-210)or quantitative RT-PCRs for miR-21,-23,-24,-26,-27,-181,-103,-107,-125,-210,and -213(see Fig.2b for a selection;the rest can be found in Fig.S1a in the supplemental material).Priority was given to HRMs that were subjected to subsequent functional analyses.A Northern blot showing a lack of induction of miR-7-3(not a member of the HRM list)served as negative control (see Fig.S1b in the supplemental material).Evidence for the role of HIFs in HRM regulation.We next addressed the role of HIFs in HRM regulation.HIFs are well-documented master regulators of the hypoxia response and transcription factors,which are stabilized during hypoxia and coordinate the transcription of a wide variety of genes that are critical for survival under conditions of low oxygen (9).In order to investigate whether HRMs exhibit a significant enrichment in predicted HIF binding sites,we performed an in silico search upstream of the genomic sequences that encode all the known microRNAs (1,039sequences,experimentally demonstrated and predicted;/sequences /ftp/genomes/hsa.gff).Since only a few microRNA promoters have been identified experimentally,a 6-kb region (5kb upstream and 1kb down-stream region of the 5Јend of the annotated microRNA)was designated as a putative promoter sequence.Indeed,most of the experimentally confirmed transcription factor binding sites are located within the kb Ϫ5region of the transcription start site.Predicted HIF binding sites were analyzed using the MATCH program and the V$HIF1_Q3(GNNK ACGTG CG GNN;boldfacing indicates the core HIF consensus),V$HIF1_Q5(NGT ACGTGC NGB),and V$HIF1_Q6(N RC GTG NGN)position weight matrices from the TRANSFAC database (version 9.1)(23).The position weight matrices de-scribe the position preferences of different nucleotides in the HIF binding site.We scanned regions around the transcription start sites of all the microRNAs from kilobase positions Ϫ5to ϩ1using the “minFP_good91.prf”profile (the profile of cutoff values with a minimum number of false-positive predictions)of MATCH,similarly to searches described previously (16,26,27).We then tested whether these consensus sequences are sig-nificantly more abundant in the promoters of the 23HRMs (target set)than in the rest of microRNA-ome.Thus,we gen-erated 50,000groups,each consisting of 23promoters ran-domly selected from the 1,039microRNAs and calculated the number of promoters that contain at least one predicted HIF binding site in both target and random sets of promoters.The search was performed separately for the HIF_Q3and HIF_Q5TABLE 1.Summary of hypoxia-regulated microRNAs aMicroRNA Symbol MicroRNA expression ratio in indicated cell lineANOVA P valueHT29HCT116MCF7MDA-MB23124h/0h48h/0h24h/0h48h/0h24h/0h48h/0h24h/0h48h/0hmiR-103-1MIRN103-1 2.33 3.46 1.11 1.71 2.56 1.72 2.38 3.190.00228miR-103-2MIRN103-2 2.31 2.91 1.22 1.70 2.58 2.07 2.6 3.760.00228miR-106a MIRN106A 1.76 2.03 1.01 1.41 2.18 1.26 1.84 1.520.00241miR-107MIRN107 1.94 2.61 1.07 1.75 2.65 1.66 2.76 3.900.00231miR-107MIRN107 2.04 3.26 1.22 1.86 2.72 1.60 2.47 3.370.00228miR-125b-1MIRN125B1 1.83 2.07 1.07 1.52 1.71 1.64 1.51 1.130.00234miR-181a-1MIRN213 2.29 3.02 1.17 1.44 1.61 1.05 1.87 2.240.00398miR-181a-2MIRN181A 2.01 2.73 1.01 1.44 1.87 1.47 2.13 2.840.00241miR-181b-1MIRN181B1 2.17 3.38 1.17 1.96 1.97 1.47 1.97 2.660.00228miR-181b-2MIRN181B2 2.17 3.33 1.04 1.66 1.73 1.28 1.95 1.980.00268miR-181c MIRN181C 2.74 3.33 1.11 1.48 1.26 1.17 1.6 1.750.0083miR-192MIRN192 1.75 2.19 1.07 1.30 2.7 1.63 1.33 1.860.00262miR-195MIRN195 1.45 2.26 1.51 1.51 1.59 1.15 1.17 1.020.00622miR-21MIRN21 2.81 4.00 1.26 2.09 2.86 1.79 2.28 1.340.00242miR-21MIRN21 2.62 3.51 1.34 1.92 2.6 1.66 1.71 1.120.00231miR-210MIRN210 2.92 3.85 2.47 3.73 4.33 3.77 1.450.940.00241miR-213-5p MIRN213 2.14 2.90 1.3 1.10 1.25 1.04 1.64 1.860.00961miR-23a MIRN23A 2.42 3.730.97 2.02 2.5 1.71 2.71 3.410.00231miR-23b MIRN23B 2.7 3.39 1.05 1.82 1.87 1.45 2.76 3.570.00241miR-24-1-3p MIRN24-1 2.44 4.090.98 1.75 