Lecture 8 Gene Regulation in Eukaryotes
- 格式:pdf
- 大小:1.22 MB
- 文档页数:59


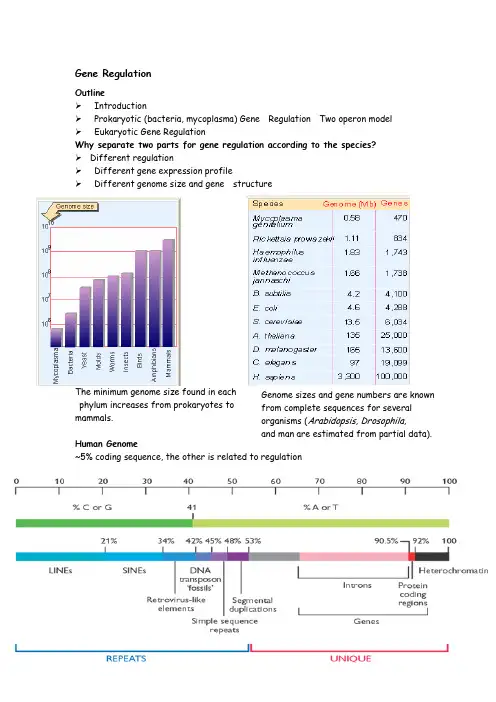
Gene RegulationOutlineIntroductionProkaryotic (bacteria, mycoplasma) Gene Regulation Two operon model Eukaryotic Gene RegulationWhy separate two parts for gene regulation according to the species? Different regulationDifferent gene expression profilephylum increases from prokaryotes to mammals.Human Genome~5% coding sequence, the other is related to regulationTwo Views of Regulation•The biochemical view: What are the specific biochemical events that regulate gene expression? E.g. how is transcription regulated?•The logical view: What is the functional outcome of a regulatory event: up-regulation (activation) or down-regulation (inhibition)?Two types of sequences in DNA for regulation:1.Sequences that code for trans-acting products;2.cis-acting sequences that function exclusively within the DNA.A gene is a sequence of DNA that codes for a diffusible product. This productmay be protein (as in the case of the majority of genes) or may be RNA (as in the case of genes that code for tRNA and rRNA).The crucial feature is that the product diffuses away from its site of synthesis to act elsewhere. Any gene product that is free to diffuse to find its target is described as trans-acting.The description cis-acting applies to any sequence of DNA that is not converted into any other form, but that functions exclusively as a DNA sequence in situ, affecting only the DNA to which it is physically linkedHow to distinguish between the components of regulatory circuits and the genes that they regulate?Structural gene Regulator gene. A structural gene is simply any gene that codes for a protein (or RNA) product. A regulator gene simply describes a gene that codes for a protein (or an RNA) involved in regulating the expression of other geneGene expression is regulated by the specificinteractions of the trans-acting products (usually proteins) with the cis -acting sequences (usually sites in DNA).Focus on the transcription level to study the regulation Operon model for procaryoteIn procaryote,Structural gene clusters are coordinately controlled Operon is a unit of bacterial gene expression and regulation, including structural genes and control elements in DNA recognized by regulator gene product(s).Components: Regulator gene, Promoter, Operotor, Structural Gene, terminatorKey terms:Promoter is a region of DNA involved in binding of RNA polymerase to initiate transcription.Terminator is a sequence of DNA, represented at the end of the transcript, that causes RNA polymerase to terminate transcription .Structural gene codes for any RNA or protein product other than a regulator.Operator is the site on DNA at which a repressor protein binds to prevent transcription from initiating at the adjacent promoter.Repressor protein binds to operator on DNA or RNA to prevent transcription or translation, respectively.A regulator gene simply describes a gene that codes for a protein involvedin regulating the expression of other genes.The core for operon regulation is:Trans-acting protein bind to cis-acting DNA Lac operonBacterial structural genes are often organized into clusters that include genes coding for proteins whose functions are related. It is common for the genes for the enzymes of a metabolic pathway to be organized into such a cluster.The cluster of the three lac structural genes, lacZYA, is typical.The protein products enable cells to take up and metabolize β-galactosides, such as lactose.Repressor and RNA polymerase bind atsites that overlap around the startpoint of the lac operon.1. The lac genes are controlledby a repressor Repressor maintains the lac operon in the inactive condition by binding to the operator; addition of inducer releases the repressor, and thereby allows RNA polymerase to initiate transcription.2.The lac operon can be inducedInductionWhen lac repressor binds to the inducer (lactose), it changes conformation and cannot bind to O lac site any more. This allows rapid induction of lacZYA transcription.2. Repressor is controlled by a small molecule inducerAddition of inducer results in rapid induction of lacthe enzymes; removal of inducer is followed by rapid cessation of synthesis.Lactose (allolactose) is a native inducer to release RNA transcription elongation from P lac .IPTG, a synthetic inducer, can rapidly stimulate transcription of the lac operon structural genes.IPTG is used to induce the expression of the cloned gene from LacZ promoter in many vectors, such as pUC19.The structure for lactose, IPTG and X-galThe core for repressor induced is that inducer repressor which binds to the operator.Question:How to identify the functional cis-acting DNA? How the repressor bind to the operator? When inducer binds to the repressor, what happened?cis-acting constitutive mutations identify the operatorOperator mutations