MicroRNAome Genome_A Treasure for Cancer Diagnosis and Therapy
- 格式:pdf
- 大小:765.23 KB
- 文档页数:26

OPENREVIEWMicroRNA regulons in tumor microenvironmentHI Suzuki 1,A Katsura,H Matsuyama 2and K MiyazonoINTRODUCTIONRecent advances in cancer therapeutics have yielded several new-generation therapeutic modalities,and cancer prognoses have improved signi ficantly.However,further advances are required before major improvements are achievable,especially in patients with advanced and/or intractable cancers,which are refractory to conventional therapeutic options.Although cancer therapies ideally seek eradication of all cancer cells in tumor tissues,recent work has emphasized that targeting of cancer cells is not equivalent to targeting of tumor tissues.The work suggests that cancer initiation and progression are de fined by not only the behavior of cancer cells per se but also the development of tumor tissues,controlled in turn by crosstalk between cancer cells and the surrounding microenvironment.1–3Recent cancer research has emphasized the signi ficance of constant evolution of the tumor microenvironment,facilitating tumor formation,metastasis and development of refractoriness to cancer therapy.3Tumor microenvironments are highly heterogeneous,contain-ing various cell types,including fibroblasts,endothelial cells,pericytes,immune cells and local and bone marrow-derived stromal stem and progenitor cells,and a surrounding extracellular matrix (ECM)(Figure 1).1–3These cells originate from normal cells but become altered during tumor development.For example,many types of solid tumor are accompanied by variable extents of stromal cell in filtration and ECM deposition,termed desmoplasia.Such cancer tissue remodeling allows tumor cells to grow and disseminate and contributes to an increase in interstitial fluid pressure,which can impede delivery of cancer drugs.2–4Cancer-associated fibroblasts (CAFs)make versatile contribution to these responses.2,3MicroRNAs (miRNAs)are small non-coding RNAs that function as major players of posttranscriptional gene regulation within diverse cell types.5Re flecting the importance in cell biology,they also have critical roles in cancer biology.In addition to various protein-coding oncogenes and tumor-suppressor genes,previous studies have demonstrated multifaceted roles of miRNAs in cancer cell behaviors,which constitute major aspects of conventional hallmarks of cancer,including sustained proliferation,resistance to cell death,evasion of growth suppressors,establishment of replicative immortality and acquisition of invasive phenotypes.1,6For example,miR-34a is associated with growth control,miR-21with cell proliferation and the miR-200family with epithelial –mesenchymal transition (EMT).7In addition,recent work has shown that miRNAs of cancer cells modulate the microenvironment via non-cell-autonomous mechanisms,and alterations in the miRNA pro files of neighboring cells that lack genetic abnormalities favor the acquisition of cancer hallmark traits,thereby unexpectedly expanding the roles of miRNAs in tumor microenvironments.8–12Such actions include regulation of tumor angiogenesis,tumor immune invasion and tumor –stromal interactions.The present review focuses on the relationship between miRNA alterations in cancer cells and tumor microenvironments,and the mechanisms of miRNA-mediated regulation of tumor microenvironments,with the perspective of possible therapeutic interventions.Department of Molecular Pathology,Graduate School of Medicine,The University of Tokyo,Tokyo,Japan.Correspondence:Professor K Miyazono,Department of Molecular Pathology,Graduate School of Medicine,The University of Tokyo,7-3-1Hongo,Bunkyo-ku,Tokyo 113-0033,Japan.E-mail:miyazono@m.u-tokyo.ac.jp 1Current address:David H.Koch Institute for Integrative Cancer Research,Massachusetts Institute of Technology,Cambridge,MA 02139,USA.2Current address:Otsuka Maryland Medicinal Laboratories,Inc.,9900Medical Center Drive,Rockville,MD 20850,USA.Received 4May 2014;revised 4June 2014;accepted 6June 2014Oncogene (2014),1–10©2014Macmillan Publishers Limited All rights reserved 0950-9232//oncDYSFUNCTION OF miRNAS IN CANCERIn mammalian cells,the primary transcripts of miRNAs (primary miRNAs;pri-miRNAs)are transcribed by RNA polymerase II or III from miRNA-coding sequences,residing principally in intergenic regions or within introns of genes.Most pri-miRNAs are processed into short hairpin RNAs (precursor miRNAs;pre-miRNAs)by the Drosha/DGCR8complex with RNase III activity in the nucleus.Pre-miRNAs are transported to the cytoplasm and further cleaved by a second RNase III Dicer to yield 21–25-nucleotide-long miRNA duplexes.Single RNA strands derived from such duplexes are incorporated as mature miRNAs within Argonaute proteins (Ago1-4)to form RNA-induced silencing complexes.In general,RNA-induced silencing complexes suppress the expression of various target genes through sequence complementarity between miRNAs and target mRNAs.5,13Various mechanisms contribute to aberrant miRNA expression in cancer.13,14As with abnormalities in oncogenes and tumor-suppressor genes,alterations in miRNAs can be explained in part by several mechanisms,including chromosomal deletion,ampli-fication,mutation,epigenetic silencing and transcriptional dysre-gulation of pri-miRNA transcripts.On the other hand,concerning the characteristic process of miRNA biogenesis,impairment of miRNA processing might explain the downregulation of both broad groups of miRNAs and certain speci fic miRNAs.13,15In addition,various miRNA-processing regulators,such as LIN28,KSRP and p53,have been identi fied to date at each step of miRNA biogenesis,suggesting multidimensional regulation of miRNA biogenesis.13,16–18Furthermore,a recent report showed that Hippo signaling pathway regulated pri-miRNA processing in a cell-density-dependent manner,linking cell –cell contact to miRNA processing.19NON-CELL-AUTONOMOUS FUNCTION OF CANCER-RELATED miRNASMost,if not all,of cancers are de fined by the evolution of genomic abnormalities.Thus,in theory,alterations of ostensibly normal cells in tumor microenvironments will be largely attributable to dysfunction of the gene regulatory network of cancer cells per se .Recent advances have shown that miRNA dysfunction in tumor cells modulates the tumor microenvironment via non-cell-autonomous mechanisms (Figure 2),thus supporting this concept.10In this context,the core targets of miRNAs,termed miRNA regulons,are currently being expanded to include various modulators of tumor microenvironments.8–12This type of regula-tion has been also described in the context of protein-coding oncogenes and tumor-suppressor genes.For example,it has been shown that KRAS,mutation in which is an early event in the development of pancreatic ductal adenocarcinoma,enhances granulocyte macrophage colony-stimulating factor (GM-CSF)production from pancreatic ductal epithelial cells and thus increases recruitment of immunosuppressive Gr1+CD11b +myeloid cells to suppress anti-tumor immunity.20,21In this section,we focus on such distal impacts of cancer-associated miRNAs onthreeFigure ponents of tumor microenvironment.Tumor micro-environment is very heterogeneous and comprised of various cell types,such as CAFs,endothelial cells,pericytes,immune cells,including various types of lymphocytes,Treg,TAMs and MDSCs,and local and bone marrow-derived stromal stem and progenitor cells,and surrounding ECM.Cancer cellMicroenvironmentFigure 2.Non-cell-autonomous roles of cancer cell miRNAs in regulation of tumor microenvironment.Alteration of miR-9(a ),miR-126(b ),miR-135b (c ),miR-29b (d ),miR-30b/d (e ),miR-34a (f )and miR-199a-5p/3p and miR-1908(g )in cancer cells elicit distal impacts on tumor microenvironment to promote tumor progression through various non-cell-autonomous mechanisms.Oncogenic miRNAs and tumor-suppressive miRNAs are represented by black and red,respectively.microRNAs in tumor microenvironmentHI Suzuki et al2Oncogene (2014),1–10©2014Macmillan Publishers Limitedmajor components of the tumor milleu;these are the vasculature,ECM and immune cells.Distal regulation of tumor angiogenesis by miR-9and miR-126 Epithelial–mesenchymal transition(EMT)is an important step incancer metastasis.Among a set of EMT-regulatory miRNAs, including miR-200,miR-103/107,miR-205,let-7and miR-9,22miR-9has been shown to simultaneously modulate tumorangiogenesis through regulation of vascular endothelial growth factor(VEGF)-A(Figure2a).23In breast cancer cells,miR-9,inducedby MYC and MYCN,targeted E-cadherin and increased cell motilityand invasiveness.E-cadherin downregulation by miR-9activated β-catenin signaling,in turn upregulating VEGF-A expression and increasing tumor angiogenesis.23This report raised the possibilitythat miRNA targets diversify onto cell-autonomous regulators andnon-cell-autonomous regulators in tumor progression.This possibility was further solidified by studies on miR-126,the expression of which is frequently suppressed in various types of human cancer,including breast,gastric and colon cancers.24–26 Tavazoie et al.24showed that miR-126non-cell-autonomously restricted metastasis via regulation of multiple targets involved in endothelial recruitment(Figure2b).27They showed that endo-genous miR-126in breast cancer cells contributed to suppression of metastatic colonization and that the metastatic nodules developing upon miR-126silencing displayed a denser vascula-ture.Mechanistically,miR-126downregulates secretion of a soluble form of c-Mer tyrosine kinase receptor(MERTK),and insulin-like growth factor binding protein2(IGFBP2),from metastatic cells,by suppressing MERTK and inhibiting IGFBP2 and a regulatory gene thereof,PITPNC1(encoding phosphatidy-linositol transfer protein,cytoplasmic1),respectively.Subse-quently,downregulation of endogenous miR-126promotes endothelial recruitment by increasing IGFBP2/IGF1/IGF1R signal-ing and attenuating GAS6/MERTK signaling,regulated by compe-tition between the MERTK ligand GAS6and MERTK,in endothelial cells.They further showed that overexpression of eight target genes of miR-126,including MERTK,IGFBP2and PITPNC1,in primary breast cancer,was associated with shorter metastasis-free survival in multiple cohorts.27This set of eight genes constitutes the core target genes of miR-126,namely the miR-126regulon,which links the anti-metastatic activity of miR-126to cancer–endothelial interactions.In addition,Wang and colleagues reported a unique function of miR-126in breast cancer metastasis.28Although the precise mechanism(s)determining asymmetric miRNA biogenesis from a single miRNA precursor have not yet been defined,the miR-126 precursor produces comparable amounts of5p and3p miRNA species.They showed that the two mature miRNAs derived from pre-miR-126,miR-126and miR-126*,cooperatively targeted expression of stromal cell-derived factor-1alpha(SDF-1α)/ chemokine(C-X-C motif)ligand12(CXCL12),and subsequently chemokine(C-C motif)ligand2(CCL2),in cancer cells.Through these inhibitory effects,they suppressed recruitment of mesench-ymal stem cells and inflammatory monocytes into tumor stroma, ultimately suppressing lung metastasis by breast cancer cells.In their report,miR-126/-126*appeared to exert metastasis-suppressive activities predominantly in primary tumor site,rather than metastatic nodules.28Regulation of tumor cell immunophenotype by miR-135b Although the roles of miRNAs in tumor microenvironments have emerged in recent reports,especially about solid tumors,8–11our group discovered a unique environmental involvement of miRNAs in hematological malignancies,where miRNA alteration accounts for regulation of immunophenotype of cancer cells per se and modulation of tumor microenvironments(Figure2c).29We showed that miR-135b,overexpressed in various types of cancer,including colon and lung cancers,30,31was highly expressed in nucleophosmin-anaplastic lymphoma kinase(NPM-ALK)-positive anaplastic large cell lymphomas(ALCLs)and mediated down-stream signaling of the NPM-ALK-STAT3(signal transducer and activator of transcription factor3)pathway.29In this tumor type, NPM-ALK oncogene strongly promotes the expression of LEMD1,ahost gene of miR-135b,and that of miR-135b,through activationof STAT3.FOXO1was identified as a target of miR-135b, suggesting contribution to the oncogenic activities of NPM-ALK.In parallel,we demonstrated immunomodulatory properties ofmiR-135b.Interestingly,miR-135b conferred interleukin(IL)-17-producing immunophenotype,which has recently been demon-strated by genome-wide expression profiling of various peripheralT-cell lymphomas,on ALCL cells.29This skewing of ALCL immunophenotype,overlapping with that of T-helper(Th)17 cells,was associated with targeting of Th2master regulators STAT6and GATA3by miR-135b,indicating that miR-135b contributes to generation of an IL-17-producing immunopheno-type by perturbing mutually antagonistic differentiation programs active during normal lymphocyte differentiation.32,33miR-135b suppression inhibited expression levels of IL-17A,IL-17F,IκBζ,IL-6and IL-8in ALCL cells.miR-135b silencing also attenuated the expression of granzyme B and perforin1,cytotoxic molecules highly expressed in ALCLs,implying that miR-135b exerts a broad range of effects on the ALCL immunophenotype.In line with theinflammatory role of IL-17,miR-135b blockade attenuated the paracrine inflammatory response in co-cultures of ALCL cells andfibroblasts and reduced tumor angiogenesis and in vivo growth. Although the mechanisms underlying lymphoma immunopheno-types related to the corresponding normal lymphocytes are largely unclear,the study illuminated the unique contribution made by an oncogenic kinase-linked miRNA to the modulation of tumor immune phenotype.Distal impacts of miR-29on angiogenesis and ECM remodelingAn association between miRNAs and GATA3transcriptional factorhas also been described in the setting of breast cancer development.34In the mammary gland,GATA3is required for luminal epithelial cell differentiation and maintenance,and its expression progressively decreases during progression of luminal breast cancer in association with a worse prognosis.A recent report demonstrated that GATA3promoted differentiation of breast cancer cells,inhibited metastasis and modified the tumor microenvironment,through induction of miR-29b(Figure2d).34 Thus the GATA3–miR29b axis functions as a tumor-suppressive arm,and loss of miR-29b in cells expressing GATA3promotes a mesenchymal phenotype and metastasis.As mechanistic insights, various targets of miR-29b,including VEGF-A,ANGPTL4,platelet-derived growth factor,LOX and matrix metalloproteinase9 (MMP9),which are involved in angiogenesis,collagen remodelingand proteolysis to promote metastasis,were identified.miR-29b introduction in a murine orthotopic breast cancer model reduced blood vessel development and the extent offibrillar collagen synthesis without a concomitant effect on primary tumor size and reduced the incidence and size of metastasis.This metastasis inhibitory effect of miR-29b was mitigated by re-introduction ofthe miR-29b regulons VEGF-A,ANGPTL4,LOX and MMP9, strengthening the importance of microenvironmental target regulation by miR-29b.34This study revealed a pleiotropic rolefor miR-29b in the modulation of the tumor microenvironment;both angiogenesis and ECM remodeling were affected.AsmiR-29b was also suppressed in NPM-ALK-positive ALCLs,in which GATA3is downregulated,35a GATA3–miR-29b axis may be operational in a range of cancer types.Additionally,in nasophar-yngeal carcinomas,miR-29c downregulation has been reported to induce the expression of ECM proteins,including COL1A2, COL3A1,COL4A1and lamininγ1,as well.36microRNAs in tumor microenvironmentHI Suzuki et al3©2014Macmillan Publishers Limited Oncogene(2014),1–10Distal modulation of tumor–immune crosstalk by miRNAsAn important aspect of the evolution of tumor microenvironments is evasion of anti-tumor immune responses.3Recent work has suggested that such immune escape of tumors can be broadly classified into two categories,depending on the characteristics of the tumor microenvironment.37One major type exhibits a T-cell-inflamed phenotype with infiltration of T cells and a broad chemokine profile.Such tumors appear to endure immune attack predominantly by engaging inhibitory effectors of the immune system,such as PD-L1,IDO and regulatory T(Treg)cells.The other major type lacks this T-cell-inflamed phenotype and