2.22 1.30 2.33 1.520.00483miR-26a MIRN26A1 1.91 2.87 1.12 1.99 2.06 1.58 2.21 2.040.00228miR-26a-1MIRN26A1 2.3 3.46 1.14 2.06 2.5 1.88 1.81 1.720.00228miR-26a-2MIRN26A2 1.94 2.62 1.1 1.57 1.37 1.07 1.54 1.660.0046miR-26b MIRN26B 2.21 3.24 1.29 2.04 3.23 2.40 2.32 2.960.00213miR-27a MIRN27A 1.77 2.48 1.13 1.51 2.14 1.65 1.18 1.440.00241miR-30b MIRN30B 2.74 3.96 1.52 2.11 2.38 1.54 1.5 1.400.00234miR-93-1MIRN93 1.84 1.79 1.02 1.34 2.18 1.54 1.86 1.560.00231aMicroRNA expression numbers represent the ratio of each microarray probe expression value at 24or 48h to that at time zero.The P value was obtained by performing an ANOVA statistical comparison between the mean expression levels at 24and 48h versus that at time zero in all cell lines.V OL .27,2007A MicroRNA SIGNATURE OF HYPOXIA1861on January 10, 2016 by guest/Downloaded fromconsensus sequences.The selection was performed using the function “sample,”which is part of the random module in the Python 2.4programming language (/),followed by data analysis using the R programming language (/).The results of the analysis are sum-marized in Fig.3and Table S1a in the supplemental material,indicating a highly significant enrichment of the HIF binding consensuses in the HRM group (P ϭ0.00294for HIF_Q3;P ϭ0.011for the HIF_Q5consensus).The search based on the HIF_Q6matrix did not yield a significant P value (not shown),which is not surprising,given the high probability of such a short and degenerate sequence arising by chance very often in the genome.The next step was to obtain experimental evidence for the role of HIF in HRM regulation.First,we transduced consti-tutively stable HIF-1and -2␣subunits versus a vector-only control (pcDNA3.1)in HT29and MCF7cell lines under nor-moxia.HIF stabilization was achieved by substituting the two prolines (at positions 564and 402in the case of HIF-1)in the alpha subunits that are subject to oxygen-dependent hydroxy-lation and proteasomal degradation via VHL-dependent ubiq-uitylation (15,16,22).The activity of exogenous HIFs was confirmed by cotransfection with an HRE-tk-luciferase re-porter (containing three hypoxia response elements)followed by standard luciferase assay (Fig.S2a in the supplemental material).Expression of miR-103,-210,and -213wasmeasuredFIG.1.Coordinated hypoxic changes of microRNA expression in colon and breast cancer cell lines.Cluster analysis of four cell lines according to the expression of microRNAs upregulated by hypoxia in at least two cell lines.Expression data were normalized to expression at time zero.1862KULSHRESHTHA ET AL.M OL .C ELL .B IOL .on January 10, 2016 by guest/Downloaded fromfollowing HIF transfection using a modified real-time RT-PCR protocol which revealed a robust and reproducible increase in the expression of all three transcripts (Fig.4).We then addressed whether HIF has a direct impact on the putative promoter regions of several HRMs (miR-24,-26,-181,and -210).To this end,we amplified 5kb and 2kb of the genomic regions upstream of miR-24-1and miR-181c,respec-tively.We also amplified a 1.2-kb region approximately 3kb upstream of the 5Јend of miR-26b (for details,see Materials and Methods).These regions were chosen based on the pre-dicted HIF binding sites identified by in silico analysis;in order to minimize the chance of missing potentially important sites,we used a “less stringent search,”by considering all theposi-FIG.2.Confirmation of HRM induction by hypoxia by Northern analysis or quantitative RT-PCR.(a)Northern blotting confirmation of miR-210induction under hypoxia (the mature form is indicated).Lanes NOR,6HY,and 30HY show results under normoxia and at 6h and 30h under hypoxia,respectively.An ethidium bromide-stained gel picture is shown as a loading control.