are constitutive because the operator is unable to bind repressor protein; this allows RNA polymerase to have unrestrained access to the promoter. The O c mutations are cis -acting, because they affect only the contiguous set of structural genes.trans-acting mutations identify the regulator geneMutations that inactivate the lacI gene cause the operon to be constitutively expressed, because the mutant repressor protein cannot bind to the operator.A lacl d mutant gene makes a monomer that has a damaged DNA binding site (shown by the red circle). When it is present in the same cell as a wild-type gene, multimeric repressors are assembled at random from both types of subunits. It only requires one of the subunits of the multimer to be of the lacl6 type to block repressor function.identifies several independent domains.Lac repressor identifies the interactionsbetween monomers in the tetramer. Eachmonomers in the tetramer. Each monomer isidentified by a different color.made of two dimmersinverted repeat of the operator binds to the DNA-binding repressor subunit.The lacnumbered relative to the startpoint for transcription at + 1. The pink arrows to left and right identify the two dyad repeats. The green blocks indicate the positions of identity.How to identify which base is crucial for binding to the repressor?repressor can be identified bychemical crosslinking or byexperiments to see whethermodification prevents binding.They identify positions on bothstrands of DNA extendingfrom +1 to +23. Constitutivemutations occur at 8 positionsin the operator between +5 and+ 17.(如上页图1)Repressor binds to three operators and interacts with RNApolymeraseIf both dimers in a repressor tetermer bind to DNA, thenthe DNA between the two binding sites is held in a loop.1.Each dimer can bind an operator sequence. Thisenables the intact repressor to bind to two operatorsites simultaneously.2. In fact, there are two further operator sites in the initialregion of the lac operon.3. The original operator, Ol, is located just at the start ofthe lacZ gene. It has the strongest affinity for repressor.sequences (sometimes called pseudo-operators) are located on either side; O2 is 410 bp downstream of the startpoint, and 03 is 83 bp upstream of it.The two dimers may be bound to DNA simultaneously,and thissimultaneous binding increases repression.The reason for this is under way ! ! !The binding of repressor actually enhances the binding of RNA polymerase to promoter!But the bound enzyme is prevented from initiating transcription.The repressor in effect causes RNA polymerase to be stored at the promoter.The complex of RNA polymerase-repressor-DNA is blocked at the closed stage. When inducer is added, the repressor is released, and the closed When inducer binds to the repressor,what happened?Inducer change the structure of the coreso that the headpieces of a repressordimer are no longer in an orientationQuestionDose the inducer bindor directly to repressorbound to operator?Repressor is always bound to DNAProteins that have a high affinity for a specific DNA sequence also have a low affinity for other DNA sequences.Every base pair in the bacterial genome is the start of a low affinity binding-site for repressor.The large number of low-affinity sites ensures that all repressor protein is bound to DNA.Lac repressor binds strongly and specifically to its what happened?Virtually all the repressor in the cell isbound to DNA.The addition of inducer abolishes theability of repressor to bind specificallyat the operator.are released,and bind to random(low-affinity) sites.ers are"stored" on random DNA sites.at the operator, while theremainingThe effect of induction is to change the distribution of repressor on DNA, rather than to generate free repressor.4. cAMP-CRP positive regulation (Glucose repression controls)E. coli uses glucose in preference to other carbon sources when it has achoice.By reducing the level of cAMP, glucose inhibits thetranscription of operons that require CRP activity.CRP bends DNA >90°be used to analyze bending.The level of cAMP is low in cell, and CRP exists as a dimer which can’t bind to DNA to regulate transcription.When glucose is absentThe level of cAMP increase and CRP bind to cAMP. The CRP-cAMP complex binds to Plac just upstream from the site for RNA polymerase. Induces a 90°bend in DNA which enhances RNA olymerase binding to the promoter and thus the ranscription by 50-fold.Mechanisms involved in Lac operon regulation1.Negative regulation: repressor inducedTrp operonAn operator at the cluster of structural genestrpEDCBA controls coordinate synthesis of theenzymes that synthesize tryptophan from chorismic acid. Corepressor is a small molecule that triggers Inducer is a small molecule that triggers gene transcription by binding toa regulator protein.Induction refers to the ability of bacteria (or yeast) to synt hesize certain enzymes