appears to prevent immunological attack through immune system ignorance and exclusion.In tumors in which the latter mechanism is operative,dense stroma and accumulation of immunosuppressive myeloid or macrophage populations might be observed,instead of T-cell infiltration.miRNA dysregulation in cancer cells appears to contribute to both types of immune escape.Regulation of SDF-1αby miR-126/126*and modulation of tumor stroma reactions by miR-29b may be associated with the latter category of escape.28,34Recent analyses have shed light on the contributions made by miRNAs to activation of the inhibitory arm of the immune system in the former category of escape.In human melanoma,high expression levels of miR-30b and miR-30d were associated with frequent metastasis,early recurrence and lower overall survival (Figure2e).38Functional analyses revealed that miR-30b/-30d directly targeted the GalNAc transferase GALNT7and subse-quently promoted the secretion of the immunosuppressive cytokine IL-10.38This reduced immune cell activation and enhanced recruitment of Treg cells,promoting metastasis.In addition,a study in hepatocellular carcinoma also revealed enhancement of Treg cell function by a miRNA-mediated non-cell-autonomous mechanism(Figure2f).39In hepatocellular carci-noma,portal vein tumor thrombus(PVTT)is associated with a poor prognosis.Persistent presence of heptitis B virus(HBV)in liver tissue enhances transforming growth factor(TGF)-βactivity, and in turn,TGF-βsuppresses miR-34a.39Although miR-34a is well known to be a transcriptional target of p53,and a representative tumor-suppressive miRNA,15miR-34a had no significant effect on cell proliferation but suppressed the production of CCL22 important for Treg cell recruitment.An inverse correlation between the expression level of miR-34a and CCL22or FoxP3 was observed in HBV+primary tumor and PVTT samples. Restoration of miR-34a inhibited the growth of murine liver tumor cells,infiltration of Treg cells and metastasis,in immune-competent mice.39Taken together,the data showed active participation of miRNAs in immune escape and also revealed one of the mechanisms of immune suppression by TGF-β,which is abundantly expressed in tumor stroma and has a versatile role in tumor development.40Furthermore,other reports have also revealed the involvement of multiple miRNAs in immune responses.In glioma,miR-124 downregulation is associated with immunosuppressive activities of glioma stem cells and suppression of the effector response of T cells.41miR-17-5p and miR-20in miR-17-92polycistronic cluster were shown to suppress cell migration and invasion by altering the secretion levels of IL-8,CK8and CXCL1from breast cancer cells.42In head and neck cancer,the tumor-suppressive miR-145 has been reported to target SOX9and ADAM17and subsequently suppress IL-6production.43Convergent modulation of environmental regulators by multiple miRNAsIndependent miRNAs potentially target a range of genes exerting various functions,including both tumor suppressors and promo-ters.The sets of miRNA regulons mentioned above serve as examples that the targets divergently biased for tumor promotion or suppression.On the other hand,another scenario has also been described:one set of miRNAs convergently target metastasis-associated genes.44In melanoma,a set of pro-metastatic miRNAs, including miR-199a-5p,miR-199a-3p and miR-1908,has been shown to combinatorially target apolipoprotein E(ApoE) (Figure2g).44Secretion of ApoE from cancer cells suppresses cancer cell invasion and endothelial cell recruitment,thereby inhibiting metastasis and angiogenesis.Regulation of SDF-1αby miR-126/126*is also categorized as such a type of regulation.28 When independent miRNAs lack the power to drive significant biological effects,a combination of alterations in multiple miRNAs may modulate the threshold to achieve metastasis. PROXIMAL ROLES OF miRNAS IN TUMOR STROMAL CELLS Various cell types,includingfibroblasts,endothelial cells,pericytes and immune inflammatory cells,contribute to the formation of tumor microenvironments favorable for cancer growth.Of these, CAFs,in contrast to normalfibroblasts,enhance ECM production and secrete cancer-activating cytokines and chemokines,signifi-cantly promoting tumorigenesis.3,45–48It remains largely unclear how CAFs arise from normalfibroblasts.In certain cancer types, CAFs are thought to be generated from normalfibroblasts by tumor-derived paracrine signals.45,46Nonetheless,it has also been known that the tumor-supporting capacity of CAFs is sustainable over multiple passages in the absence of cancer cells.45Although early studies suggested the presence of genetic abnormalities in tumor-suppressors p53and phophatase and tensin homolog (PTEN)in CAFs,49detailed genetic analyses have indicated that genetic alterations are in fact rare,50,51suggesting that other epigenetic mechanisms stabilize the CAF phenotype.In this context,recent studies have revealed that the miRNA profiles of normalfibroblasts and CAFs differ and that several features of the CAF phenotype are attributable to miRNA dysregulation.9In addition,miRNA function has been linked to variations in other tumor-associated cell types,including osteoclasts,myeloid-derived suppressor cells(MDSCs),and tumor-associated macro-phages(TAMs).We next focus on the dysregulation of miRNAs in stromal cells and the proximal effects of such changes in tumor microenvironments(Figure3).Reprogramming of CAFs by miR-31,miR-214,and miR-155Mitra et al.52recently conducted miRNA expression profiling of primary CAFs vs adjacent normalfibroblasts in ovarian cancer patients,and induced CAFs(iCAFs),generated from normal fibroblasts upon co-culture with tumor cells,vs normalfibroblasts(Figure3a).This study showed downregulation of miR-214and miR-31and upregulation of miR-155in CAFs.Interestingly, introduction of miR-214and miR-31and silencing of miR-155 converted the CAF phenotype to normality,and vice versa, suggesting a direct involvement of miRNAs in reversible conver-sion of normalfibroblasts into CAFs.Patient CAFs,iCAFs and miRNA-reprogrammed CAFs(miR-CAFs)all exhibited high expres-sion levels of several pro-tumorigenic chemokines,including CCL5, CCL20and CXCL8/L5was shown to be a direct target of miR-214.Moreover,patient CAFs and miR-CAFs enhanced the growth of co-injected ovarian cancer cells and increased invasion by co-cultured cancer cells;these effects were abrogated upon neutralization of CCL5,indicating that CCL5is a key tumor modulator in miR-CAFs.52The results indicated that ovarian cancer cells reprogramfibroblasts to CAFs via the action of miRNAs. In addition,downregulation of miR-31has been reported in CAFs isolated from endometrial cancers,and miR-31downregulation upregulated the target SATB2(special AT-rich sequence-binding protein2),enhancing tumor cell migration.53Downregulation of miR-148a in CAFs of endometrial cancer has also been reported.54microRNAs in tumor microenvironmentHI Suzuki et al4Oncogene(2014),1–10©2014Macmillan Publishers LimitedProximal roles of PTEN-miR-320axis in CAFCAFs frequently show characteristic upregulation of CAF-related genes,such as α-smooth muscle actin,fibroblast-speci fic protein,platelet-derived growth factor-B and fibroblast activation protein.47,55In addition,alterations in the expression levels of several other markers,including periostin,p53,PTEN and podoplanin,have been reported to correlate with the prognostic outcomes of solid tumors.55–57Accordingly,abrogation of PTEN in stromal cells has been shown to promote the development of epithelial mammary tumors.57A recent study found that a speci fic miRNA was responsible for the tumor-suppressive activity of PTEN (Figure 3a).58Of several miRNAs exhibiting distinct expression patterns in PTEN-depleted fibroblasts,miR-320was identi fied as a tumor suppressor acting downstream of PTEN.miR-320targets ETS2(v-ets erythroblastosis virus E26oncogene homolog 2),MMP9and Emilin2,and downregulation thereof induces a tumor-speci fic secretome,composed of MMP9,MMP2,BMP1and LOXL2,leading to enhanced angiogenesis and tumor cell invasion.Expression of the miR-320-Ets2-related secretory regulons sepa-rated human normal breast stroma from tumor stroma and correlated with the outcomes of breast cancer patients.58Co-downregulation of miR-15a and miR-16-1in tumor cells and CAFsAnother report demonstrated that certain miRNAs exhibit a pan-tumor-suppressive function in both cancer cells and CAFs.59miR-15a and miR-16-1are well-characterized tumor-suppressive miRNAs encoded in the chromosomal region 13q14and are frequently lost in chronic lymphocytic leukemia and prostate cancer.60,61miR-15a and miR-16-1function as tumor suppressors in prostate cancer by targeting BCL2,cyclin D1and Wnt3a.61Musumeci et al .59reported that miR-15and miR-16were also downregulated in the CAFs of most of prostate tumor patients (Figure 3a).Restoration of miR-15a and miR-16-1suppressed proliferation and migration of CAFs as similarly observed in cancer cells.Furthermore,miR-15a and miR-16suppressed two novel targets,fibroblast growth factor 2(FGF2)and its receptor FGFR1,which enhance the proliferation and migration of both stromal and cancer cells.Reconstitution of miR-15and miR-16suppressed the tumor-promoting activity of stromal cells in vivo .59These findings suggest that some miRNAs can function as pan-tumor suppressors in cancer cells and tumor microenvironments,although the mechanisms of deregulation may be distinctbetween cancer cells and microenvironments.From the stand-point of therapeutic intervention,simple restoration of this type of miRNA might aid co-targeting cancer cells and tumor microenvironment.miRNA-mediated osteoclast differentiation in bone metastasis Osteolytic bone metastasis is frequently observed in many cancer types,including breast and lung cancers,and is associated with aberrant osteoclast activation.48,62,63In a recent study,treatment of preosteoclast cells with conditioned media from highly bone metastatic breast and bladder cancer cell lines was shown to induce characteristic changes in miRNA expression,similar to those noted upon stimulation with RANKL (receptor activator of nuclear factor κB ligand),a major stimulator of osteoclast differentiation (Figure 3b).64Bone-metastatic cancer cells trig-gered such miRNA expression changes partly through soluble intercellular adhesion molecule 1(sICAM1)-mediated NF-κB activation.Overexpression of multiple miRNAs that were down-regulated during osteoclastogenesis suppressed various osteo-clast genes,including Calcr (encoding the calcitonin receptor)and TRAF6,and inhibited osteoclast differentiation.Intravenous delivery of these miRNAs such as miR-141and miR-219reduced osteolytic bone metastasis.Interestingly,the serum levels of two miRNAs miR-16and miR-378,which increased during osteoclas-togenesis,were associated with bone metastasis,suggesting that the miRNAs might serve as useful biomarkers.64These findings emphasize the key roles played by miRNAs in aberrant osteoclastogenesis during bone metastasis.Proximal functions of miRNAs in MDSCsIn tumor-in filtrating immune cells,MDSCs have key roles in immune suppression,in turn allowing tumor progression.48,62,65–68MDSCs are myeloid-related cells characterized by the expression of Gr-1and CD11b markers and the ability to suppress T lymphocyte activation.MDSCs are thought to arise upon deviation of immature myeloid progenitor cells continually generated in bone marrow from the normal differentiation programs toward macrophages,dendritic cells and granulocytes.67,68MDSC num-bers increase in the bone marrow and blood of tumor-bearing mice and cancer patients and accumulate in tumor microenvironments.62,65MDSCs are heterogenous in nature and are mainly divided into Ly6G low /Ly6C high monocytic MDSCs and Ly6G high /Ly6C low granulocytic or polymorphonuclearmiR-214miR-31miR-155CCL5CCL20IL-8miR-320Ets2MMP9PTEN FGFR1FGF2miR-15miR-16CAFCCL5CCL20, IL-8MMP2, MMP9BMP1, LOXL2FGF2Calcr TRAF6miR-141miR-219sICAM1TGF-βmiR-494arginaseMMP2MMP13MMP14SHIP1, PTENmiR-21miR-155STAT3OsteoclastMDSCMicroenvironmentimmunosuppressionbone metastasisMDSC expansiontumor progressionCancer cellCancer cellFigure 3.Proximal functions of miRNAs in tumor stromal cells.Speci fic changes of multiple miRNAs in CAFs (a ),osteoclasts (b )and MDSCs (c )induce phenotypic changes of these cell types,leading to tumor progression.Oncogenic miRNAs and tumor-suppressive miRNAs are represented by black and red,respectively.microRNAs in tumor microenvironment HI Suzuki et al5©2014Macmillan Publishers LimitedOncogene (2014),1–10。

microRNA :一种新型肿瘤分子标志物MicroRNA:A Novel Biomarker of CancerLIANG Dong -yu ,HOU Yan -qiang梁冬雨综述,侯彦强审校(上海交通大学附属第一人民医院松江分院中心实验室,上海201600)摘要:早期诊断、早期治疗是降低肿瘤死亡率的有效途径。
microRNA 是一类内源性的长约18~22个核苷酸的非编码小RNA ,其在肿瘤组织以及循环中的特异表达,为肿瘤的早期诊断带来新的希望。
肿瘤中microRNA 的表达模式不但与肿瘤诊断有关,与肿瘤的分期、进展以及预后也密切相关。
全文就microRNA 在肿瘤早期诊断以及预后判断中的作用作一综述。
主题词:肿瘤;microRNA ;分子标志物中图分类号:R730.4文献标识码:A 文章编号:1671-170X (2011)07-0485-04基金项目:上海市卫生局局级项目(2010104)通讯作者:侯彦强,副主任技师,博士;上海交通大学附属第一人民医院松江分院中心实验室,上海市松江区中山中路746号(201600);E -mail:houyanqiang@ 。
收稿日期:2011-04-14肿瘤标志物是反映肿瘤存在和生长的化学物质,临床上通过对这些化学物质的检测,可以对相应肿瘤作早期诊断、疗效评估和预后判定。
目前,有多种肿瘤标志物应用于临床,但大多数标志物仍然缺乏足够的组织特异性和肿瘤特异性。
microRNA 是一组长约22个核苷酸的非编码小分子RNA ,其通过转录后水平调控参与细胞的增殖、分化、凋亡等生命活动。
近几年的研究发现mi -croRNA 表达具有明显的组织细胞特异性,且与肿瘤的发生和发展有着密切联系。
最近,Lawrie 等[1]在外周血中检测到microRNA 的存在,且发现循环mi -croRNA 的表达与肿瘤也有一定的相关性,这提示microRNA 可作为一种新型的肿瘤标志物。


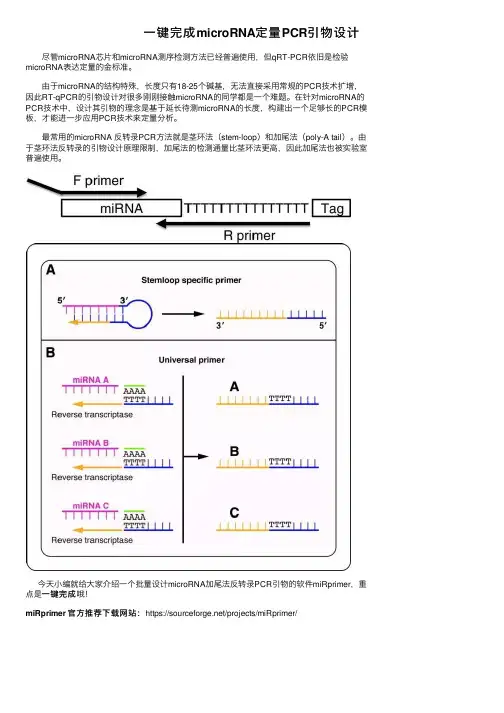
⼀键完成microRNA定量PCR引物设计尽管microRNA芯⽚和microRNA测序检测⽅法已经普遍使⽤,但qRT-PCR依旧是检验microRNA表达定量的⾦标准。
由于microRNA的结构特殊,长度只有18-25个碱基,⽆法直接采⽤常规的PCR技术扩增,因此RT-qPCR的引物设计对很多刚刚接触microRNA的同学都是⼀个难题。
在针对microRNA的PCR技术中,设计其引物的理念是基于延长待测microRNA的长度,构建出⼀个⾜够长的PCR模板,才能进⼀步应⽤PCR技术来定量分析。
最常⽤的microRNA 反转录PCR⽅法就是茎环法(stem-loop)和加尾法(poly-A tail)。
由于茎环法反转录的引物设计原理限制,加尾法的检测通量⽐茎环法更⾼,因此加尾法也被实验室普遍使⽤。
今天⼩编就给⼤家介绍⼀个批量设计microRNA加尾法反转录PCR引物的软件miRprimer,重⼀键完成哦!点是⼀键完成miRprimer 官⽅推荐下载⽹站:https:///projects/miRprimer/⽹站后台⽂件直接下载miRprimer地址:https:///project/miRprimer/miRprimer2_installer.zipmiRprimer2_installer.zip,整个软件的压缩包只有2.68M (2848kb)⼤⼩。
提醒:提醒1. 软件⽀持在Windows XP或更⾼系统中运⾏,还没在苹果电脑Mac OS系统测试(原因是⼩编的钱包羞涩~~)2. 经⼩编测试,最新版本miRprimer顺利运⾏,不需在电脑中安装Ruby脚本环境。
第⼀步miRprimer2_installer.zip压缩包2.68M⼤⼩,下载后解压缩⽣成同名⽂件夹,内有三个⽂件:input_miRs.txt、miRprimer2.exe、README.txt。
特别提⽰:不要更改这些⽂件的⽂件名。
特别提⽰:第⼆步input_miRs.txt,fasta格式储存的是需要设计引物的microRNA名字和序列。
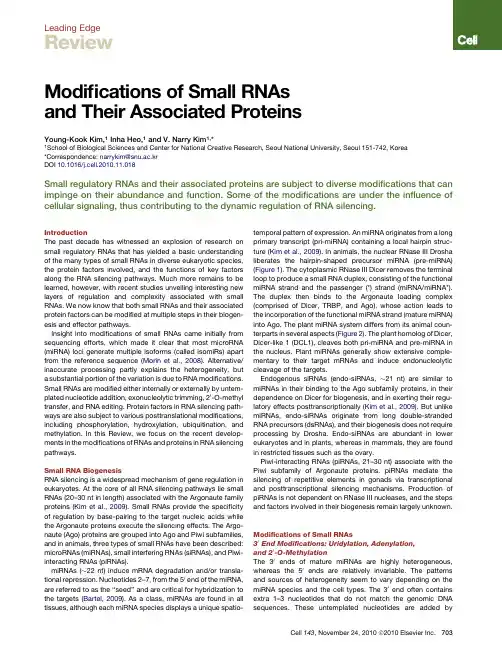