(b)Quantitative RT-PCR con-firmation of HRM induction by 24-hour hypoxia (H)compared with HRM induction for normoxic controls (N).Bars indicate means from two independent experiments.I bars indicate standarddeviations.FIG.3.Distribution of random 23-microRNA groups (samples)based on HIF1_Q3(a)or HIF1_Q5(b)binding sites.The arrow indicates where the experimental data (HRMs)fall within the random sample population,with the corresponding P value.V OL .27,2007A MicroRNA SIGNATURE OF HYPOXIA 1863on January 10, 2016 by guest/Downloaded fromtion weight matrices (HIF_Q3,_Q5,and _Q6)(see Table S1b in the supplemental material).The promoter fragments were subcloned into the pGL3-luciferase (for miR-24and -181)or,as an enhancer,into the pGL3-tk-luciferase (for miR-26)vec-tor.HT29cells were cotransfected with these constructs or the control construct pGL3-luc/pGL3-tk-luciferase and constitu-tively active HIF constructs (HIF1P/A or HIF2P/A)or the empty vector pcDNA3.1(PC),followed by incubation under normoxia or hypoxia for 24h.Standard luciferase assays showed that HIF transduction or hypoxia induced a robust activation of all the promoter-luciferase constructs,supporting a direct role of HIF in HRM upregulation (Fig.5).Interest-ingly,HIF-1had a more robust impact on the promoters of miR-24and miR-181c than HIF-2,which was more efficient in activating miR-26.This is unlikely to be attributed to differ-ences in transfection efficiency,as parallel controls using a three-HRE (Epo)-luciferase reporter cotransfected with the HIF plasmids showed comparable levels of luciferase induc-tion in response to both isoforms (Fig.S2b in the supplemental material).While these differences between HIF-1and HIF-2were not tested at the level of full HRM induction,recent data argue for specific targets and biological effects of the two forms (12).Additionally,for miR-26a-2and miR-210,we confirmed the dynamic recruitment of HIF to the promoter by ChIP.As shown in Fig.6,the HIF-1␣antibody (but not the control IgG antibody)immunoprecipitated the miR-210and miR-26a-2promoter fragments in hypoxic HT29cells but very little in the normoxic controls.A similar assay performed for a region upstream of miR-130b (which is not an HRM),did not reveal measurable recruitment of HIF upon hypoxia exposure.Roles of select HRMs in cell survival.The impact of HRMs on cell survival was addressed by direct transfection of mature microRNAs or their antisense inhibitors (4)in human cell lines under hypoxic conditions.We transduced miR-24,-26,-107,-210,and -213into MCF7cells and then incubated them under hypoxia for 48h.The proapoptotic response was interrogated by a caspase-3/7assay (Fig.7a).Efficient trans-fer and blockade of microRNAs were confirmed byNorth-FIG.4.Effect of HIF on specific microRNA expression.The im-pact of exogenous constitutively active HIF-1␣(HIF1P/A)and HIF-2␣(HIF2P/A)subunits on miR-103,-210,and -213expression was deter-mined by quantitative RT-PCR in HT29(a)and MCF7(b)cells.The control was the pcDNA3.1empty vector (PC).Bars indicate means from two independent experiments.I bars indicate standard devia-tions.FIG.5.Direct effect of HIF in the up-regulation of select HRMs.Relative luciferase activities of HRM promoter reporter constructs in HT29cells.(a)miR-24-1and -181c promoters in a pGL3context;(b)miR-26b fragment in a pGL3-tk context.The constructs were cotrans-fected with constitutively active HIF-1␣(HIF1P/A),HIF-2␣(HIF2P/A),or the empty vector pcDNA3.1(PC)and incubated under nor-moxia (NOR).The effect of hypoxia (HYP)on the reporter is also shown.Bars indicate means from three independent experiments.I bars indicate standarddeviations.FIG.6.Direct recruitment of HIF on the miR-210and miR-26promoters under hypoxia.Chromatin was immunoprecipitated from HT29cells using a HIF-1␣antibody or an IgG control,and the en-riched genomic fragment was amplified using primers spanning the candidate HREs located at positions Ϫ1720and Ϫ1822(210-A);Ϫ1166(210-B)of the miR-210promoter;or Ϫ2860for the miR-26a-2promoter (26-A).A fragment of the miR-130b promoter was used as a negative control.1864KULSHRESHTHA ET AL.M OL .C ELL .B IOL .on January 10, 2016 by guest/Downloaded from。