only when their substrates are present; applied to gene expression, refers to switching on transcription as a result of interaction of the inducer with the regulator protein.Repression is the ability of bacteria to prevent synthesis of certain enzymes when their products are present; more generally, refers to inhibition of transcription (or translation) by binding of repressor protein to a specific site on DNA (or mRNA).In bacteria, an mRNA is in principle available for translation as soon as it is synthesizedAttenuation can be controlled by translationThe trp operon consists of five contiguous structural genesleader peptide coding region, and attenuatorThe alternatives for RNA polymerase at theattenuatorTermination can be controlled via changes inRNA secondary structure that aredetermined by ribosome movement.The mechanisms involved in the Trp operonattenuationTrp repressorThe attenuationAttenuation: 10-fold repressiontrp repressor:70-fold repressionTotal regulatory effect:700-fold repressionAttenuation occurs in at least 6 operonsrelated amino acid biosynthesisHow to distinguishing positive and negative controlInduction can be achieved by inactivating a repressor or activating anactivator.Repression can be achieved by activating a repressor or inactivating anactivator.Control circuits are versatile and can be designed to allow positive or negative control of induction or repression.The stringent response produces (p)ppGppPoor growth conditions cause bacteria to produce the small molecule regulators ppGpp and pppGpp.When bacteria find themselves in such poor growth conditions that they l ack a sufficient supply of amino acids to sustain protein synthesis, they shut down a wide range of activities. This is called the stringent response.The stringent factor RelA is a (p)ppGpp synthetase that is associatedwith ribosomes.5.Repressor is controlled by a small molecule inducer6.cis-acting constitutive mutations identify the operator7.trans-acting mutations identify the regulator gene8.Repressor protein binds to the operator10.The repressor monomer has several domains11. Repressor is a tetramer made of two dimers12. Repressor binds to three operators and interacts with RNA polymerase13. Repressor is always bound to DNA14. Binding of inducer releases repressor from the operator15. The operator competes with low-affinity sites to bind repressor16. Camp-CRP involves positive regulation at the promoter17. The E. co//tryptophan operon is controlled by attenuation18. Attenuation can be controlled by translation19. Termination can be controlled via changes in RNA secondary structure that are determined by ribosome movement.。

The English-Chinese Bilingual Teaching and Learning Outline of Biochemistry Ⅱ(English V ersion)Chapter one Nucleotide and Nucleic acidsPropose:1.Grasp of the nucleotide structure, including the component of the nucleotide and the chemicalbonds linking the components, and the names and abbreviations of bases, nucleosides and nucleotides.2.Profound grasp of the key knowledge of the structure and function of DNA and RNA.3.Familiarity with the physio-chemical properties of DNA and the relation between theseproperties and the DNA structure.4.To know the technique of isolation and purification of nucleic acids and DNA sequencing. Contents:1.General concepts of nucleic acids.(1)Nucleotides are the building blocks of nucleic acids.(2)Nucleotide is composed of base, pentose, and phosphate.(3)The 3’,5’phosphodiester bond links nucleotides, many nucleotides are linked by the 3’,5’phosphodiester bond to form nucleic acids.(4)Nucleic acids are divided into DNA and RNA.(5)Other functions of the nucleotides.2.the three dimensioned structure of DNA(1)Double helix structure is the secondary structure of DNA.①The double helix DNA model put forth by Waston and Crick.②The features of double helix DNA.③A- DNA and Z- DNA.④Triple strand DNA.(2)Supercoils of DNA①Prokaryotic circular supercoiled DNA.②Eukaryotic mtDNA and ctDNA.③Topoisomerases.(3)Eukaryotic chromosome①Nucleosome.②From chromosome to chromatin and chromosome.(4)DNA is the genetic material①A very’s experiment.②The genome.③Euchromatin and heterochromatin.3.Structure and functions of RNA(1)Four main kinds of RNA(2)mRNA: structure and function.(3)tRNA: structure and function.(4)rRNA: structure and function.(5)Other small RNAs.4.Physico-chemical properties of nucleic acids(1)Hydrolysis.(2)UV absorbance.(3)Denaturation, renaturation, and hybridization.5. Nuclease(1) Ribonuclease.(2) Dexoyribonuclease.6. Techniques of isolation and purification of nucleic acids and DNA sequencing.(1) Methods of isolation and purification of nuceic acids.