Leading EdgeReviewModifications of Small RNAsand Their Associated ProteinsYoung-Kook Kim,1Inha Heo,1and V.Narry Kim1,*1School of Biological Sciences and Center for National Creative Research,Seoul National University,Seoul151-742,Korea*Correspondence:narrykim@snu.ac.krDOI10.1016/j.cell.2010.11.018Small regulatory RNAs and their associated proteins are subject to diverse modifications that can impinge on their abundance and function.Some of the modifications are under the influence of cellular signaling,thus contributing to the dynamic regulation of RNA silencing.IntroductionThe past decade has witnessed an explosion of research on small regulatory RNAs that has yielded a basic understanding of the many types of small RNAs in diverse eukaryotic species, the protein factors involved,and the functions of key factors along the RNA silencing pathways.Much more remains to be learned,however,with recent studies unveiling interesting new layers of regulation and complexity associated with small RNAs.We now know that both small RNAs and their associated protein factors can be modified at multiple steps in their biogen-esis and effector pathways.Insight into modifications of small RNAs came initially from sequencing efforts,which made it clear that most microRNA (miRNA)loci generate multiple isoforms(called isomiRs)apart from the reference sequence(Morin et al.,2008).Alternative/ inaccurate processing partly explains the heterogeneity,but a substantial portion of the variation is due to RNA modifications. Small RNAs are modified either internally or externally by untem-plated nucleotide addition,exonucleolytic trimming,20-O-methyl transfer,and RNA editing.Protein factors in RNA silencing path-ways are also subject to various posttranslational modifications, including phosphorylation,hydroxylation,ubiquitination,and methylation.In this Review,we focus on the recent develop-ments in the modifications of RNAs and proteins in RNA silencing pathways.Small RNA BiogenesisRNA silencing is a widespread mechanism of gene regulation in eukaryotes.At the core of all RNA silencing pathways lie small RNAs(20–30nt in length)associated with the Argonaute family proteins(Kim et al.,2009).Small RNAs provide the specificity of regulation by base-pairing to the target nucleic acids while the Argonaute proteins execute the silencing effects.The Argo-naute(Ago)proteins are grouped into Ago and Piwi subfamilies, and in animals,three types of small RNAs have been described: microRNAs(miRNAs),small interfering RNAs(siRNAs),and Piwi-interacting RNAs(piRNAs).miRNAs( 22nt)induce mRNA degradation and/or transla-tional repression.Nucleotides2–7,from the50end of the miRNA, are referred to as the‘‘seed’’and are critical for hybridization to the targets(Bartel,2009).As a class,miRNAs are found in all tissues,although each miRNA species displays a unique spatio-temporal pattern of expression.An miRNA originates from a long primary transcript(pri-miRNA)containing a local hairpin struc-ture(Kim et al.,2009).In animals,the nuclear RNase III Drosha liberates the hairpin-shaped precursor miRNA(pre-miRNA) (Figure1).The cytoplasmic RNase III Dicer removes the terminal loop to produce a small RNA duplex,consisting of the functional miRNA strand and the passenger(*)strand(miRNA/miRNA*). The duplex then binds to the Argonaute loading complex (comprised of Dicer,TRBP,and Ago),whose action leads to the incorporation of the functional miRNA strand(mature miRNA) into Ago.The plant miRNA system differs from its animal coun-terparts in several aspects(Figure2).The plant homolog of Dicer, Dicer-like1(DCL1),cleaves both pri-miRNA and pre-miRNA in the nucleus.Plant miRNAs generally show extensive comple-mentary to their target mRNAs and induce endonucleolytic cleavage of the targets.Endogenous siRNAs(endo-siRNAs, 21nt)are similar to miRNAs in their binding to the Ago subfamily proteins,in their dependence on Dicer for biogenesis,and in exerting their regu-latory effects posttranscriptionally(Kim et al.,2009).But unlike miRNAs,endo-siRNAs originate from long double-stranded RNA precursors(dsRNAs),and their biogenesis does not require processing by Drosha.Endo-siRNAs are abundant in lower eukaryotes and in plants,whereas in mammals,they are found in restricted tissues such as the ovary.Piwi-interacting RNAs(piRNAs,21–30nt)associate with the Piwi subfamily of Argonaute proteins.piRNAs mediate the silencing of repetitive elements in gonads via transcriptional and posttranscriptional silencing mechanisms.Production of piRNAs is not dependent on RNase III nucleases,and the steps and factors involved in their biogenesis remain largely unknown. Modifications of Small RNAs30End Modifications:Uridylation,Adenylation,and20-O-MethylationThe30ends of mature miRNAs are highly heterogeneous, whereas the50ends are relatively invariable.The patterns and sources of heterogeneity seem to vary depending on the miRNA species and the cell types.The30end often contains extra1–3nucleotides that do not match the genomic DNA sequences.These untemplated nucleotides are added by Cell143,November24,2010ª2010Elsevier Inc.703terminal nucleotidyl transferases that preferentially introduce uridyl or adenyl residues to the 30terminus of RNA.The first indication of 30end modification of small RNA came from a hen1mutant of Arabidopsis (Li et al.,2005).HEN1is a methyl transferase that adds a methyl group to the 20-OH at the 30end of RNA (Yu et al.,2005).In hen1mutants,miRNAs are reduced in abundance and become heterogeneous in size due to uridylation at the 30end.Because U tailing correlates with the exonucleolytic degradation of mRNAs (Shen and Goodman,2004),it was postulated that uridylation induces degradation of plant miRNAs and that the 20-O-methyl moiety is required to protect small RNAs from uridylation and decay (see below).Consistent with this notion,in green algae Chlamy-domonas ,a nucleotidyl transferase,MUT68,uridylates the 30end of small RNA,and the RRP6exosome subunit facilitates small RNA decay in a manner dependent on MUT68in vitro (Ibra-him et al.,2010).Deletion of MUT68results in elevated miRNA and siRNA levels,indicating that MUT68and RRP6collaborate in the turnover of mature small RNAs in plants.Similar links between 20-O-methylation,uridylation,and decay appear to exist in animals.A recent study on the zebrafish Hen1homolog shows that piRNAs are uridylated and adenylated andthat piRNA levels are reduced in hen1mutant germ cells (Kam-minga et al.,2010).In flies and mice,piRNAs are methylated by HEN1orthologs,but the connection to stability control remains unclear (Horwich et al.,2007;Kirino and Mourelatos,2007;Ohara et al.,2007;Saito et al.,2007).In flies,dAgo2-bound RNAs (mostly siRNAs)are protected by 20-O-methylation from being uridylated/adenylated,which in turn induces 30exonucleolytic trimming (Ameres et al.,2010).In nematode worms,the role of 20-O-methylation has yet to be determined.However,a subset of endo-siRNAs associated with an Ago homolog CSR-1is uridylated at the 30end,and the uridyl trans-ferase CDE-1(also known as CID-1or PUP-1)negatively regu-lates these siRNAs,indicating that uridylation serves as a trigger for decay (van Wolfswinkel et al.,2009).Although mature miRNAs lack methylation in animals,uridyla-tion plays a significant role in the control of miRNA biogenesis.In mammalian embryonic stem cells,let-7biogenesis is sup-pressed by the Lin28protein that binds to the terminal loop of the let-7precursors (Heo et al.,2008;Newman et al.,2008;Rybak et al.,2008;Viswanathan et al.,2008).Of interest,Lin28induces 30uridylation of pre-let-7by recruiting the terminal nucleotidyl transferase TUT4(also known as ZCCHC11)(HaganFigure 1.Modifications in the AnimalMicroRNA Pathway(Left)MicroRNAs (miRNAs)are subject to diverse modifications.Pri-miRNAs are edited by ADARs,which convert adenosine to inosine (I).RNA editing inhibits processing and/or alters target specificity.Pre-let-7is regulated through uridylation.Lin28recognizes pre-let-7and,in turn,recruits a nucleo-tidyl transferase TUT4(mammal)or PUP-2(worms),which adds an oligo-uridine tail at the 30end of RNA.The uridylated pre-miRNA is resistant to Dicer processing and subject to decay.TUT4also uridylates mature miRNA (miR-26),which reduces miRNA activity.Another nucleotidyl trans-ferase GLD-2adenylates mature miRNAs,which reduces the activity of miRNA and/or increases the stability of specific miRNAs (such as miR-122).(Bottom)Mature miRNAs are degraded through several mechanisms.In worms,a 50/30exonu-clease XRN-2degrades miRNAs that are released from Ago.In flies and humans,extensive pairing between miRNA/siRNA and target RNA triggers tailing as well as 30/50trimming of miRNA/siRNA.(Right)Protein factors,which are involved in the miRNA pathway,are also subject to various post-translational modifications.Human Drosha is phosphorylated at two serine residues,S300/S302,by an unknown kinase.Phosphorylation localizes Drosha to the nucleus,where the pri-miRNA processing occurs.MAP kinases Erk1/2phosphorylate human TRBP at S142,S152,S283,and S286,which increases the protein stability of TRBP and Dicer.Ago2is regulated by multiple modifications.A prolyl hydroxylase C-P4H(I)hydroxylates P700in human Ago2,which enhances stability of Ago2and increases P body localization.Phosphorylation of human Ago2at S387by MAPKAPK2,which is induced by p38pathway,also promotes P body localization of Ago2.However,the biological significance of P body localization of Ago2remains unclear.In mice,a stem cell-specific E3ligase,mLin41,ubiq-uitinates Ago2and targets it for proteosome-dependent degradation.704Cell 143,November 24,2010ª2010Elsevier Inc.et al.,2009;Heo et al.,2009).The oligo U-tail added by TUT4blocks Dicer processing and facilitates the decay of pre-let-7.The homologs of TUT4may have related functions in other organisms.In nematode worms,PUP-2uridylates pre-let-7in vitro and suppresses the let-7function in vivo (Lehrbach et al.,2009).Let-7is unlikely to be the only miRNA uridylated at the pre-miRNA level.In support of this notion,untemplated 30uridine is frequently found in other mature miRNAs originating from the 30arm of pre-miRNAs (but significantly less frequently in those from the 50arm)(Burroughs et al.,2010;Chiang et al.,2010).Because untemplated uridylation is observed in cells lacking Lin28,it will be interesting to determine which pre-miRNAs other than pre-let-7are controlled by uridylation and to identify addi-tional factors required for pre-miRNA uridylation.Although uridylation is generally thought to induce the decay of small RNAs,adenylation may have the opposite conse-quence.In cottonwood P.trichoacarpa ,many miRNA families are adenylated at their 30ends,and adenylation prevents miRNA degradation in in vitro decay assay (Lu et al.,2009).In the case of mammalian miR-122,which is adenylated by cytoplasmic poly (A)polymerase GLD-2(or TUTase2),30end adenylation is also implicated in its stabilization (Katoh et al.,2009).In the liver of Gld-2knockout mice,the steady-state level of mature miR-122is reduced,and the abundance of target mRNAs of miR-122increases.However,a recent study indicates that GLD-2adenylates most miRNAs,and the adenylation may affect their activity rather than stability (Burroughs et al.,2010).Deep sequencing of Ago-associated small RNAs shows that adenylated miRNAs are relatively depleted in the Ago2and Ago3complexes,suggesting that adenylation may interfere with Ago loading.Similarly,it has been reported that uridylation of mature miR-26by TUT4results in the reduction of miR-26’s activity without altering the miRNA levels (Jones et al.,2009).Therefore,it remains an inter-esting but yet unresolved issue whether or not uridylation/adenylation affects the stability of miRNAs in animals.One may speculate that 30modified miRNAs enter the silencing complex with altered frequencies,which in turn affects the small RNA’s sensitivity to nucleases.Further examination is needed to iden-tify the players involved in these processes,particularly the nucleases that recognize a U/A tail,and to dissect their action mechanisms.miRNA DecaySeveral nucleases degrade small RNAs (Figures 1and 2).An Arabidopsis enzyme SDN1(small RNA degrading nuclease,a 30-to-50exonuclease)degrades single-stranded miRNAs in vitro (Ramachandran and Chen,2008).miRNAs accumulate in a mutant lacking SDN1and its related nucleases SDN2and SDN3,indicating that the SDN proteins may act redundantly to degrade plant miRNAs.The 20-O-methyl group at the 30end of miRNAs,which is a general feature of plant miRNAs,has a protective effect against SDN1in in vitro assays.Of note,uridy-lation causes a small but detectable protective effect in the same in vitro assay,indicating that SDN1is unlikely to be the nuclease responsible for U-tail-promoted degradation.Given that RRP6(a 30-to-50exonuclease)facilitates decay of small RNAs in a MUT68-dependent manner in Chlamydomonas extracts,multi-ple enzymes may be involved in small RNA decay in plants,playing partially overlapping but differential roles (Ibrahim et al.,2010).In C.elegans ,XRN-2(a 50-to-30exonuclease)is involved in the degradation of mature miRNAs (Chatterjee and Grosshans,2009).Because miRNAs are tightly bound to and protected by Ago,it is unclear how XRN-2accesses the 50end of an miRNA for decay.Of interest,larval lysate promotes efficient release of miRNA in vitro,implicating an as yet unknown factor that assists the release of miRNA from the otherwise tightly associated Argo-naute protein (Chatterjee and Grosshans,2009).In Arabidopsis ,two XRN-2homologs,XRN2and XRN3,degrade the loop of miRNA precursor following processing,but they do not affect mature miRNA levels (Gy et al.,2007).In mammals,a general nuclease for miRNAs has yet to be identified.Knockdown of XRN-1or an exosome subunit in human cells results in only partial upregulation of miR-382,and XRN-2depletion does not have a significant effect (Bail et al.,2010).Thus,it awaits further investigation whether or not there is one major conserved pathway for miRNA decay in mammals.There have been intriguing reports of regulated decay of miRNAs.For instance,miR-29b is degraded in dividing cells more rapidly than in mitotically arrested cells (Hwang et al.,2007).In the central nervous system of Aplysia ,the levels of miR-124and miR-184decrease in 1hr after treatment with the neurotransmitter serotonin (Rajasethupathy et al.,2009).Figure 2.RNA Modifications in the Plant miRNA PathwayIn plants,both pri-miRNA and pre-miRNA are cleaved by DCL1/HYL1complex.After cleavage,30ends of miRNA duplex are 20-O-methylated by a methyl transferase HEN1.The methylation protects miRNAs from uridylation and exonucleolytic degradation.In the green algae Chlamydomonas ,the nu-cleotidyl transferase MUT68attaches uridine residues at the 30end of mature miRNA lacking a methyl group.Then,the RRP6exosome subunit,a 30-to-50exonuclease,degrades the uridylated miRNAs.In Arabidopsis ,a 30/50exonuclease SDN1is reported to degrade mature miRNAs.Cell 143,November 24,2010ª2010Elsevier Inc.705Because U0126,an inhibitor of mitogen-activated protein kinase (MAPK),blocks the reduction of miR-124,the decay process may be dependent on the MAPK pathway.Of interest,a study on mammalian neuronal cells shows that most miRNAs turn over more rapidly in neurons than in other cell types (Krol et al.,2010).Neuronal activation accelerates decay of the miRNAs,whereas blocking neuronal activity stabilizes the miRNAs.It will be exciting to discover the nuclease(s)and the upstream signals for miRNA degradation in these systems.Recently it has been shown that a polynucleotide phosphory-lase (PNPase,a type I interferon-inducible 30-to-50exonuclease)binds specifically to several miRNAs (miR-221,miR-222,and miR-106b)and induces rapid turnover in a human melanoma cell line (Das et al.,2010).Because there is no apparent commonality in terms of the sequences,it is unclear how PNPase recognizes the miRNAs specifically.As mentioned above,there is substantial evidence linking uri-dylation/adenylation and exonucleolytic attack on small RNAs.A recent study provides evidence that extensive complemen-tarity between a small RNA and its target RNA triggers uridyl/adenyl tailing as well as 30/50trimming in flies and humans (Figure 1)(Ameres et al.,2010).Animal small RNAs with high complementarity to the targets,such as piRNAs and fly endo-siRNAs,appear to be generally protected by 20-O-methylation at the 30end like plant small RNAs.It has been postulated that animal miRNAs,which do not carry methylation,maintain only partial complementarity with their targets so as to avoid tailing and trimming of miRNAs.Of note,viruses seem to exploit a related miRNA decay pathway to invade host cells more effec-tively.Herpesvirus saimiri ,a family of primate-infecting herpesvi-ruses,expresses viral noncoding RNAs called HSURs (H.saimiri U-rich RNAs).A recent report reveals that HSURs rapidly down-regulate host miR-27and that base-pairing between HSUR and miR-27is required for the degradation (Cazalla et al.,2010).These discoveries imply an additional layer of stability controlof small RNAs,which is influenced by the interaction with the target RNA.miRNA EditingAdenosine deaminases acting on RNAs (ADARs)convert adenosine to inosine on the dsRNA region of small RNA precur-sors (Figure 1and Figure 3A).Because inosine (I)pairs with cytosine instead of uridine,such edits could alter the structure of small RNA precursor,thereby interfering with processing.For instance,editing of pri-miR-142by ADAR1and ADAR2suppresses Drosha processing (Yang et al.,2006),whereas that of pre-miR-151by ADAR1interferes with Dicer processing (Kawahara et al.,2007a ).Because hyperedited dsRNAs can be targeted by the nuclease Tudor-SN,RNA editing may also desta-bilize small RNA precursors (Scadden,2005).In rare cases,RNA editing occurs in the seed sequence of miRNA,changing the targeting specificity.In the brain,where ADAR is abundant,miR-376cluster miRNAs are frequently edited in the seed region and are redirected to repress a different set of mRNAs (Kawahara et al.,2007b ).High-throughput sequencing of the fly endo-siRNA pool also reveals evidence for RNA editing (Kawamura et al.,2008).The precursors of endo-siRNAs (long hairpins and sense-antisense pairs)may be targeted by ADARs,although the functional significance of this siRNA modification is unknown.Posttranslational Protein Modifications Phosphorylation of RNase III EnzymesHuman Dicer interacts with two related dsRNA-binding proteins,TRBP and PACT.Although they do not influence Dicer process-ing itself,TRBP and PACT stabilize Dicer and may also function in RISC assembly (Chendrimada et al.,2005;Haase et al.,2005;Lee et al.,2006).A recent study indicates that four serine residues of human TRBP (S142,S152,S283,and S286)are phosphorylated by the MAP kinase Erk,which controls cell proliferation,survival,and differentiation (Figure 1)(Paroo etal.,Figure 3.Modifications in the Endo-siRNA and piRNA Pathways(A)Endogenous small interfering RNAs (endo-siRNAs)are processed from long dsRNAs in a Dicer-dependent manner and are loaded onto Ago proteins.High-throughput sequencing data show that the adenosine-to-inosine (I)editing occurs in fly endo-siRNAs,likely by ADAR,although the role of RNA editing is unknown.Fly endo-siRNAs bound to dAgo2are 20-O-methyl-ated by HEN1homolog,which protects RNAs from uridyl/adenyl tailing and degradation.In worms,a subset of endo-siRNAs,which are asso-ciated with an Ago homolog CSR-1,is uridylated at the 30end by the nucleotidyl transferase CDE-1.