(2) DNA sequencing.Chapter two Nucleotide metabolismPurpose1.To know digestion and absorption of nucleic acids and anabolism pathway of nucleic acids inthe human body.2.Profound grasp of the raw material of purine and pyrimidine synthesis, the end products ofpurine and pyrimidine breakdown.3.Grasp of the formation of deoxynucleotide from ribonucleotide.4.To know the function of antimetabolites.Contents1.Nucleotide functions.2.Anabolism and catabolism of purine.3.Anabolism and catabolism of pyrimidine.4.From ribonucleotides to deoxynucleotide.5.antimetabolites.Chapter Three DNA biosynthesis and DNA damage repairPurpose1.Familiarity with the central dogma.2.Grasp DNA replication and reverse transcription.3.To know DNA damage repair systems.Contents1.General feature of DNA replication.(1)Semiconservative.(2)Bidirectional.(3)Replication fork(4)Semidiscontinuous(5)The Ori.(6)The primer(7)Enzymes participating in DNA replication.2.Features of DNA polymerase.(1)The substrates.(2)The active center.(3)The enzyme activation.3.DNA replication in E.coli.(1)DNA replication initiation.(2)Chain elongation.(3)Replication termination.4.Eukaryotic DNA replication.(1)Eukaryotic DNA polymerase.(2)Process of DNA replication.(3)Telomerase and telomere.5.DNA damage.(1)Causes of DNA damage.(2)Mismatch repair system.(3)Other DNA repair systems.6.Reverse transcription.(1)Development of the central dogma put forth by Crick.(2)Retrovirus and reverse transcription.Chapter Four RNA biosynthesis and post-transcriptional processingPurpose1.Grasp of the concept of RNA biosynthesis.2.Grasp the feature of transcription and post-transcriptional processing of pre-mRNA pre-tRNAof pre-rRNA.3.Familiarity with ribozyme and self-splic ing.4.To know RNA replicase.Contents1.DNA dependent RNA synthesis.(1)RNA polymerases.①Prokaryotic RNA polymerase.②Eukaryotic RNA polymerase.(2)Transcriptional initiation.①Prokaryotic transcriptional initiation.②Eukaryotic transcriptional initiation.(3)Transcriptional termination.2.Pre-RNA processing.(1)Eukaryotic pre-mRNA processing.①5’ end capping.②3’ end polyadenylation.③Splicing.(2)Pre-rRNA and pre-tRNA processing.(3)mRNA edition.3.RNA dependent RNA synthesis.Chapter Five Protein biosynthesis, folding and processing Purpose1.Grasp of the features of the genetic code.2.Familiarity with the structure and function of mRNA, tRNA, and rRNA.3.Graspthe process of protein synthesis in prokaryotes.4.Familiarity with the process of protein biosynthesis in eukaryotes.5.To know protein folding, and clinical relatives in protein synthesis.Contents1.The protein biosynthesis system.(1)mRNA, the template.(2)The genetic code.(3)tRNA, the adapter.(4)Ribosome, and rRNAs.2.The five stages of protein biosynthesis.(1)Activation of amino acids.(2)Translation initiation.(3)Translation elongation.(4)Translation termination.(5)Protein folding and post-translational processing.3.Clinical relations in protein synthesis.(1)Antibiotics and protein synthesis.(2)Diphtheria and protein synthesis.(3)Interferon and protein synthesis.Chapter Six Regulation of gene expression in prokaryotesPurpose1.Grasp of the basic principle of regulation of prokaryotic gene expression.2.Grasp of the concept of operon and familiarity with lac operon.3.To know other important E.coli operons and regulation of gene expression in phage λ.4.To know translational regulation in prokaryotes.Contents1.Basic principle of regardless of gene expression.(1)The common feature of gene expression is both prokaryotes and eukaryotes.(2)Regulation of transcriptional initiation is the key step in regulation of gene expression.(3)Induction and repression are the basic models of prokaryotic gene expression.2.Bacterial operons.(1)Concept of operon.(2)Lac operon.(3)Other important operons.3.Regulation of gene expression in phage λ.(1)The life cycle of phage λ.(2)Regulation of gene expression in lysogen and lysis pathway.4.Translational regulation in prokaryotes.(1)Attenuation.(2)Regulon.(3)Coordinate synthesis of ribosomal proteins and rRNA.(4)mRNA stability.Chapter Seven Regulation of gene expression in eukaryotesPurpose1.Familiarity with regulation of transcriptional initiation in eukaryotes.2.Grasp of the structure and function of the transcription factors.3.To know post-translational regulation of gene expression.4.Familiarity with RNAi.5.To know translational regulation.Contents1.Regulation of transcriptional initiation.(1)Eukaryotic promoter.(2)Transcription factors.(3)Molecular mechanism of transcriptional initiation.2.Post-transcriptional regulation.(1)5’ end capping and 3’ end polyadenylation of mRNA.(2)Splicing.(3)mRNA edition.(4)mRNA transporting from nucleus to cytoplasm.(5)RNAi.3.Translational regulation.(1)eIFs.(2)5’ and 3’ UTR binding protein.(3)Translational initiation sites.Chapter Eight Recombinant DNA technologyPurpose1.Grasp of concepts of recombinant DNA and recombinant DNA technology.2.Familiarity with the basic principles of recombinant DNA technology.3.To know the relation between recombinant DNA technology and medicine. Contents1.Introduction to recombinant DNA technology(1)Molecular cloning.(2)DNA manipulation.2.Molecular tools.(1)Tool enzymes.(2)V ectors.(3)Host cells.3.DNA cloning.(1)Process of DNA cloning.(2)Application of DNA cloning.4.Analysis of cloned DNA.(1)RE mapping.(2)DNA sequencing.(3)Bioinformatics in DNA cloning.(4)Molecular hybridization.5.Application of recombinant DNA technology in biology and medicine.(1)To produce bioactive proteins.(2)To get site-specific mutation.(3)RNAi.(4)Transgenic animals and gene knock out.(5)Gene diagnosis and gene therapy.Chapter Nine Signal transducer and cellular signal transductionPurpose1.Grasp of concept of cellular signal transduction.2.Familiarity with types of signal transducers and secondary messengers.3.To know the basic structure of cellular signal transduction network.4.Familiarity with general signal pathways mediated by receptors.Contents1.The general of signal transduction.(1)Soluble and membrane binding chemical signal molecules.(2)Receptors.(3)The working models of cellular signal transduction.2.Small chemicals as transduction messengers.(1)The features.(2)cAMP and cGMP as secondary messengers.(3)Lipids as the secondary messengers.(4)Calc ium ion.3.Proteins families as signal transducers.(1)Protein kinases and phosphotases.(2)MAPK pathway members.(3)PTK pathway members.(4)G-proteins and small G-proteins.4.The structural basis of signal transduction network.(1)The features of transducer compound.(2)Protein-protein interaction.(3)The adapter protein and the scaffolding proteins.5.General signal pathways mediated by membrane receptors.(1)Pathways mediated by ligand-gated receptor channel.(2)Pathways mediated by G-proteins.(3)Pathways mediated by RTKs and PSTKs.。


The English-Chinese Bilingual Teaching and Learning Outline of Biochemistry Ⅱ(English Version)Chapter one Nucleotide and Nucleic acidsPropose:1.Grasp of the nucleotide structure, including the component of the nucleotide and the chemicalbonds linking the components, and the names and abbreviations of bases, nucleosides and nucleotides.2.Profound grasp of the key knowledge of the structure and function of DNA and RNA.3.Familiarity with the physio-chemical properties of DNA and the relation between theseproperties and the DNA structure.4.To know the technique of isolation and purification of nucleic acids and DNA sequencing. Contents:1.General concepts of nucleic acids.(1)Nucleotides are the building blocks of nucleic acids.(2)Nucleotide is composed of base, pentose, and phosphate.(3)The 3’,5’phosphodiester bond links nucleotides, many nucleotides are linked by the 3’,5’phosphodiester bond to form nucleic acids.(4)Nucleic acids are divided into DNA and RNA.(5)Other functions of the nucleotides.2.the three dimensioned structure of DNA(1)Double helix structure is the secondary structure of DNA.①The double helix DNA model put forth by Waston and Crick.②The features of double helix DNA.③A- DNA and Z- DNA.④Triple strand DNA.(2)Supercoils of DNA①Prokaryotic circular supercoiled DNA.②Eukaryotic mtDNA and ctDNA.③Topoisomerases.(3)Eukaryotic chromosome①Nucleosome.②From chromosome to chromatin and chromosome.(4)DNA is the genetic material①Avery’s experiment.②The genome.③Euchromatin and heterochromatin.3.Structure and functions of RNA(1)Four main kinds of RNA(2)mRNA: structure and function.(3)tRNA: structure and function.(4)rRNA: structure and function.(5)Other small RNAs.4.Physico-chemical properties of nucleic acids(1)Hydrolysis.(2)UV absorbance.(3)Denaturation, renaturation, and hybridization.5. Nuclease(1) Ribonuclease.(2) Dexoyribonuclease.6. Techniques of isolation and purification of nucleic acids and DNA sequencing.(1) Methods of isolation and purification of nuceic acids.(2) DNA sequencing.Chapter two Nucleotide metabolismPurpose1.To know digestion and absorption of nucleic acids and anabolism pathway of nucleic acids inthe human body.2.Profound grasp of the raw material of purine and pyrimidine synthesis, the end products ofpurine and pyrimidine breakdown.3.Grasp of the formation of deoxynucleotide from ribonucleotide.4.To know the function of antimetabolites.Contents1.Nucleotide functions.2.Anabolism and catabolism of purine.3.Anabolism and catabolism of pyrimidine.4.From ribonucleotides to deoxynucleotide.5.antimetabolites.Chapter Three DNA biosynthesis and DNA damage repair Purpose1.Familiarity with the central dogma.2.Grasp DNA replication and reverse transcription.3.To know DNA damage repair systems.Contents1.General feature of DNA replication.(1)Semiconservative.(2)Bidirectional.(3)Replication fork(4)Semidiscontinuous(5)The Ori.(6)The primer(7)Enzymes participating in DNA replication.2.Features of DNA polymerase.(1)The substrates.(2)The active center.(3)The enzyme activation.3.DNA replication in E.coli.(1)DNA replication initiation.(2)Chain elongation.(3)Replication termination.4.Eukaryotic DNA replication.(1)Eukaryotic DNA polymerase.(2)Process of DNA replication.(3)Telomerase and telomere.5.DNA damage.