(B)piRNAs are generated from single-stranded RNA precursors that are processed by primary processing and/or secondary processing (ping-pong amplification cycle).piRNAs are associated with Piwi subfamily proteins (PIWI).Animal piRNAs are 20-O-methylated by HEN1orthologs.In zebra-fish,depletion of hen1induces uridylation of piRNAs and facilitates decay,suggesting that methylation stabilizes piRNAs.However,the phys-iological significance of piRNA methylation in fliesand mammals remains unclear.PIWI proteins are methylated at arginine residues (sDMA,symmetrical dimethyl arginine)at their N termini by orthologs of the methyl transferase PRMT5.In flies and mice,TDRD proteins interact with PIWI proteins through sDMA and may play important roles in piRNA metabolism.706Cell 143,November 24,2010ª2010Elsevier Inc.2009).Phosphorylation enhances protein stability of TRBP, consequently elevating Dicer protein levels.Intriguingly,TRBP phosphorylation preferentially increases growth-promoting miRNAs such as miR-17,whereas tumor-suppressive let-7is reduced.The mechanism of selective downregulation of let-7 is unclear,but it may be an indirect effect.An interesting implica-tion of thesefindings is that the MAPK/Erk pathway exerts its effects,in part,by regulating miRNA biogenesis.Drosha,a nuclear enzyme for pri-miRNA processing(Lee et al.,2003),has recently been shown to be a direct target of posttranslational modification(Tang et al.,2010).Mass spec-trometry and mutagenesis studies reveal that human Drosha is phosphorylated at serine300(S300)and serine302(S302) (Figure1).Phosphorylation of these residues is essential for the nuclear localization of Drosha and is required for pri-miRNA processing.Because both endogenous and overexpressed Drosha localize to the nucleus constitutively,it is unclear whether or not the phosphorylation at S300/S302is a regulated process. Understanding the physiological significance of this regulation will require the identification of the kinase that phosphorylates Drosha.Argonaute2Is a Target of Multiple ModificationsAgo2is subject to multiple posttranslational modifications (Figure1).Human Ago2binds to the type I collagen prolyl-4-hydroxylase(C-P4H(I))that hydroxylates Ago2at proline700 (Qi et al.,2008).Depletion of C-P4H(I)reduces the stability of the Ago2protein and,accordingly,downregulates siRNA-medi-ated silencing.Furthermore,hydroxylation is required for Ago2 localization to the processing body(P body),a cytoplasmic granule that is thought to be a site for RNA storage and degrada-tion.P body localization of Ago2is also enhanced by phosphor-ylation at serine387,which is mediated by the p38MAPK pathway(Zeng et al.,2008).However,given the controversy over the direct role of P body in small RNA-mediated silencing, the biological significance of P body localization of Ago2remains unclear.Ubiquitination also plays a part in the control of Ago2.Mouse Lin41(mLin41or Trim71),a stem cell-specific Trim-NHL protein, inhibits the miRNA pathway(Rybak et al.,2009).As an E3ubiq-uitin ligase,mLin41ubiquitinates Ago2and targets it for protea-some-dependent degradation.Of interest,mLin41is a target of let-7miRNA,suggesting that mLin41and let-7may be engaged in a reciprocal negative feedback loop.Recently,other Trim-NHL proteins have been reported to associate with the Argonaute proteins and affect miRNA pathway.Mei-P26(fly)inhibits miRNA biogenesis,whereas TRIM32(mouse)and NHL-2(worm)acti-vate the miRNA pathway(Hammell et al.,2009;Neumu¨ller et al.,2008;Schwamborn et al.,2009).Their mechanism of action appears to be different than that of mLin41because the E3ligase activity of Mei-P26and TRIM32is dispensable for their effects and because NHL-2enhances miRNA activity without a change in miRNA levels.Tudor Regulates PIWI ProteinsThe PIWI(P element-induced wimpy testis)clade proteins bind to Piwi-interacting RNAs(piRNAs)and silence transposable elements in gonads.Mouse has three PIWI homologs(MILI, MIWI,and MIWI2),and there are three PIWI proteins inflies (Aubergine[Aub],AGO3,and Piwi)(Kim et al.,2009).Recent studies have revealed that PIWI proteins carry symmetrical dimethyl arginine(sDMA)at their N termini.Arginine methylation of PIWI is mediated by a methyl transferase PRMT5(dPRMT5/ capsuleen[csul]/dart5in Drosophila)(Figure3B)(Heo and Kim, 2009;Siomi et al.,2010).sDMA is recognized by Tudor domain-containing proteins(TDRDs),which are critical for germ-line development.In bothflies and mice,deletion of TDRDs alters piRNA abundance and/or composition,indicating that TDRDs play important roles in the piRNA metabolism through specific binding to the sDMAs of PIWI proteins.How TDRDs act in the piRNA pathway at a molecular level awaits further investigation. PerspectivesAs we delve deeper and wider into the small RNA world,the emerging landscape becomes ever more complex on both the RNA and protein sides.High-throughput analyses have uncov-ered a considerable heterogeneity in small RNA populations. Some isomiRs are expressed differentially in certain tissues, suggesting that these variations may be associated with specific regulatory functions(Chiang et al.,2010).Biochemical and genetic studies also provide substantial evidence for the regula-tory roles of the modifications discussed in this Review.Thus,it is likely that at least some of the observed heterogeneity reflects multiple layers of regulation.We should be cautious,however,in extrapolating the current evidence because it is unclear how much fraction of the small RNA and protein modifications trans-late into functional consequences and whether certain modifica-tions simply reflect the noise of RNA metabolism.In addition to the functionality issue,a number of key questions remain to be answered.Are there conserved pathways and enzymes for RNA and protein modifications?If so,what are the similarities and differences?20-O-methylation is applied to many small RNA pathways,but the details differ significantly in different systems.For instance,plant HEN1acts on dsRNA duplexes,whereas animal HEN1homologs methylate ssRNA loaded on Argonaute proteins.Uridylation/adenylation is carried out by a family of ribonucleotidyl transferases.How each member selectively recognizes its substrates is largely unknown. RNA stability is likely to play important roles in RNA silencing pathways.Decay pathways of small RNA are beginning to be unraveled,but there is no consensus between different species as yet.One possibility is that multiple enzymes act in parallel as in the mRNA decay pathway,which involves several30exonucle-ases,50exonucleases,and endonucleases.Some of the decay enzymes may function redundantly,and it remains one of the major challenges in thefield to identify them.Protein modifica-tion is also emerging as one of the key regulatory layers. Outstanding questions include which enzymes are involved, what the in vivo significance of such modifications is,and whether the protein modifications are developmentally regu-lated.Future studies will reveal new types of modifications,addi-tional regulatory factors,and their biological relevance.The RNA silencing machinery should respond accurately to developmental and environmental cues.Most signaling path-ways are thought to be connected to RNA silencing,but we are just beginning to understand the molecular links between RNA silencing and cell signaling.What the upstream signals are,how certain RNAs and proteins get specifically recognized, Cell143,November24,2010ª2010Elsevier Inc.707and what the downstream effects of the modifications are await elucidation.We also need to understand the interplay between different modifications.There appears to be a crosstalk between certain modifications of RNA(such as methylation,uridylation, and decay),which may influence their fate and function.It is likely that there is a crosstalk between the different posttranslational modifications in the proteins involved in the biogenesis and effector functions of small RNA silencing pathways.Under-standing these networks will undoubtedly provide ample oppor-tunities to manipulate RNA silencing and will reveal new lessons about gene regulation.ACKNOWLEDGMENTSWe thank members of V.N.K.’s laboratory for helpful discussions and comments.This work was supported by the Creative Research Initiatives Program(20100000021)and the National Honor Scientist Program (20100020415)through the National Research Foundation of Korea(NRF) and the BK21Research Fellowships(I.H.)from the Ministry of Education, Science and Technology of Korea.We apologize to authors whose work has not been covered because of space limitations.REFERENCESAmeres,S.L.,Horwich,M.D.,Hung,J.H.,Xu,J.,Ghildiyal,M.,Weng,Z.,and Zamore,P.D.(2010).Target RNA-directed trimming and tailing of small silencing RNAs.Science328,1534–1539.Bail,S.,Swerdel,M.,Liu,H.,Jiao,X.,Goff,L.A.,Hart,R.P.,and Kiledjian,M. (2010).Differential regulation of microRNA stability.RNA16,1032–1039.Bartel,D.P.(2009).MicroRNAs:target recognition and regulatory functions. Cell136,215–233.Burroughs,A.M.,Ando,Y.,de Hoon,M.J.,Tomaru,Y.,Nishibu,T.,Ukekawa, R.,Funakoshi,T.,Kurokawa,T.,Suzuki,H.,Hayashizaki,Y.,and Daub,C.O. (2010).A comprehensive survey of30animal miRNA modification events and a possible role for30adenylation in modulating miRNA targeting effectiveness. Genome Res.20,1398–1410.Cazalla,D.,Yario,T.,Steitz,J.A.,and Steitz,J.(2010).Down-regulation of a host microRNA by a Herpesvirus saimiri noncoding RNA.Science328, 1563–1566.Chatterjee,S.,and Grosshans,H.(2009).Active turnover modulates mature microRNA activity in Caenorhabditis elegans.Nature461,546–549.Chendrimada,T.P.,Gregory,R.I.,Kumaraswamy,E.,Norman,J.,Cooch,N., Nishikura,K.,and Shiekhattar,R.(2005).TRBP recruits the Dicer complex to Ago2for microRNA processing and gene silencing.Nature436,740–744.Chiang,H.R.,Schoenfeld,L.W.,Ruby,J.G.,Auyeung,V.C.,Spies,N.,Baek, D.,Johnston,W.K.,Russ,C.,Luo,S.,Babiarz,J.E.,et al.(2010).Mammalian microRNAs:experimental evaluation of novel and previously annotated genes. Genes Dev.24,992–1009.Das,S.K.,Sokhi,U.K.,Bhutia,S.K.,Azab,B.,Su,Z.Z.,Sarkar,D.,and Fisher, P.B.(2010).Human polynucleotide phosphorylase selectively and preferen-tially degrades microRNA-221in human melanoma cells.Proc.Natl.Acad. A107,11948–11953.Gy,I.,Gasciolli,V.,Lauressergues,D.,Morel,J.B.,Gombert,J.,Proux,F., Proux, C.,Vaucheret,H.,and Mallory, A.C.(2007).Arabidopsis FIERY1, XRN2,and XRN3are endogenous RNA silencing suppressors.Plant Cell19, 3451–3461.Haase,A.D.,Jaskiewicz,L.,Zhang,H.,Laine´,S.,Sack,R.,Gatignol,A.,and Filipowicz,W.(2005).TRBP,a regulator of cellular PKR and HIV-1virus expression,interacts with Dicer and functions in RNA silencing.EMBO Rep. 6,961–967.Hagan,J.P.,Piskounova,E.,and Gregory,R.I.(2009).Lin28recruits the TUTase Zcchc11to inhibit let-7maturation in mouse embryonic stem cells. Nat.Struct.Mol.Biol.16,1021–1025.Hammell,C.M.,Lubin,I.,Boag,P.R.,Blackwell,T.K.,and Ambros,V.(2009). nhl-2Modulates microRNA activity in Caenorhabditis elegans.Cell136, 926–938.Heo,I.,and Kim,V.N.(2009).Regulating the regulators:posttranslational modifications of RNA silencing factors.Cell139,28–31.Heo,I.,Joo,C.,Cho,J.,Ha,M.,Han,J.,and Kim,V.N.(2008).Lin28mediates the terminal uridylation of let-7precursor MicroRNA.Mol.Cell32,276–284.Heo,I.,Joo,C.,Kim,Y.K.,Ha,M.,Yoon,M.J.,Cho,J.,Yeom,K.H.,Han,J., and Kim,V.N.(2009).TUT4in concert with Lin28suppresses microRNA biogenesis through pre-microRNA uridylation.Cell138,696–708.Horwich,M.D.,Li,C.,Matranga,C.,Vagin,V.,Farley,G.,Wang,P.,and Zamore,P.D.(2007).The Drosophila RNA methyltransferase,DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC.Curr.Biol. 17,1265–1272.Hwang,H.W.,Wentzel,E.A.,and Mendell,J.T.(2007).A hexanucleotide element directs microRNA nuclear import.Science315,97–100.Ibrahim,F.,Rymarquis,L.A.,Kim,E.J.,Becker,J.,Balassa,E.,Green,P.J., and Cerutti,H.(2010).Uridylation of mature miRNAs and siRNAs by the MUT68nucleotidyltransferase promotes their degradation in A107,3906–3911.Jones,M.R.,Quinton,L.J.,Blahna,M.T.,Neilson,J.R.,Fu,S.,Ivanov,A.R., Wolf,D.A.,and Mizgerd,J.P.(2009).Zcchc11-dependent uridylation of micro-RNA directs cytokine expression.Nat.Cell Biol.11,1157–1163.Kamminga,L.M.,Luteijn,M.J.,den Broeder,M.J.,Redl,S.,Kaaij,L.J., Roovers,E.F.,Ladurner,P.,Berezikov,E.,and Ketting,R.F.(2010).Hen1is required for oocyte development and piRNA stability in zebrafish.EMBO J. 29,3688–3700.Katoh,T.,Sakaguchi,Y.,Miyauchi,K.,Suzuki,T.,Kashiwabara,S.,Baba,T., and Suzuki,T.(2009).Selective stabilization of mammalian microRNAs by30 adenylation mediated by the cytoplasmic poly(A)polymerase GLD-2.Genes Dev.23,433–438.Kawahara,Y.,Zinshteyn,B.,Chendrimada,T.P.,Shiekhattar,R.,and Nishi-kura,K.(2007a).RNA editing of the microRNA-151precursor blocks cleavage by the Dicer-TRBP complex.EMBO Rep.8,763–769.Kawahara,Y.,Zinshteyn,B.,Sethupathy,P.,Iizasa,H.,Hatzigeorgiou,A.G., and Nishikura,K.(2007b).Redirection of silencing targets by adenosine-to-inosine editing of miRNAs.Science315,1137–1140.Kawamura,Y.,Saito,K.,Kin,T.,Ono,Y.,Asai,K.,Sunohara,T.,Okada,T.N., Siomi,M.C.,and Siomi,H.(2008).Drosophila endogenous small RNAs bind to Argonaute2in somatic cells.Nature453,793–797.Kim,V.N.,Han,J.,and Siomi,M.C.(2009).Biogenesis of small RNAs in animals.Nat.Rev.Mol.Cell Biol.10,126–139.Kirino,Y.,and Mourelatos,Z.(2007).The mouse homolog of HEN1is a poten-tial methylase for Piwi-interacting RNAs.RNA13,1397–1401.Krol,J.,Busskamp,V.,Markiewicz,I.,Stadler,M.B.,Ribi,S.,Richter,J., Duebel,J.,Bicker,S.,Fehling,H.J.,Schu¨beler,D.,et al.(2010).Characterizing light-regulated retinal microRNAs reveals rapid turnover as a common property of neuronal microRNAs.Cell141,618–631.Lee,Y.,Ahn,C.,Han,J.,Choi,H.,Kim,J.,Yim,J.,Lee,J.,Provost,P., Ra˚dmark,O.,Kim,S.,and Kim,V.N.(2003).The nuclear RNase III Drosha initi-ates microRNA processing.Nature425,415–419.Lee,Y.,Hur,I.,Park,S.Y.,Kim,Y.K.,Suh,M.R.,and Kim,V.N.(2006).The role of PACT in the RNA silencing pathway.EMBO J.25,522–532.Lehrbach,N.J.,Armisen,J.,Lightfoot,H.L.,Murfitt,K.J.,Bugaut,A.,Balasu-bramanian,S.,and Miska,E.A.(2009).LIN-28and the poly(U)polymerase PUP-2regulate let-7microRNA processing in Caenorhabditis elegans.Nat. Struct.Mol.Biol.16,1016–1020.708Cell143,November24,2010ª2010Elsevier Inc.。


Cell:哺乳动物microRNA表达图谱-生物研究-生物谷生物谷报道:在6月29日著名国际杂志Cell上发表的一篇文章中,发布了一张microRNA表达图谱,该结果显示,哺乳动物为数众多的microRNA只是起源于少数几个microRNA基因,而且组织或细胞特异的microRNA很少。
该图谱对来自人类和啮齿类的26种正常、肿瘤组织的细胞的microRNA表达进行了定量测定。
“他们掌握了一种极为有用的资源,”Dartmouth医学院Victor Ambros说(未参与实验),他们在研究中收集的组织种类非常多,并对这些组织中的microRNA的特征进行了系统性分析。
新绘制的microRNA表达图谱含有许多关于microRNA结构、序列、表达方式和进化的有用信息。
Ambros认为这是一个基本数据库,为研究发育和疾病有关的microRNA世界提供了重要线索。
microRNA是动物、植物和病毒中普遍存在的非编码小RNA,通过干扰mRNA的功能调节基因表达。
科研人员已经鉴定出多种哺乳动物microRNA,但对这些microRNA中大多数的表达水平和特征都是未知的。
洛克菲勒大学Pablo Landgraf率领的研究小组克隆并测序了256个哺乳动物小RNA数据库中的30多万个序列,找到了大约400个在任何组织都表达的microRNA——比之前预期的要少。
约翰霍普金斯大学Joshua Mendell说,这个结果提示在其它研究中找到的几个百microRNA是未知的。
Landgraf等还发现他们测序的microRNA克隆中,97%的来自不到300个microRNA。
文章共同作者、瑞士Basel大学Mihaela Zavolan认为许多microRNA在各种组织间是低背景水平表达的(low background level),现在不能确定这些“痕量”microRNA的功能,但很有可能大多数microRNA的功能是不相关的。

m ic ro RNA的生物形成参与细胞生长/分化和肿瘤形成徐广峰,付海龙△(综述),赵亚萍※(审校)(解放军第82医院检验科,江苏淮安223001)中图分类号:R318 文献标识码:A 文章编号:1006-2084(2012)03-0368-03摘要:micr oRNA(miRNA)作为一类非编码小分子调控RNAs,参与调节多种生理和病理过程。
在动物细胞中,miRNA基因的转录初产物pri-miRNA被RNaseⅢ家族酶成员Drosa和DGCR8催化加工成为m iRNA前体(pr e-miRNA),然后由细胞核转运至细胞质中,经另一种核糖核酸酶ⅢDicer识别剪切为成熟m iRNA。
在pri-miRNA向成熟miRNA加工转化过程中发生的改变参与细胞生长、分化以及肿瘤形成。