(1)Causes of DNA damage.(2)Mismatch repair system.(3)Other DNA repair systems.6.Reverse transcription.(1)Development of the central dogma put forth by Crick.(2)Retrovirus and reverse transcription.Chapter Four RNA biosynthesis and post-transcriptional processing Purpose1.Grasp of the concept of RNA biosynthesis.2.Grasp the feature of transcription and post-transcriptional processing of pre-mRNA pre-tRNAof pre-rRNA.3.Familiarity with ribozyme and self-splicing.4.To know RNA replicase.Contents1.DNA dependent RNA synthesis.(1)RNA polymerases.①Prokaryotic RNA polymerase.②Eukaryotic RNA polymerase.(2)Transcriptional initiation.①Prokaryotic transcriptional initiation.②Eukaryotic transcriptional initiation.(3)Transcriptional termination.2.Pre-RNA processing.(1)Eukaryotic pre-mRNA processing.①5’ end capping.②3’ end polyadenylation.③Splicing.(2)Pre-rRNA and pre-tRNA processing.(3)mRNA edition.3.RNA dependent RNA synthesis.Chapter Five Protein biosynthesis, folding and processing Purpose1.Grasp of the features of the genetic code.2.Familiarity with the structure and function of mRNA, tRNA, and rRNA.3.Graspthe process of protein synthesis in prokaryotes.4.Familiarity with the process of protein biosynthesis in eukaryotes.5.To know protein folding, and clinical relatives in protein synthesis.Contents1.The protein biosynthesis system.(1)mRNA, the template.(2)The genetic code.(3)tRNA, the adapter.(4)Ribosome, and rRNAs.2.The five stages of protein biosynthesis.(1)Activation of amino acids.(2)Translation initiation.(3)Translation elongation.(4)Translation termination.(5)Protein folding and post-translational processing.3.Clinical relations in protein synthesis.(1)Antibiotics and protein synthesis.(2)Diphtheria and protein synthesis.(3)Interferon and protein synthesis.Chapter Six Regulation of gene expression in prokaryotes Purpose1.Grasp of the basic principle of regulation of prokaryotic gene expression.2.Grasp of the concept of operon and familiarity with lac operon.3.To know other important E.coli operons and regulation of gene expression in phage λ.4.To know translational regulation in prokaryotes.Contents1.Basic principle of regardless of gene expression.(1)The common feature of gene expression is both prokaryotes and eukaryotes.(2)Regulation of transcriptional initiation is the key step in regulation of gene expression.(3)Induction and repression are the basic models of prokaryotic gene expression.2.Bacterial operons.(1)Concept of operon.(2)Lac operon.(3)Other important operons.3.Regulation of gene expression in phage λ.(1)The life cycle of phage λ.(2)Regulation of gene expression in lysogen and lysis pathway.4.Translational regulation in prokaryotes.(1)Attenuation.(2)Regulon.(3)Coordinate synthesis of ribosomal proteins and rRNA.(4)mRNA stability.Chapter Seven Regulation of gene expression in eukaryotes Purpose1.Familiarity with regulation of transcriptional initiation in eukaryotes.2.Grasp of the structure and function of the transcription factors.3.To know post-translational regulation of gene expression.4.Familiarity with RNAi.5.To know translational regulation.Contents1.Regulation of transcriptional initiation.(1)Eukaryotic promoter.(2)Transcription factors.(3)Molecular mechanism of transcriptional initiation.2.Post-transcriptional regulation.(1)5’ end capping and 3’ end polyadenylation of mRNA.(2)Splicing.(3)mRNA edition.(4)mRNA transporting from nucleus to cytoplasm.(5)RNAi.3.Translational regulation.(1)eIFs.(2)5’ and 3’ UTR binding protein.(3)Translational initiation sites.Chapter Eight Recombinant DNA technology Purpose1.Grasp of concepts of recombinant DNA and recombinant DNA technology.2.Familiarity with the basic principles of recombinant DNA technology.3.To know the relation between recombinant DNA technology and medicine. Contents1.Introduction to recombinant DNA technology(1)Molecular cloning.(2)DNA manipulation.2.Molecular tools.(1)Tool enzymes.(2)Vectors.(3)Host cells.3.DNA cloning.(1)Process of DNA cloning.(2)Application of DNA cloning.4.Analysis of cloned DNA.(1)RE mapping.(2)DNA sequencing.(3)Bioinformatics in DNA cloning.(4)Molecular hybridization.5.Application of recombinant DNA technology in biology and medicine.(1)To produce bioactive proteins.(2)To get site-specific mutation.(3)RNAi.(4)Transgenic animals and gene knock out.(5)Gene diagnosis and gene therapy.Chapter Nine Signal transducer and cellular signal transduction Purpose1.Grasp of concept of cellular signal transduction.2.Familiarity with types of signal transducers and secondary messengers.3.To know the basic structure of cellular signal transduction network.4.Familiarity with general signal pathways mediated by receptors.Contents1.The general of signal transduction.(1)Soluble and membrane binding chemical signal molecules.(2)Receptors.(3)The working models of cellular signal transduction.2.Small chemicals as transduction messengers.(1)The features.(2)cAMP and cGMP as secondary messengers.(3)Lipids as the secondary messengers.(4)Calcium ion.3.Proteins families as signal transducers.(1)Protein kinases and phosphotases.(2)MAPK pathway members.(3)PTK pathway members.(4)G-proteins and small G-proteins.4.The structural basis of signal transduction network.(1)The features of transducer compound.(2)Protein-protein interaction.(3)The adapter protein and the scaffolding proteins.5.General signal pathways mediated by membrane receptors.(1)Pathways mediated by ligand-gated receptor channel.(2)Pathways mediated by G-proteins.(3)Pathways mediated by RTKs and PSTKs.。