关键词:microRNA生物形成;细胞生长;细胞分化;肿瘤形成;调节MicroRNA Bio genes is Participating in Cell G ro wth,Different iat ion and Tumorigenesis XU Guang-feng1,FU Hai-long2,ZH AO Ya-ping1.(Department Laboratory,the82nd Hos pital of the People′s L iberation Ar my,H uaian223001,China)Abst rac t:Micr oRNA(miRNA),as a kind of noncoding,small,regulatory RNAs,par ticipates in r egulatinga variety of physiolog ical and pathological process.In anim al cells,prim ary tra nscript of miRNA gene(pr i-miR-NA)is processed by RNaseⅢfamily members Drosa and DGCR8to produce the miRNA pr ecursor(pre-miR-NA).S ubsequently,the pre-miRNA is tr ansported from the nucleus to the cytoplasm,w her e it is shar ed by a second RNaseⅢ-like enzyme,ter med Dicer,into mature miRNA.The changes that take place in miRNA pro-cessing ar e involved in cell g rowth,differ entiation and tumor igenesis.Key words:MicroRNA biog enesis;Cell g rowth;Cell differentia tion;Tumorigenesis;Regulationmicr oRN A(miRN A)是真核生物中一类参与基因转录后水平调控的非编码小分子RNAs。


microRNA的基因组学研究技术与应用北京基因组研究所米双利研究员2011-11-9microRNA•microRNA(miRNA)是一类广泛存在于生物体中的非编码、小分子、单链RNA,大约含有18-25nt,能够结合在靶基因3’UTR 区,通过降解mRNA、影响mRNA的稳定性或抑制蛋白合成,在转录后水平上调控靶基因的表达。
小分子非编码RNA•Small noncoding RNA,包括:–miRNA(microRNA);–siRNA(small interfering RNA);–piRNA(piwi-interacting RNA);–esiRNA(Endoribonuclease-prepared siRNAs)–等等miRNA的发现RNAi•1998年2月,华盛顿卡耐基研究院的Andrew Fire 和马萨诸塞大学癌症中心的Craig Mello发现RNAi 现象。
2006年获得诺贝尔医学/生理学奖。
miRNA的发现线虫秀丽隐杆线虫Caenorhabditis elegans,3d L1-L4, 2-3w, 2n=12, genome 8x107bp, 13500 genes. 1965, 1998 sequencing,2002Sydney Brenner, John Sulston, H. Robert HorvitzmiRNA的发现•1993年,哈佛大学Rosalind C. Lee 、Rhonda L. Feinbaum和Victor Ambros等人发现在线虫体内存在一种RNA(lin-4),不编码蛋白,但可以生成一对小的RNA转录本,每一个转录本能在翻译水平通过抑制核蛋白lin-14的表达而调节了线虫的幼虫发育进程,在第一幼虫阶段的末期降低lin-14的表达将启动发育进程进入第二幼虫阶段。
•lin-14的mRNA的3’UTR区独特的重复序列和lin-4之间有部分的序列互补。
MiRNAome基因组:癌症诊断和治疗的宝藏Ioana Berindan-Neagoe PhD,Paloma del C. Monroig BS,Barbara Pasculli MS,George A. Calin MD, PhD介绍:小分子核糖核酸基因组中陌生人的星系分子生物学的中心法则的解释遗传信息的流动在一个生物系统,总结了“DNA使RNA,它“然而,在过去的几年中,DNA片段已经被证明产生不编码蛋白质的RNA转录。
编码的蛋白质。
这些非蛋白这些记录被命名的非编码RNA基因,他们认为是一个“黑暗”的一部分未被探索的人类基因组。
小分子核糖核酸(MiRNA)是一类小ncRNAs 19到25核苷酸(nt)的长度,可以通过各种机制调节基因的表达,还没有被完全调查。
他们代表了大多数探索的“黑暗”的基因组,和已知的全部(克隆)MiRNA出现在一个名叫MiRNAome基因组。
最初,含有miRNA编码基因的DNA片段的转录的RNA聚合酶II或III(RNA Pol II III)发起的生物合成。
初级转录物(pri-miRNA)可以成百上千个核苷酸长,但它进一步加工,形成一个100 nt的前体的转录,褶皱本身(图1)的前体序列然后出口到细胞质,它经历了一系列的催化步骤实现成熟以前。
在细胞质中,成熟的单链miRNA都集成到一个数量的蛋白质组成RNA诱导沉默复合体(RISC),此后他们相互作用的互补序列的信使RNA(mRNA)的MiRNA加工。
RNA聚合酶II负责MiRNA(miRNA)基因的初始转录成长,封顶,和多聚腺苷酸(polyA)的前体,称为原发性miRNA(pri-miRNA)。
双链RNA核糖核酸酶,Drosha,与它的结合伙伴DGCR8连词(DiGeorge综合征临界区基因8、总督),进一步处理pri-miRNA成70到100核苷酸的RNA前体(pre-miRNA)。
pre-miRNA从细胞核向细胞质易位的形成/RanGTP,和裂解成18到24个核苷酸的双工的核糖核蛋白复合体组成的核糖核酸酶III(Dicer)和TRBP(人类免疫缺陷病毒1型反应的RNA结合蛋白反式激活)。
Cancer is a complex genetic disease involving structural and expression abnormalities of both coding and non-coding genes. For almost three decades, the alteration of protein-coding oncogenes and/or tumour-suppres-sor genes (for reviews see REFS 1–3) have been thought to be the causes of tumorigenesis. With the discovery in the past few years of thousands of genes that pro-duce non-coding RNA (ncRNA) transcripts with no significant open reading frame, it has become evident that the genomic complexity of the cancer cell is far greater than expected. Although the identification of long ncRNAs was quite a slow process owing to the difficulties of experimentally cloning non-coding cDNAs of tens or hundreds of thousands of bases4, the cloning of small members of the ncRNA class was recently extremely successful5–9.These microRNAs (miRNAs) are small non-coding RNAs of 20–22 nucleotides, typically excised from 60–110 nucleotide foldback RNA precursor structures (for reviews see REFS 10–12). The biogenesis of miRNAs involves a complex protein system, including members of the Argonaute family, Pol II-dependent transcription and the RNase IIIs Drosha and Dicer13. MiRNAs are involved in crucial biological processes, including devel-opment, differentiation, apoptosis and proliferation11,14, through imperfect pairing with target messenger RNAs (mRNAs) of protein-coding genes and the transcriptional or post-transcriptional regulation of their expression15–17. The number of human miRNAs reported so far (the July 2006 release of miRBase, at the Sanger Institute18) is in excess of 450, twice as many as initial calculations indicated19, and more than1,000 predicted miRNA genes20–22 are awaiting experimental confirmation.Initially identified in B-cell chronic lymphocytic leukaemia (CLL)23, changes in the expression level of miRNAs have subsequently been detected by differ-ent groups in many types of human tumours (several reviews give various details of the link between miRNAs and cancer, such as REFS 24–34). MiRNAs have been proposed to contribute to oncogenesis because they can function either as tumour suppressors (as is the case for miR-15a and miR-16-1) or oncogenes (as is the case for miR-155 or members of the miR-17–92 cluster). The genomic abnormalities found to influence the activity of miRNAs are the same as those previously described for protein-coding genes, such as chromosomal rearrangements, genomic amplifications or deletions and mutations. In a specific tumour, abnormalities both in protein-coding genes and miRNAs can be identified29. Homozygous mutations or the combination of deletion plus mutation in miRNA genes are rare events35, and the functional consequences of heterozygous sequence variations of miRNAs in human cancers have not been identified36. Furthermore, the role of polymorphisms in the complementary sites of target mRNAs in cancer patients37 or individuals with a predisposition to other hereditary diseases38 have just started to be understood. Therefore, at the present time, the main mechanism that underlies changes in the function of miRNAs in cancer cells seems to be aberrant gene expression, char-acterized by abnormal levels of expression for mature and/or precursor miRNA sequences compared with the corresponding normal tissues.Department of Molecular Virology, Immunology and Medical Genetics and Comprehensive Cancer Center, Ohio State University, Columbus, Ohio 43210, USA. Correspondence to C.M.C.e-mail:Carlo.Croce@doi:10.1038/nrc1997MicroRNA signatures inhuman cancersGeorge A. Calin and Carlo M. CroceAbstract | MicroRNA (miRNA ) alterations are involved in the initiation and progression of human cancer. The causes of the widespread differential expression of miRNA genes in malignant compared with normal cells can be explained by the location of these genes in cancer-associated genomic regions, by epigenetic mechanisms and by alterations in the miRNA processing machinery. MiRNA-expression profiling of human tumours has identified signatures associated with diagnosis, staging, progression, prognosis and response to treatment. In addition, profiling has been exploited to identify miRNA genes that might represent downstream targets of activated oncogenic pathways, or that target protein-coding genes involved in cancer.Bead-based flow cytometry A high-throughput microRNA (miRNA)-profiling technique in which individual beads are coupled to miRNA complementary probes and marked with fluorescent tags. After hybridization with size-fractioned RNAs and staining, the beads are analysed using a flow cytometer.Hierarchical clusteringA computational method that groups genes (or samples) into small clusters and then groups these clusters into increasingly higher level clusters. As a result, a dendrogram (that is, a tree) of connectivity emerges.MiRNA profiling of tumour versus normal tissuesOligonucleotide miRNA microarray analysis is the mostcommonly used high-throughput technique for theassessment of cancer-specific expression levels forhundreds of miRNAs in a large number of samples(for reviews see REFS 13,39,40). Another method todetermine miRNA expression levels involves the useof a bead-based flow-cytometric technique41(FIG. 1). Otherdevelopments include quantitative real-time PCR forprecursor miRNAs42 or active miRNAs43,44, miRAGE— a genome-wide miRNA analysis with serial analysisof gene expression (SAGE)45 — or the high-throughputarray-based Klenow enzyme (RAKE) assay46. Each ofthese techniques has its strengths and weaknesses, butafter several years of study using these technologies, andthe analysis of more than 1,000 primary tumours, com-mon characteristics of miRNA deregulation in humantumours have emerged.MiRNA-expression profiles classify human cancers.To date, every type of tumour analysed by miRNA pro-filing has shown significantly different miRNA profiles(for mature and/or precursor miRNAs) compared withnormal cells from the same tissue (TABLE 1,FIG. 2).Two large profiling studies using various tumoursand two distinct technologies to investigate genome-wide miRNA expression have been published41,47.Lu et al. developed a bead-based flow-cytometricprofiling technology that is highly specific. Theyperformed a systematic analysis of 334 leukaemiasand solid cancers, and found that miRNA-expressionprofiles classify human cancers according to thedevelopmental lineage and differentiation state ofthe tumours. Furthermore, they observed patterns ofgene expression for each type of tumour that reflecteddistinct mechanisms of transformation. For exam-ple, the acute leukaemia samples were divided intothree clusters that corresponded to the BCR–ABL,TEL-AML1 (acute myeloid leukeamia 1, also knownas runt-related transcription factor 1; RUNX1) andMLL (mixed lineage leukaemia) rearrangements,respectively. An interesting observation was thatmiRNA expression in tumour samples seems globallylower than in normal tissue, as 129 out of 217 meas-ured miRNAs showed this pattern of expression. Oneexplanation could be that global miRNA levels reflectthe state of cellular differentiation. In fact, severalprofiling studies, including those by Lu et al.41 and byGarzon et al.48, showed that distinct miRNA profilescharacterize different haematopoietic differentiationstages.Using a large-scale microarray analysis of 540samples, including 363 from six of the most frequenthuman solid tumour types and 177 normal controls,Volinia et al. also found that cancer cells showed dis-tinct miRNA profiles compared with normal cells47(FIG. 2). Out of the 228 miRNA genes analysed, 36 wereoverexpressed and 21 were downregulated in cancercells versus normal cells. Hierarchical clustering analysesshowed that this miRNA signature enabled the tumoursamples to be grouped on the basis of their tissue oforigin. Several other genome-wide profiling studieshave been performed on various cancer types, includingCLL49, breast cancer50, glioblastoma51, thyroid papillarycarcinoma37, hepatocellular carcinoma52, lung cancer53,colon cancer45 and endocrine pancreatic tumours54. Forexample, in a study of 104 matched pairs of primarycancerous and non-cancerous lung tissue, Yanaiharaet al. found a set of 43 differentially expressed miRNAs;28 were downregulated and 15 were overexpressed inthe tumour cells53. Murakami et al., using a microar-ray platform, found that in hepatocellular carcinomafive out of eight differentially expressed miRNAswere downregulated in cancer cells52. All these studiessupport the same view: the alterations seen in cancercells that express miRNAs consist of both overexpressedand downregulated microRNAs.One key issue is the level of differential expression thatcan be considered biologically significant. 10–20-folddifferences in miRNA expression levels have been foundto be important in papillary thyroid carcinomas37,however, we share the view that much smaller changes(less than twofold) could also be significant35,49–51. Thisreflects the fact that specific miRNAs target many genesand, as a large number of miRNAs seem to be alteredin human tumours, one can postulate that the disrup-tion of two or more cooperating oncogenes and/ortumour suppressors is a likely event29. An opposite viewis that in some cases the described changes in miRNAexpression between normal and tumour cells could bebiologically irrelevant: the main interactions with varioustargets could have antagonistic rather than accumulatingbiological consequences (for example, the repression ofboth pro-apoptotic and anti-apoptotic genes).Significance analysis of microarrays (SAM)A statistical method used in microarray analyses that calculates a score for each gene, and therefore identifies genes that are significantly associated with an outcome variable, such as the type of analysed tissue (normal or cancerous).Common miRNA genes are differentially expressed invarious cancers, suggesting common altered regulatorypathways. Assembling the statistical analyses of microar-ray data obtained by two different methods, significanceanalysis of microarrays (SAM) and prediction analysis ofmicroarrays (PAM) from six solid tumours (lung, breast,colon, gastric and prostate carcinomas and endocrinepancreatic tumours), Volinia et al. described a com-mon signature composed of 21 miRNAs differentially47. At the top ofmiR-21, which was overexpressed in six typesmiR-17-5p and miR-191, which were47. Furthermore, miR-21, the55,51 and cholangiocarcinomas56, was shown by Mengto directly target the tumour suppressor PTEN inPTEN encodes a phosphatase57. In allPTEN locus is relatively small (less thanPTEN is shownmiR-21 in the other tumour types thenPTEN inactivation.miR-21 in cultured gliob-55. All these data suggest that miR-21 is anFurther examples of miRNAs that are frequentlylet-758. Another example of fine functionalet al., which shows the oncogenic func-tion of miR-372 and miR-373 in testicular germ-celltumours of adolescents and adults. These miRNAsfacilitate the proliferation and transformation of cellsthat harbour both oncogenic HRAS and functionalwild-type TP53(REF. 59).Additional support for the function of miRNAs inhuman cancers has come from transgenic and knockoutmouse models, which have been important for under-standing the tumorigenic pathways in which deregulatedmiRNAs are involved. He et al. showed that enforcedexpression of the miR-17-92 cluster accelerated MYC-induced lymphomagenesis60, suggesting the oncogenicpotential of some of the miRNAs that are overexpressedin B-cell lymphoma samples owing to genomic ampli-fication. Recently, Costinean et al. produced the firsttransgenic mouse that specifically overexpresses amiRNA gene, miR-155,in B cells61. This gene is overex-pressed in various types of B-cell malignancy62–64. Thetransgenic mice developed polyclonal pre-leukaemicpre-B-cell proliferation followed by B-cell malignancy.These findings suggest that miR-155 deregulation couldbe an early event in oncogenesis that needs additionalgenetic alterations for the development of the fullymalignant phenotype61. The same was also shown forstudies on primary tumours. The bead-based miRNA profiling technique was developed by Lu et al41. Both strategies involve four main steps (shown on the left side): target labelling, DNA–DNA hybridization, staining and signal detection. The different replicates of the spots on the glass slide represent different oligonucleotide sequences corresponding to sequences from the precursor miRNA or active miRNA molecule, whereas the different beads are marked with different fluorescent tags (each colour represents a specific miRNA using up to 100 different colours). The main advantage of the microarray-based miRNA profiling technique is the high standardization of the procedure (which enables tens of samples to be processed in parallel), whereas the main advantage of the bead-based miRNA profiling is that, because hybridization takes place in solution (instead of a glass surface), it offers a higher specificity for closely related miRNAs. The described polyacrylamide gel (PAA) fractionation is an additional processing step in the bead-based system. This figure is modified with permission from REF. 49© (2004) National Academy of Science, USA.Prediction analysis of microarrays (PAM)A statistical technique used in microarray analyses that identifies a subgroup of genes that best characterizes a predefined class, and uses this gene set to predict the class of new samples.the MYC oncogene, which is activated by chromosomaltranslocation in human B-cell neoplasia65. In an avianmodel, oncogenic cooperation in lymphomagenesisand erythroleukaemogenesis between the non-protein-coding BIC gene, which was later shown to include themiR-155 sequence, and MYC has been observed66, indi-cating that MYC and BIC might cooperate in humantumours. A microarray study