Similarity of regulation between eukaryotes and prokaryote1、Principles are the same: signals, activators and repressors, recruitment and allostery (变构), cooperative binding2、Expression of a gene can be regulated at the similar steps, and the initiation of transcription is the most pervasively regulated step.Difference in regulation between eukaryotes and prokaryote1. Pre-mRNA splicing adds an important stepfor regulation.2. The eukaryotic transcriptional machineryis more elaborate than its bacterial counterpart.3. Nucleosomesand their modifiers influenceaccess to genes.4. Many eukaryotic genes have more regulatory binding sites and are controlled by more regulatory proteins than are bacterial genes.cis-regulatory elementA cis-regulatory element or cis-element is aregion of DNA or RNA that regulates the expression of Genes located on that same strand. This term is constructed from the Latin word cis, which means "on the same side as". These cis-regulatory elements are often binding sites of one or more trans-acting factors. A cis-element may be located in the promoter region 5' to the gene it controls, in an intron, or in the 3'untranslated region. Trans-regulatory elementsTrans-regulatory elements are species which may modify the expression of genes distant from the gene that was originally transcribedto create them. To demonstrate the concept (this is not a specific example), a transcription factor which regulates a geneon chromosome 6 might itself have been transcribed from a gene on chromosome 11. This term is constructed from the Latin root -trans, which means "across from". Expound the general mechanism of control of gene expression of Eukaryotes。



Gene Regulation in PlantsGene regulation in plants is a complex process that involves the control of gene expression, which is essential for their growth, development, and response to environmental stresses. The regulation of gene expression in plants is tightly controlled at various levels, including transcriptional, post-transcriptional, translational, and post-translational levels. This complex regulatory network ensures that genes are expressed at the right time, in the right place, and in the right amount, thereby enabling plants to adapt to changing environmental conditions.At the transcriptional level, gene expression is regulated through the interaction of transcription factors with specific DNA sequences in the promoter region of genes. Transcription factors are proteins that bind to specific DNA sequences and either activate or repress gene expression. The binding of transcription factors to DNA is regulated by various signaling pathways that are activated in response to environmental cues, such as light, temperature, and stress. For example, the binding of the transcription factor HY5 to the promoter region of the gene encoding the chlorophyll biosynthetic enzyme, Lhcb1, is regulated by light signaling pathways, which enable plants to adjust their photosynthetic capacity in response to changes in light intensity.At the post-transcriptional level, gene expression is regulated through the processing of messenger RNA (mRNA) molecules, which are transcribed from DNA and serve as templates for protein synthesis. mRNA processing involves various modifications, including capping, splicing, and polyadenylation, which affect the stability and translatability of mRNA molecules. The regulation of mRNA processing is mediated by RNA-binding proteins and small RNA molecules, which bind to specific sequences in mRNA and regulate their processing and stability. For example, the RNA-binding protein, FCA, regulates the splicing of the mRNA encoding the flowering time regulator, FLC, in response to environmental cues, such as temperature and photoperiod.At the translational level, gene expression is regulated through the control of protein synthesis, which is mediated by ribosomes and associated factors. The regulation of protein synthesis is mediated by various signaling pathways that are activated in response toenvironmental cues, such as nutrient availability and stress. For example, the Target of Rapamycin (TOR) signaling pathway regulates the translation of mRNA molecules encoding proteins involved in nutrient uptake and metabolism in response to changes in nutrient availability.At the post-translational level, gene expression is regulated through the modification and degradation of proteins, which affect their activity and stability. The regulation of protein modification and degradation is mediated by various signaling pathways that are activated in response to environmental cues, such as stress and pathogen attack. For example, the ubiquitin-proteasome system regulates the degradation of proteins involved in plant immunity in response to pathogen attack.Overall, the regulation of gene expression in plants is a complex and dynamic process that enables them to adapt to changing environmental conditions. The regulatory network that controls gene expression involves multiple levels of regulation, including transcriptional, post-transcriptional, translational, and post-translational levels, which are mediated by various signaling pathways and regulatory factors. Understanding the mechanisms underlying gene regulation in plants is essential for developing strategies to improve crop productivity and enhance plant resilience to environmental stresses.。
Genetic Regulation of DevelopmentGenetic regulation of development is a complex process that involves the activation and repression of genes in response to various signals. This process is critical for the proper development of an organism, as it controls the formation of different tissues and organs, and ultimately determines the overall body plan. In this essay, I will explore the various mechanisms of genetic regulation of development, as well as the factors that influence this process.One of the key mechanisms of genetic regulation of development is the control of gene expression through transcription factors. Transcription factors are proteins that bind to specific DNA sequences and activate or repress the transcription of nearby genes. They play a crucial role in determining cell fate and differentiation during development. For example, the transcription factor Oct4 is essential for maintaining the pluripotency of embryonic stem cells, while the transcription factor MyoD is required for the differentiation of muscle cells.Another important mechanism of genetic regulation of development is epigenetic modification. Epigenetic modifications are changes to the DNA molecule or its associated proteins that do not involve changes to the underlying DNA sequence. These modifications can be heritable and can affect gene expression in a variety of ways. For example, DNA methylation, which involves the addition of a methyl group to a cytosine base in DNA, can silence gene expression by blocking the binding of transcription factors to the DNA. Histone modifications, such as acetylation and methylation, can also affect gene expression by altering the structure of chromatin and making it more or less accessible to transcription factors.In addition to transcription factors and epigenetic modifications, genetic regulation of development is also influenced by signaling pathways. Signaling pathways involve the transmission of signals between cells, often through the binding of ligands to receptors on the cell surface. These signals can activate or repress gene expression, leading to changes in cell behavior and differentiation. For example, the Notch signaling pathway is involved in the differentiation of many cell types, including neurons, muscle cells, and blood cells.The genetic regulation of development is also influenced by environmental factors, such as nutrition, temperature, and exposure to toxins. For example, studies have shown that maternal nutrition during pregnancy can affect the expression of genes related to metabolism and growth in the developing fetus. Similarly, exposure to certain toxins, such as lead or alcohol, can lead to epigenetic modifications that affect gene expression and lead to developmental abnormalities.Finally, it is worth noting that genetic regulation of development is a highly dynamic process that can be influenced by feedback mechanisms. These feedback mechanisms can involve the regulation of gene expression by other genes or by the products of gene expression themselves. For example, the product of a gene may act as a transcription factor that activates or represses the expression of other genes in a feedback loop.In conclusion, genetic regulation of development is a complex process that involves the activation and repression of genes in response to various signals. This process is critical for the proper development of an organism, and is influenced by a variety of mechanisms, including transcription factors, epigenetic modifications, signaling pathways, environmental factors, and feedback mechanisms. Understanding the mechanisms of genetic regulation of development is essential for understanding the basis of developmental disorders and for developing new treatments for these disorders.。