of the malignant B cellsisolated from miR-155 transgenic mice compared withnon-transgenic controls resulted in a complete list ofthe deregulated protein-coding genes in these cells, andtherefore potentially identified direct and indirect targetsof miR-155. Of particular note, the expression of 16 dif-ferent miRNAs was altered in pre-malignant cells fromthe transgenic mice61, supporting the view that complexrelationships exist between protein-coding genes andmiRNAs that affect specific signalling pathways and,probably, direct or indirect interactions exist betweenmiR-155 and some of the other deregulated miRNAs.Onco-miRNAs and tumour-suppressor-miRNAs: twodifferent views of the same gene?Genome-wide expressionstudies also offer intriguing new perspectives on theinvolvement of miRNAs in human cancer. The firstperspective comes from the studies performed byO’Donnell et al. on cell models, which showed that in thehuman B-cell line P493-6, which overexpresses MYC,miRNA members of the miR-17–92 locus have tumour-suppressor activity because their expression decreasesthe expression of E2F1, and so inhibits MYC-mediatedcellular proliferation67. However, a different perspec-tive is offered by the results from B-cell lymphomas.The same cluster of miRNAs function as a potentialoncogene by cooperating with MYC and blockingapoptosis60. One possible explanation is that the samemiRNA can participate in distinct pathways, havingdifferent effects on cell survival, growth and proliferationthat are dependent on the cell type and the pattern ofgene expression. The combinatorial nature of miRNA–mRNA interactions means that the same miRNA couldhave different targets and the same mRNA could betargeted by different miRNAs in different cell types. Forexample, Cimmino et al. found that in leukaemic cellswith a deletion of the locus on chromosome 13 that con-tains miR-15a and miR-16-1 miRNA genes, apoptosisis induced as a result of the exogenous re-expressionof these miRNAs. MiR-15a and miR-16-1 interact withthe anti-apoptotic gene BCL2 when added back to thesecells, however, a similar response is not seen in 293 fetalkidney cells, which normally express miR-15a, miR-16-1and BCL2 (REF. 68). Therefore, it is probable that the lossof miR-15a and miR-16-1 induces the overexpressionof BCL2 in B cells, giving rise to B-cell malignancies.In a different tissue, the overexpression of miR-15a andmiR-16-1 might cause the loss of expression of a tumoursuppressor that is important for growth control and/orapoptosis. In support of this hypothesis are reports thatshow that, although miR-15a and miR-16-1 are down-regulated in B lymphocytes from patients with CLL23,the same miRNAs are overexpressed in endocrinepancreatic tumours of neuroectodermal origin47,54.The second perspective comes from the finding thatthe consistent abnormal expression of the precursormiRNAs, but not the active molecules, can be identifiedin various types of cancer42,47,49,52,53. Y anaihara et al. foundthat in their 43 miRNA gene signature, seven precursormiRNAs were differentially expressed in matched lungcancer versus normal lung tissue. In addition, Jiang et al.,using a real-time PCR method specifically designed forprecursor profiling42, found significant variations in theTable 1 |Facts about microRNA-expression profiling in human cancersH MiRNA, microRNA. ZAP70, 70 kDa zeta-associated protein.expression of nine precursors in a panel of 32 cancer celllines69. These data raise the possibility that the ‘non-active’part of the miRNA molecule could have ‘independent’and as yet unknown functions that could be important intumorigenesis. Putative interaction partners might be theRNA-binding proteins (such as the heterogenous nuclearribonucleoproteins, hnRNPs), a class of proteins that areabnormally expressed in all types of human cancers70.These proteins bind RNA molecules to regulate theirstability, and therefore it is possible that in the sequenceof several precursor miRNAs such hnRNP-interactingmotifs exist. However, there are no data so far that directlyattribute regulatory function to a miRNA precursor.Causes of abnormal miRNA expressionThe causes of the widespread disruption of miRNAexpression in tumour cells are only partially known and,very probably, various abnormalities in each tumourcould contribute to the global miRNA-expression pro-file. So far, at least three different mechanisms (that couldfunction independently or together) have been described.Such alterations correlate with and could explain theabnormal expression of specific miRNA genes.The location of miRNAs at cancer-associated genomicregions (CAGRs).Soon after the identification of hun-dreds of new members of the miRNA family, it was shownthat more than half of the known human miRNAs residein particular genomic regions that are prone to alterationin cancer cells71. Such regions include minimal regions ofLOH, which are thought to harbour tumour-suppressorgenes, minimal regions of amplification, which mightcontain oncogenes, common breakpoint regions in ornear possible oncogenes or tumour-suppressor genes andfragile sites (FRA). FRA are preferential sites of sister-chromatid exchange, translocation, deletion, amplificationor integration of plasmid DNA, and tumour-associatedviruses such as human papilloma virus (HPV). Of note,miRNA loci were significantly associated with insertionsites of tumorigenic HPV strains linked to endometrialcancers, leading to the possibility that viral insertions inthe human genome might disturb small ncRNAs71. Figure 2 | Examples of microRNA profiles in human solid and liquid cancers. The general consensus is that profiling enabled the signatures that are associated with diagnosis — the process of identifying the nature of a disease — and prognosis — a prediction of the probable course and outcome of a disease—to be defined. a |The profiling of 836 solid tissue and haematological primary cancers (and corresponding normal tissues samples) using the microarray technology (as described in REF. 39) and reported to date is shown. The exact number of analysed samples from each tumour type (corresponding normal controls included) is shown.The data were collected from the references cited in the text. b |The common expression signature in six solid cancers indicates common altered regulatory pathways involving the same microRNA (miRNA) genes (named on the right side of the dendrogram)47. These miRNAs could be considered as ‘cancer markers’, because variations of their expression could identify the cancerous state. c |The distinct clusters identified by miRNA profiles in chronic lymphocytic leukaemia (CLL)49 were further confirmed to be associated with prognostic factors and disease progression35. Note also that various normal haematopoietic samples cluster in a group that is distinct from all CLL samples. MNC, mononuclear cells; Ly, B lymphocytes; CD5, a subset of B lymphocytes largely accepted to represent the equivalent of malignant cells in CLL93. Panel b is modified with permission from REF. 47© (2006) National Academy of Science, USA.Several reports show that this computationally identified association is significant for miRNA expression. Zhang et al. showed, by using high-resolution array com-parative genomic hybridization (aCGH), that the miRNA loci have a high frequency of genomic alteration in human cancers 72. Furthermore, about 75% of the miRNAs ana-lysed showed concordance between mature miRNA levels and DNA copy number, and, importantly, their genomic data were in agreement (81% concordance for amplified and/or overexpressed miRNAs, and 60% concordance for deleted and/or downregulated miRNAs, respectively) with the expression data published with an independent set of breast cancers by Iorio et al 50. Other examples of concordance by two independent reports, involve two miRNA clusters located at chromosome 13 (FIG. 3). The cluster miR-15a–miR-16-1, located at 13q14.3 in a region deleted in B-cell chronic lymphocytic leukaemia and pituitary adenomas was shown by northern blots to be downregulated in most patients with these tumours 23,73. The cluster miR-17–92, located at chromosome 13q31, in a region amplified in B-cell lymphomas and lung cancers, was found to be overexpressed using microarray studies by He et al.59, and by Hayashita and colleagues and Tagawa and Seto using northern blot analyses in lung cancers and malignant lymphomas, respectively 74,75.Epigenetic regulation of miRNA expression. DNA hypomethylation, CpG island hypermethylation and histone-modification losses represent epigenetic markers of malignant transformation 76. Therefore, several groups have investigated whether such events also affect miRNA expression. Scott et al. showed that in breast cancer cells histone deacetylase inhibition is followed by the exten-sive and rapid alteration of miRNA levels 77. Furthermore, Saito et al. found that the combined treatment of human bladder cancer cells with 5-aza-2′-deoxycytidine (5-Aza-CdR) and the histone deacetylase (HDAC) inhibitor 4-phenylbutyric acid (PBA) has a significant effect on the expression of miRNAs 78. Seventeen miRNAs (out of 313 screened by a microarray assay) were upregulated more than threefold, and miR-127 was the most differentially expressed . This miRNA is located in a CpG island at chromosome 14q32, a region that is involved in several types of translocations identified in haematological can-cers and deleted by LOH in solid tumours 71. Furthermore, the combined treatment was accompanied by a decreasein DNA methylation and an increase in active histone markers around the transcription start site of miR-127. The investigators showed that miR-127 can translation-ally repress the transcription of the zinc-finger repressor BCL6 (REF . 78). Therefore, the induction of miR-127 by 5-Aza-CdR and PBA treatment in cancer cells might have an anticancer effect by downregulating anti-apoptotic factors, such as BCL6, and inducing apoptosis 78.The effects of DNA demethylation and histone deacetylation on miRNA expression could be tissue-specific, as the treatment of non-small cell lung cancer (NSCLC) cells with demethylating agents and/or HDAC inhibitors does not have a significant influence on miRNA expression 36,53. Supporting this assumption is the fact that miR-127 was not found to be expressed in normal lung cells by microarray analysis 39, and wasnot differentially expressed between cancerous and non-cancerous lung tissue 53.Abnormalities in miRNA-processing genes and proteins. The protein machinery that is involved in the biogenesis of miRNAs is complex. Theoretically, alterations of these proteins (for a comprehensive description see REF. 34) should have dramatic effects on miRNA expression. In fact, it was shown that a failure of the Drosha processing step could explain the downregulation of miRNAs observed in primary tumours 79. Furthermore, in a large study of patients with NSCLC, Karube et al. found that the expression levels of Dicer, but not Drosha, were reduced in a fraction of lung cancers. This correlated with reduced post-operative survival and poor tumour differentiation status, both of which are associated with a poor prog-nosis 80,81. A similar correlation with reduced survival in patients with NSCLC was found for the expression of miR-155 and let-7 (REFS 53,82). These results suggest that patient survival could be a consequence of both aber-rant miRNA processing and aberrant expression. The conditional knockout Dicer mouse has developmental defects, including lung epithelium abnormalities 81,83,but no reports on tumour formation are available, Figure 3 | Chromosomal alterations at microRNA loci. The main chromosomalalterations at microRNA (miRNA) loci, loss of heterozygosity and amplification, areidentified at two separate regions of chromosome 13. a | Shows the 13q14.3 deletionfound in chronic lymphocytic leukaemia (CLL) as described in REF . 23. The downregulation of miR-15a and miR-16-1 induces overexpression of the anti-apoptotic BCL2 protein inleukaemia cells 68. The genomic regions are not drawn to scale, and the cluster on 13q13 is simplified. The functional consequences are shown on the right of the figure. b | Shows the 13q13 amplification identified in high-grade lymphomas, as described in REF . 74. The functional consequences are presented on the right side of the figure.The overexpression of miR-17-5p and miR-20a from the same cluster induces the downregulation of the transcription factor E2F1. It was also shown that the MYConcogene increases the expression of the miR-17–92 cluster and activates E2F1 (REF . 67),and that the MYC-activated cluster increases tumour angiogenesis 113. Therefore, theoverexpression of the miR-17–92 cluster is correlated with hyperproliferation, theblocking of apoptosis and neovascularisation. The arrows and the bars representstimulatory and inhibitory signals, respectively. Orf, open reading frame.Richter transformation Denotes the development of high-grade non-Hodgkin lymphoma, prolymphocytic leukaemia, Hodgkin disease or acute leukaemia in patients with chronic lymphocytic leukaemia. Current treatments are aggressive, but prognosis is poor.Univariate versus multivariate analyses Univariate analysis explores each variable in a data set separately and describes the pattern of response to the variable. It describes each variable on its own. Multivariate statistical analysis describes a collection of procedures that involve the observation and analysis of more than one statistical variable at a time. It describes all variables together.and Dicer-deficient embryonic stem cells injected intonude mice do not form tumours84.The influence of genetic abnormalities in small-RNA-processing proteins could have a more significant influ-ence than is recognized at present. Recently, a new classof germline-specific small ncRNAs (26–31 nucleotideslong), named PIWI-interacting RNAs (piRNAs), was dis-covered and found to bind the testis-specific murine PIWIprotein orthologues85–89. Intriguingly, the human homo-logue of this subgroup of the Argonaute proteins, namedHIWI, is located in the genomic region 12q24.33 thatis genetically linked with the development of testiculargerm-cell tumours in adolescents and adults. In addition,HIWI is overexpressed in most testicular seminomasanalysed so far90. The expression of HIWI in humangastric cancer is associated with proliferating cells91. Thesecorrelations raise the possibility that piRNAs could rep-resent another example of cancer-altered small ncRNAs,specifically in relation to PIWI overexpression.MiRNA profiling as a new clinical toolAs active players in important human oncogenic signal-ling pathways, miRNAs should affect cancer diagnosisand prognosis (TABLE 1). As discussed above, severalreports have already shown this for several tumourtypes49,50,62. But other reports indicate that miRNAprofiles can be used to diagnose tumour types that cannotbe determined on the basis of tumour biopsy samples.MiRNA profiling as a diagnostic tool. Metastatic cancerof unknown primary site (CUP) is one of the 10 most fre-quent cancer diagnoses worldwide, and constitutes 3–5%of all human malignancies92. Patients with CUP presentwith metastases (late-stage disease) without an establishedprimary tumour (that is, a site at which the tumour hasinitially developed and from which it has metastasized).The study of Lu et al. produces an important advance inthe diagnosis of this peculiar type of cancer. Analysing17 poorly differentiated tumours with non-diagnostichistological appearance, they showed that the miRNA-based classifier was much better at establishing the correctdiagnosis of the samples than the mRNA classifier41. Thisresult is exciting — profiling a few hundred miRNAs hasa much better predictive power for CUP diagnosis thanprofiling several tens of thousands of mRNAs. As miRNAexpression changes with differentiation, the poorlydifferentiated tumours have lower global expression levelsof miRNAs compared with well-differentiated tumoursfrom control groups41. The reduced expression levels ofmiRNAs in poorly differentiated tumours is probably whymiRNA profiling is effective in the diagnosis of CUP.CLL is the most common adult leukaemia in theWestern world93,94. During its natural history, this indolentdisease can evolve into an aggressive type of lymphomaand/or leukaemia, including Richter syndrome95.Furthermore, as the prognosis of most cases of CLL isrelatively good, treatment after diagnosis is started only ifpoor prognostic markers are evident96. Few such markershave been found, but those that have include high expres-sion levels of the 70 kDa zeta-associated protein (ZAP70)and an absence of mutations in the immunoglobulinheavy-chain variable-region gene (IgVH). By performinga miRNA-profiling screen on 144 patients with CLL, aunique signature of 13 miRNAs (out of the 190 analysed)was shown to differentiate cases on the basis of a good orbad prognosis, or on the presence or absence of diseaseprogression35. Among them, miR-16-1 and miR-15a wereexpressed at lower levels in patients with a good progno-sis, in agreement with early reports that 13q14.3 genomicdeletions at the locus that harbours these genes correlatewith a favourable course of the disease97. Among the genesfound to be downregulated in the group of patients with abad prognosis were members of the miR-29 family. Thiscould be explained by the recent findings that TCL1, anoncogene that is overexpressed in CLL cells from patientswith aggressive disease98, is a target of miR-29 genes(Y. Pekarsky, unpublished observations). Therefore, thesefindings support a model in which two different molecu-lar types of CLL with opposite clinical behaviours exist:one with a good prognosis and slow disease progressioncharacterized by deletions of chromosome 13q14.3 andlow levels of miR-15a and miR-16-1, and a second thatis aggressive and prone to develop Richter syndrome,characterized by low levels of expression of miR-29 genesand high expression levels of TCL1 protein. As BCL2 isconsistently overexpressed in CLL samples99, miR-15aand miR-16-1 might have additional targets importantfor progression; furthermore, other mechanisms of BCL2deregulation (such as the minority of cases that have aBCL2 translocation100), and other interacting miRNAsmight be involved.MiRNA profiling as a prognostic tool.Lung cancer is theleading cause of death from cancer in males worldwide101.Therefore, the identification of new prognostic markers(markers that correlate with disease evolution) could be asignificant advance for the identification of patients thatwould benefit from more aggressive therapy. In univariateanalyses, the expression of both miR-155 (high levels) andlet-7a-2 (low levels)has been shown to correlate with poorBox 1 | Putative mechanisms of cancer predisposition by microRNA alterations In most familial cases of cancer, culprit genes have not been identified despite several decades of investigation. So it is possible that members of the new category of non-coding RNAs (ncRNAs) are involved in cancer predisposition. Germline sequence abnormalities were identified in microRNA (miRNA) genes or transcripts35, and in targeted sequences in messenger RNAs (mRNAs) that interact with miRNAs37. Predisposing mutations in miRNAs, although rare events, can affect miRNA activity in at least three distinct ways: first, variation in the expression levels of miRNAs, by abnormal processing, if located in the miRNA themselves (as shown in REF. 112) or very close to the gene (as shown in REF. 35); second, an alteration of the spectrum of interactor mRNAs when present in the active molecule; and third, an alteration of putative interactions with proteins (such as heterogenous nuclear ribonucleoproteins),if located in the precursor molecule. A destabilizing effect of the miRNA–mRNA interaction was described for germline single nucleotide polymorphisms found in the two 3′UTR recognition sequences in the KIT oncogene that determine the interaction with miR-221, miR-222 and miR-146, as reported by He et al37. As each miRNA has many targets, and, as a large number of miRNAs seem to be altered in human tumours, onecan postulate that the disruption of two or more cooperating oncogenes and/or tumour suppressors is a likely event29. The accumulation of additional somatic events that occur in genes or ncRNAs, including miRNAs, is necessary for the development of the malignant phenotype (see also FIG. 4).。
新型分子标志物microRNA 在肿瘤诊断中的应用主讲人:刘敏中山大学附属第一医院目录content 123microRNA概述循环microRNAmicroRNA的产业应用4总结1 microRNA概述什么是microRNA?microRNA是一类长约22nt的单链小RNA,由细胞内源产生的发卡(hairpin)结构转录本加工而来1。
pri-miRNApre-miRNAmiRNA1. Huang L, et al: Hsa-miR-19a is associated with lymph metastasis and mediates the TNF-alpha induced epithelial-to-mesenchymal transition in colorectal cancer. Sci Rep 2015.的发现microRNAVictor Ambros Gary Ruvkun200019932001lin-4let-7miRNA 1.Ambros V,et al: The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 19932.Ruvkun G,et al: Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 19933.Ruvkun G,et al: The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 2000microRNA的功能转录抑制|mRNA切割|mRNA降解1. Cellura D, et al: miR-19-Mediated Inhibition of Transglutaminase-2 Leads to Enhanced Invasion and Metastasis in Colorectal Cancer. Mol Cancer ResmicroRNA参与调节各种病理生理过程miRNA的功能涉及到各种生理病理过程,包括发育过程调节、抵抗病毒入侵、动物免疫功能调节、各器官/系统疾病以及肿瘤等。
MicroRNAome Genome:A Treasure for Cancer Diagnosis and TherapyIoana Berindan-Neagoe,PhD1;Paloma del C.Monroig,BS2;Barbara Pasculli,MS3;George A.Calin,MD,PhD4The interplay between abnormalities in genes coding for proteins and noncoding microRNAs(miRNAs)has been among the most exciting yet unexpected discoveries in oncology over the last decade.The complexity of this network has redefined cancer research as miRNAs,produced from what was once considered“genomic trash,”have shown to be crucial for cancer initiation, progression,and dissemination.Naturally occurring miRNAs are very short transcripts that never produce a protein or amino acid chain,but act by regulating protein expression during cellular processes such as growth,development,and differentiation at the transcriptional,posttranscriptional,and/or translational level.In this review article,miRNAs are presented as ubiquitous players involved in all cancer hallmarks.The authors also describe the most used methods to detect their expression,which have revealedthe identity of hundreds of miRNAs dysregulated in cancer cells or tumor microenvironment cells.Furthermore,the role of miRNAsas hormones and as reliable cancer biomarkers and predictors of treatment response is discussed.Along with this,the authors explore current strategies in designing miRNA-targeting therapeutics,as well as the associated challenges that research envisionsto overcome.Finally,a new wave in molecular oncology translational research is introduced:the study of long noncoding RNAs.CA Cancer J Clin2014;64:311-336.V C2014American Cancer Society.Keywords:microRNA,diagnosis,therapy,cancer.IntroductionMicroRNAs Are Strangers in the Genomic GalaxyThe central dogma of molecular biology is an explanation of theflow of genetic information within a biological system andit is summarized in the fact that“DNA makes RNA,which encodes protein.”However,over the past few years,DNA seg-ments have been shown to produce RNA transcripts that do not encode proteins.These transcripts were named noncoding RNAs(ncRNAs),and they were considered to be part of a“dark”unexplored segment of the human genome.MicroRNAs (miRNAs)are a class of small ncRNAs19to25nucleotides(nt)in length that can regulate gene expression by various mechanisms that have still not been fully investigated.They represent the most explored side of the“dark”matter of the genome,1-3and the full complement of known(cloned)miRNAs present in a genome is named microRNAome(for a glos-sary of terms,see Table1).Initially,the DNA segments containing miRNA-coding genes are transcribed by an RNA polymerase II or III(RNAPol II and III)to initiate their biogenesis.The primary transcript(pri-miRNAs)can be hundreds or thousands of nt long,but it is further processed,forming a$100-nt precursor transcript that folds upon itself(Fig.1).4-6The precursor sequenceis then exported to the cytoplasm,where it undergoes a series of catalytic steps prior to achieving maturation.In the cyto-plasm,mature single-stranded miRNAs are integrated into a number of proteins that compose the RNA-induced silencing complex(RISC),and thereafter they interact by sequence complementarity with the messenger RNA(mRNA)(Fig.1).4-6,71Department of Functional Genomics,The Oncology Institute,Research Center for Functional Genomics,Biomedicine and Translational Medicine, Department of Immunology,Iuliu Hatieganu University of Medicine and Pharmacy,Cluj-Napoca,Romania;2Department of Experimental Therapeutics,The University of Texas MD Anderson Cancer Center,Houston,TX,University of Puerto Rico School of Medicine,San Juan,Puerto Rico;3Visiting PhD Student,Department of Experimental Therapeutics,The University of Texas MD Anderson Cancer Center,Houston TX;PhD Student,Department of Bio-sciences,Biotechnology and Biopharmaceutics,University of Bari,Bari,Italy;4Department of Experimental Therapeutics,Department of Leukemia,Cen-ter for RNA Interference and Non-Coding RNAs,The University of Texas MD Anderson Cancer Center,Houston,TX.Corresponding author:George A.Calin,MD,PhD,Department of Experimental Therapeutics,The University of Texas MD Anderson Cancer Center,1881EastRd,Unit1950,3SCR4.3424,Houston,TX77030;gcalin@DISCLOSURES:George A.Calin,MD,PhD,is The Alan M.Gewirtz Leukemia&Lymphoma Society Scholar.Work in Dr.Calin’s laboratory is supported in part by National Institutes of Health/National Cancer Institute grants1UH2TR00943-01and1R01CA182905-01;Developmental Research Awards in prostate cancer, multiple myeloma,leukemia(P50CA100632),and head and neck(P50CA097007)Specialized Programs of Research Excellence(SPORE)grants;a Sister Institu-tion Network Fund(SINF)University of Texas MD Anderson Cancer Center DKFZ grant in chronic lymphocytic leukemia(CLL);a SINF grant in colon cancer;a Kidney Cancer Pilot Project;The University of Texas MD Anderson Cancer Center Duncan Family Institute for Cancer Prevention and Risk Assessment;The Blanton-Davis Ovarian Cancer-2013Sprint for Life Research Award;the Laura and John Arnold Foundation;the RGK Foundation and the Estate of C.G.Johnson,Jr;and by the CLL Global Research Foundation.Dr.Berindan-Neagoe’s work was financed by a POSCCE709/2010grant.doi:10.3322/caac.21244.Available online at VOLUME64_NUMBER5_SEPTEMBER/OCTOBER2014311As part of the new ncRNA-based dogma,miRNAs have been known to bind to mRNA at the30-untranslated region(UTR)and cause the downregulation of protein-coding genes(named targets)in the cytoplasm.They do so by inhibiting the ability of the ribosome to“translate”the mRNA.Alternatively,miRNAs can increase mRNA deg-radation,also reducing the possibility of the mRNA being translated into proteins.8The level of complementarity between miRNA and the target mRNA may determine the mechanism by which the translation of mRNA to pro-tein is blocked.Perfect or near-perfect complementarity has been found to induce mRNA degradation by RISC, and partial complementarity has been found to repress mRNA translation by blocking ribosomal access to the mRNA.9Due to the fact that each miRNA has hundreds or thousands of mRNA targets,a broad segment of the protein-coding genome is under their control.The regula-tion of miRNA-coding genes is of critical interest because they might be involved in any type of pathophysiological process and/or pathway,such as B-cell survival(miR-15a and miR-16-1),B-cell lineage fate(miR-181),brain patterning(miR-430),pancreatic cell insulin secretion (miR-375),adipocyte development(miR-145),cell prolifer-ation control(miR-125b and let-7),or cell survival(let-7 family).10The understanding of the mechanisms of action of miRNAs has significantly expanded in the last few years, with discoveries demonstrating unexpected complexities of their regulative manner,such as promoter binding,protein binding,or direct interaction with other ncRNAs(Fig.2). miRNAs can be relocalized in the nucleus;for example, human miR-29b has been found predominantly in the nucleus,demonstrating that despite their small size,specific miRNAs contain specific nucleotide sequences that control their subcellular localization.11This localization supports the already proven hypothesis that miRNAs can alternatively regulate transcriptional processes at a DNA level.For exam-ple,human miR-373binds to the E-cadherin(CDH1)pro-moter,thereby inducing its expression.12Futhermore, miRNAs target other genetic regions at the mRNA level besides the30-UTRs,such as50-UTR and coding regions.13-15Finally,besides mRNAs,miRNAs can targetTABLE1.Glossary of Terms Used in the MicroRNA WorldASO:An antisense oligonucleotide is a single-stranded,chemically modified DNA-like molecule that is17to22nucleotides in length and designed to becomplementary to a selected messenger RNA(mRNA)or noncoding RNA and thereby specifically inhibits expression of that gene.Exome sequencing(targeted exome capture):An efficient strategy to selectively sequence the coding regions of the genome.Messenger RNA(mRNA):The form of RNA that mediates the transfer of genetic information from the DNA in the cell nucleus to ribosomes in the cytoplasm,in which it serves as a template for protein synthesis.MicroRNAome:The full complement of known(cloned)microRNAs(miRNAs)present in a genome.Due to multiple cloning efforts,it is growing constantly and by May2014contains2578mature human miRNAs(release20of miRBase available at /index.shtml).Noncoding RNAs(ncRNAs):Any RNA molecule that is not translated into a protein.Oncogenic miRNA:A miRNA that when expressed at higher levels than normal initiates or favors the development of a tumor.Open reading frame(ORF):A section of an mRNA that begins with an initiation(methionine ATG)codon and ends with a nonsense codon.ORFs all have the potential to encode a protein or polypeptide;however,many may not actually do so.Pol II:RNA polymerase II(also called RNAP II)catalyzes the transcription of DNA to synthesize precursors of miRNAs and most small nuclear RNAs.Pol III:RNA polymerase III(also called RNAP III)transcribes DNA to synthesize ribosomals RNA,transfer RNAs,and other small RNAs.The genes transcribed by RNA Pol III fall in the category of“housekeeping”genes whose expression is required by all cell types and most environmental conditions.Pseudogene:A copy of a gene that usually lacks introns and other essential DNA sequences necessary for function.A pseudogene has been mutated into an inactive form over the course of evolution,but contains the majority of interactor sites with miRNAs as the original functional gene.Sense/antisense:Refers to the strand of a nucleic acid that directly specifies the product or refers to the strand of a double-stranded molecule that does not directly encode the product but is complementary to it(antisense).Single nucleotide polymorphism(SNP):A variation at a single position in a DNA sequence among individuals.If a SNP occurs within a gene,then the gene has more than one allele.The Cancer Genome Atlas(TCGA):A project started in2005to catalogue genetic alterations responsible for cancer,using gene expression profiling,copy number variation profiling,single nucleotide polymorphism genotyping,genome-wide DNA methylation profiling,miRNA profiling,and exome sequencing.Transcription:The process by which RNA is synthesized from a DNA template.Translation:The process of ribosome-mediated production of a protein by which the primary structure of the protein is determined by the codon nucleotide sequence of an mRNA.Tumor suppressor miRNA:A miRNA that when expressed normally blocks the initiation or development of a tumor;at lower levels of expression,this braking effect disappears and the tumor can develop.The same miRNA can behave as an oncogene in one type of tissue and as a tumor suppressor in another type.Untranslated region(UTR):5’UTR is the portion of an mRNA from the5’end to the position of the first codon used in translation.The3’UTR is the portion of an mRNA from the3’end of the mRNA to the position of the last codon used in translation.312CA:A Cancer Journal for Cliniciansdifferent types of ncRNAs,some of them being highly conserved among species such as the ultraconserved genes 4or poorly conserved such as the pseudogenes (see Table 1for the definition of terms).5In the past few years,miRNAs have also been found to favor protein expression (in addition to downregulating it).MiR-369-3p was shown to interact with adenylate uridy-late-rich elements in the tumor necrosis factor-alpha (TNF-alpha)mRNA,consequently recruiting proteins thatincreased the process of translation during cell cycle arrest.6Similarly,miR-328was revealed to increase translation by interacting with hnRNPE2,a translational regulator,lead-ing to the release of CCAAT/enhancer-binding protein alpha mRNA.Conversely,it decreased the translation of PIM1kinase by binding specifically to its mRNA.Both mechanisms are active during the acute transformation of the chronic phase of chronic myeloid leukemia (CML)and are independent of patients’response to imatinib.Together,these data reveal that miRNAs possess the ability to control the fate of protein-coding genes by attaching through base pairing complementary to mRNA sequences or by interfering with regulatory proteins directly.16MiRNAs also work as secreted molecules that trigger a receptor-mediated response in a different cell or tissue.They have the ability to be released into the extracellular environment within exosomes (cell-derived vesicles origi-nated by the inward budding in the plasma membrane gen-erating multivesicular bodies)that are present in many and perhaps all biological fluids.In this way,they can act as “hormones.”17For example,it has been shown that macro-phages influence breast cancer cell invasion through the exosome-mediated delivery of oncogenic miR-223;more-over,pretreatment of mice with tumor-derived exosomes accelerates lung metastasis formation.18Similarly,exosomes have been shown to modulate tumor microenvironments by releasing miRNAs in a coordinated manner.In this regard,exosomes derived from hypoxic cul-tures of a leukemic cell line in vitro were found to carry and release miR-210among other angiogenic miRNAs,increasing tube formation by endothelial cells.19Along this line,it was also shown that miR-21and miR-29a can be transported through exosomes and can act as direct agonists of Toll-like receptors (TLRs).By binding as ligands to receptors of the TLR family in immune cells,these miRNAs were shown to trigger a TLR-mediated prometastatic inflammatory response (such as secretion of interleukins)that could favor tumor growth and metastasis.20,21Under-standing such newly recognized ways in which the miRNAs are working is important for both scientists and physicians as novel therapeutic approaches can be designed,such as blocking of miR-328interaction with hnRNP E2in patients with CML or blocking the miR-21and miR-29a agonistic effects on TLRs in patients with metastatic diseases.MiRNAs as Ubiquitous Players in CancerMiRNA alterations have been identified in many human diseases such as autoimmune and cardiac disorders,schizo-phrenia,and cancer (where they have been found to be highly dysregulated).22MiRNAs have been found to be differentially expressed between all types of analyzed human tumors and normal tissues,including benignandFIGURE 1.MicroRNA Processing.RNA polymerase II is responsible for theinitial transcription of the microRNA (miRNA)gene into a long,capped,and polyadenylated (poly-A)precursor,called primary miRNA (pri-miRNA).4,5A double-stranded RNA-specific ribonuclease,Drosha,in conjunction with its binding partner DGCR8(DiGeorge syndrome critical region gene 8or Pasha),further processes pri-miRNA into a 70-to 100-nucleotide hairpin RNA precursor (pre-miRNA).6Pre-miRNA is translocated from the nucleus to the cytoplasm by Exportin-5/RanGTP,and cleaved into an 18-to 24-nucleotide duplex by a ribonucleoprotein complex composed of ribonucle-ase III (Dicer)and TRBP (human immunodeficiency virus-1transactivating response RNA-binding protein).Finally,the duplex interacts with the RNA-induced silencing complex (RISC),which includes proteins of the Argonaute family (Ago1to Ago4in humans).One strand of the miRNA duplex remains stably associated with RISC and becomes the mature miRNA,which pri-marily,but not exclusively,guides the RISC complex to the 30-untranslated region of target messenger RNAs (mRNA).The other strand named “star”has also been found to be functional.Although the interaction between miRNA and mRNA usually results in translation inhibition,some cleavage of target mRNAs has also been observed.VOLUME 64_NUMBER 5_SEPTEMBER/OCTOBER 2014313malignant tumors 23such as leukemias,lymphomas,lung cancers,breast cancers,colorectal cancers (CRCs),papillary thyroid carcinomas,glioblastomas and other brain tumors,hepatocellular carcinomas,pancreatic tumors,cervical cancers,prostate cancers,kidney and bladder cancers,or pituitary adenomas.23,24MiRNAs can act as oncogenes and/or tumor suppressors.MiRNAs have been proven to work as oncogenes (Table 1),such as miR-21or miR-155,both of which are among the most frequently overexpressed miRNAs in human cancers.23For example,in a transgenic miR-155mouse model,the rodents initially exhibited a preleukemic pre-B-cell prolifera-tion evident in the spleen and bone marrow,followed by frank B-cell malignancy.25Overexpression of miR-21in mice similarly leads to a pre-B-cell malignant lymphoid-like phenotype,demonstrating that this gene is also a genuine oncogene.When miR-21was inactivated in vivo,the tumors regressed completely within a few days,partly as a result of enhanced apoptosis.26MiRNAs can act also as tumor suppressors,such as the miR-15a/16-1cluster,which is frequently deleted in chronic lymphocytic leukemia (CLL)27and prostate cancers.28The induced deletion of these miRNAs in a knockout mouse model causes the development of indolent B-cell,autono-mous,clonal lymphoproliferative disorders,recapitulating the spectrum of CLL-associated phenotypes observed in humans.The miR-15a/16-1-deletion has been demonstrated to accelerate the proliferation of both humanand mouse B cells by modulating the expression of genes controlling cell cycle progression.29In some instances,the same miRNA can have dual activ-ities,thereby acting as an oncogene in one specific cell type and as a tumor suppressor in another.For example,miR-221is overexpressed in liver cancers,in which it targets the PTEN tumor suppressor,and in this way it promoted liver tumorigenicity in a miR-221mouse transgenic model.30Similarly,in CRC,this same miRNA promotes cell invasion and metastasis by targeting RECK (which normally inhibits invasion and metastasis).31However,in other tumor types such as gastrointestinal stromal tumors,miR-221is downre-gulated and the consequent derepression of c-KIT and ETV1(target oncogenes)promotes this malignancy.32The genetic basis for abnormal miRNA expression in cancer cells is complex.The widespread disruption of miRNA expression in malignant cells is only beginning to be understood,and a variety of abnormalities could contrib-ute to the miRNAome expression profile in each tumor.Transcription factors are involved in the regulation of spe-cific miRNAs,such as the miR-34family and miR-15/16cluster modulated by TP53,33,34the miR-17-92cluster regulated by MYC,34,35or miR-210regulated by hypoxia-inducible factor-1alpha (HIF-1a ).36The aberrant expres-sion causes abnormal levels of mature and/or precursor miRNAs in comparison with the corresponding levels in normal tissues.37At least 3different mechanisms (that could act independently or together)have been describedtoFIGURE 2.Mechanisms of Action of MicroRNAs.MicroRNAs can work through a variety of mechanisms,including (A)direct binding by complementarity tomultiple regions at the messenger RNA (mRNA)including the 30-untranslated region (UTR),50UTR,and coding regions and decreasing protein expression (majority of miRNAs);(B)positive regulation of translation (miR-369-3p)through the increased recruitment of processing proteins;(C)direct interaction with promoter sequences (miR-373);(D)direct agonism of receptors such as Toll-like receptors (miR-21,miR-29a,and let-7);and (E)direct interaction with pro-tein and enhancing protein expression through a decoy mechanism (miR-328).For gene abbreviations please see /gene.314CA:A Cancer Journal for Clinicianscause this:the location of miRNA-coding genes at cancer-associated genomic regions frequently deleted or ampli-fied38;epigenetic regulation of miRNA expression by methylation(which adds methyl groups to the cytosine or adenine DNA nucleotides,thereby suppressing genetic expression)or histone modification(which are proteins that package and order the DNA)39;andfinally,abnormal-ities in miRNA processing genes or proteins(required for miRNA biogenesis and maturation).40,41Germline and Somatic Mutations in miRNAs Variations in the sequences of miRNAs located in the mature,precursor,or primary transcript may contribute to cancer predisposition and initiation.42For example,germ-line(passed from parental germ cell)or somatic(acquired by the somatic cell)mutations of some miRNA genes were found in patients with CLL.In the initial analyses of sequence variations in miRNAs,it was reported that in2 patients diagnosed with CLL,a nucleotide substitution (C for T)was associated with lower levels of mature miR-16(a tumor suppressor miRNA).43This mutation proved to affect the SRp20site,which is an RNA-splicing protein implicated in processing the primary miR-16transcript, and this was hypothesized to be contributing to the devel-opment of CLL.44Findings such as this point out that some patients with CLL may have a genetic predisposition toward this type of cancer.Furthermore,a mouse model also supported the role of certain miRNAs in the pathoge-nesis of CLL.Similarly,these mice harbor a point muta-tion(one nt)adjacent to miR-16,which results in its reduced overall expression.45On a separate note,single nucleotide polymorphisms (SNP,see Table1for definition)in the protein-coding mRNAs targeted by miRNAs have shown to influence can-cer risk as well.For example,let-7,a tumor suppressor miRNA known to target the mRNA of the KRAS onco-gene,has been proven to be unable to bind to a KRAS SNP variant found to be disproportionately enriched in non-small cell lung carcinoma(NSCLC)(present in18%-20% of cases).46This same KRAS variant was also associated with an increased risk of developing epithelial ovarian can-cer,afinding that was consistent among3independent cohorts and2case-control studies.46Moreover,it was pres-ent in61%of patients with a history of hereditary breast and ovarian cancer who were previously genetically uninfor-mative(BRCA1-2negative).46This suggests that the SNP variant may be a new independent cancer risk biomarker for hereditary breast and ovarian cancer families.To date,most studies of miRNA binding-site SNPs have followed the case-control study design,and therefore they are centered on cancer risks.Consequently,the majority of our knowl-edge to date centers on cancer risk.For more information regarding miRNA binding-site SNPs considered to be can-cer risk biomarkers,see the recent review by Preskill and Weidhaas.46MiRNAs as HormonesA large amount of data have accumulated over the last few years describing the participation of miRNAs and micro-environment cells in malignant processes.47,48Similar to hormones,miRNAs are released by a donor cell as“free”molecules or in secreted vesicles,and delivered from bodilyfluids into receptor cells located in other areas.17For exam-ple,the tumor suppressor miR-143was found to be released by normal epithelial prostate cells,inducing growth inhibition of prostate cancer cells in vitro and in vivo.49Similarly,intercellular transfer of miR-142andmiR-223from immune cells to malignant cells(hepatocel-lular carcinoma cells)inhibited proliferation and destabi-lized microtubule regulation in vitro.50In addition,in a breast cancer study,miR-210was shown to be released from tumor cells to endothelial cells,thereby promoting angiogenesis and metastasis through an exosomal-mediated transfer.51MiR-210was also proven to be traf-ficked through exosomes in a leukemia cell line(under hypoxic conditions).52Along with other oncogenic miR-NAs,the delivery of miR-210resulted in increased angio-genesis through the promotion of tube formation by endothelial cells in vitro.19MiRNAs Are Involved in All Cancer HallmarksThe hallmarks of cancer comprise the biological capabilities acquired during the stepwise process of developing human tumors(Fig.3).MiRNAs have the potential to influence these hallmarks,and the recognition of the applicability of these interactions will increasingly influence the develop-ment of new therapeutic alternatives for patients with can-cer.53,54Herein are some representative examples of miRNAs that act as regulators of tumor biology;more detailed information on the roles of miRNAs in cancer canbe found in several reviews.22,24,40,55,56Self-Sufficiency in Growth SignalsActivation of the RAS oncogene is a common event that allows tumor cells to escape growth factor dependency and become“oncogene addicted.”All3RAS genes(K-RAS, NRAS,and H-RAS)have been proven to be directly modu-lated by the tumor suppressor miRNA family of let-7’s in a posttranscriptional manner.57Alternatively,let-7also tar-gets high mobility group A2(HMGA2),a pleiotropic tran-scription factor.The characteristic downregulation of let-7 accompanying tumor development results in an increased expression of the RAS oncogenes along with their down-stream effects.It also derepresses HMGA2,therebyVOLUME64_NUMBER5_SEPTEMBER/OCTOBER2014315facilitatinganchorage-independent growth in cell cul-tures,58as well as cell proliferation and differentiation.59 Evidently,both of these mechanisms have significant rele-vance regarding tumorigenesis and cancer development. Furthermore,reconstituting the levels of let-7in an experi-mental approach demonstrated that this precursor miRNA family can inhibit cell proliferation,resulting in tumor regression as shown in lung cancer models in mice.60 Insensitivity to Antigrowth SignalsE2F is a group of genes that encode a family of transcrip-tion factors that tightly regulate cell cycle progression and DNA synthesis.Three of these,E2F1,E2F2,and E2F3a, are known as the cell cycle“activators,“and they represent attractive factors to target in cancer because they contrib-ute to uncontrolled cell growth.Several miRNAs have been demonstrated to have the potential to modulate the translation of the mRNAs of these transcription factors. For example,miR-20a,miR-17-5p,miR-93,and miR-106b have been shown to negatively regulate E2F1.61,62 Moreover,the miR-17-92cluster has been shown to decrease the levels of E2F1-3.63Therefore,in this sense,it is likely that the downregulation of these miRNAs in dif-ferent types of cancer could favor a proliferative transcrip-tional network contributing to tumorigenesis.Thus, reconstituting the basal levels of expression of these miRNAs(in E2Fs-dependent tumors)could serve as a future therapeutic alternative with clinical relevance.Some of these miRNAs are part of positive or negative feedback mechanisms,and thus the end result of modulating their levels still remains to be explored.Finally,in separate studies,the transcription factor FOXO1(tumor suppressor that controls proliferation and regulates apoptosis)was found to be decreased in classic Hodgkin lymphoma.In classic Hodgkin lymphoma cell lines,the levels of FOXO1were proven to be repressed by 3upregulated microRNAs:miR-96,miR-182,and miR-183.64This repression highly increased proliferation,and at the same time inhibited apoptosis in vitro.Evasion From ApoptosisApoptosis is a physiological self-destructive cellular mecha-nism that leads to the removal of unwanted cells.MiR-25 was identified as being elevated in cholangiocarcinoma cell lines as well as patient samples,and its increased levels were shown to contribute to the evasion of TNF-related apoptosis. In experiments that reduced levels of miR-25(in vitro),cells in culture were sensitized to apoptotic death,thereby impli-cating the miRNA in the control of tumor cell apoptosis.65 The cancer-associated genomic region1p36(frequently lost or rearranged in many types of leukemias)contains a crucial tumor suppressor,miR-34a.In neuroblastoma,loss of miR-34a synergizes with MYCN oncogene amplification, and miR-34a has been shown to be a MYCN-negative regulator.66In addition,miR-34a is known to induce cell cycle arrest and subsequent caspase-dependentFIGURE3.Examples of MicroRNAs Involved in the Cancer Hallmarks.The8biological capabilities acquired during the multistep development of human tumors include sustaining proliferative signaling,evading growth suppressors,resisting cell death,enabling replicative immortality,inducing angiogenesis,activating invasion and metastasis,reprogramming of energy metabolism,and evading immune destruction.For each,one representative example of a microRNA is presented.316CA:A Cancer Journal for Clinicians。