Effect of genetic divergence in identifying ancestral origin using HAPAA
- 格式:pdf
- 大小:549.32 KB
- 文档页数:7

Advances in Analytical Chemistry 分析化学进展, 2017, 7(2), 131-138Published Online May 2017 in Hans. /journal/aachttps:///10.12677/aac.2017.72018Determination of IAA and ABA in PlantTissue by the GPC-HPLC/MS/MSLiu Yang1, Shuai Liu1, Zongyi Wang2, Qingqin Cao3, Jianli Wang1, Yu Xing1, Ling Qin1*1Beijing Key Laboratory of New Technology in Agricultural Application, College of Plant Science and Technology, Beijing University of Agriculture, Beijing2Beijing Key Laboratory of Safety Detection and Control on Harmful Microbes and Pesticide Residues inAgricultural Products, Beijing University of Agriculture, Beijing3College of Biological Science and Engineering, Beijing University of Agriculture, Beijing CollaborativeInnovation Center for Eco-Environmental Improvement with Forestry and Fruit Trees, Beijing University ofAgriculture, BeijingReceived: May 8th, 2017; accepted: May 24th, 2017; published: May 27th, 2017AbstractThe effectiveness and efficiency of the plant hormone detection is a prerequisite for plant re-search. The sample was purified with the GPC system, and the method employed HPLC-MS/MS for multiple reaction monitoring of concentrations of IAA, ABA, and deuterated IAA and ABA analogs. The condition of extraction and purification of hormones were optimized by orthogonal design. The results show that the composite of 0.5 g samples, 80% methanol extraction solvent, concentration temperature 35˚C and C18 cartridge column for solid-phase extraction (SPE), were optimal extraction procedure for each type of plant tissue. The contents of IAA and ABA as high as 20.34 ng/g and 789.3 ng/g were achieved by this method, respectively; the detectable limits were 2.36 ng/g and 31.95 ng/g; the recoveries were 70.43% and 80.17%; the RSD were1.87% and2.26%.KeywordsLiquid Chromatography-Mass Spectrometry, Purifying-Quantitative-Concentrated More OnlineSystem, Internal Standard Method, Indole-3-Acetic, Abscisic AcidGPC-HPLC-MS/MS测定植物组织中IAA与ABA的方法杨柳1,刘帅1,王宗义2,曹庆芹3,王建立1,邢宇1,秦岭1**通讯作者。
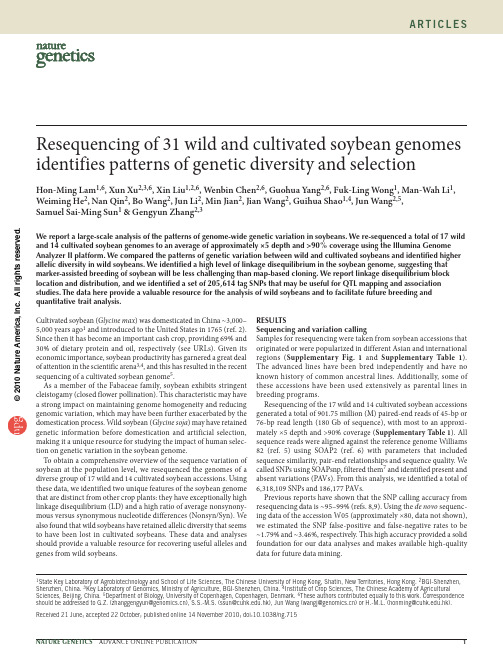


生物多样性 2005, 13 (5): 376-386 doi: 10.1360/biodiv.050070 Biodiversity Science http: //——————————————————收稿日期: 2005-03-15; 接受日期: 2005-04-19基金项目: 国家重点基础研究发展规划项目(G2000046805)和科技部国际科技合作重点项目计划(2001CB711103)资助* 通讯作者Author for correspondence. E-mail: gesong@第5期孙海芹等: 濒危植物独花兰的形态变异及其适应意义 377物种的遗传变异是长期进化的产物, 是物种生存适应和发展的前提(Soltis & Soltis, 1991; 葛颂, 1997)。
遗传变异体现在不同水平上, 如居群水平、个体水平、组织细胞水平以及分子水平等(Moritz & Hillis, 1996)。
因此, 检测遗传变异可以在不同层次利用不同的手段和方法进行:既包括传统的形态学和细胞学方法, 也包括新近发展起来的同工酶和DNA技术, 这些方法相互补充、相互印证, 为我们全面、深入认识遗传变异及其生物学意义提供有价值的资料(葛颂, 1997)。
从形态学或表型性状上来检测遗传变异是最古老也是最简便易行的方法(Schaal et al., 1991; 葛颂和洪德元, 1994)。
通常, 度量遗传变异的形态学性状主要有两类: 一是符合孟德尔遗传规律的单基因性状, 如质量形态性状、稀有突变等; 另一类是由多基因决定的数量性状, 如大多数形态性状和生活史性状等(葛颂和洪德元, 1994)。
在自然居群中, 植物的大多数形态性状是多基因决定的数量性状, 往往具有适应和进化意义, 故对其进行研究可以更清楚地揭示植物与其环境之间的关系, 有助于我们认识植物适应和进化的方式、机制及其影响因素, 加深对自然选择、基因流和遗传漂变等进化因素的理解(葛颂和洪德元, 1994; Schaal et al., 1991; Schwaegerle et al., 1986;葛颂, 1997)。

Documentation for structure software:Version2.3Jonathan K.Pritchard aXiaoquan Wen aDaniel Falush b123a Department of Human GeneticsUniversity of Chicagob Department of StatisticsUniversity of OxfordSoftware from/structure.htmlApril21,20091Our other colleagues in the structure project are Peter Donnelly,Matthew Stephens and Melissa Hubisz.2Thefirst version of this program was developed while the authors(JP,MS,PD)were in the Department of Statistics,University of Oxford.3Discussion and questions about structure should be addressed to the online forum at structure-software@.Please check this document and search the previous discus-sion before posting questions.Contents1Introduction31.1Overview (3)1.2What’s new in Version2.3? (3)2Format for the datafile42.1Components of the datafile: (4)2.2Rows (5)2.3Individual/genotype data (6)2.4Missing genotype data (7)2.5Formatting errors (7)3Modelling decisions for the user73.1Ancestry Models (7)3.2Allele frequency models (12)3.3How long to run the program (13)4Missing data,null alleles and dominant markers144.1Dominant markers,null alleles,and polyploid genotypes (14)5Estimation of K(the number of populations)155.1Steps in estimating K (15)5.2Mild departures from the model can lead to overestimating K (16)5.3Informal pointers for choosing K;is the structure real? (16)5.4Isolation by distance data (17)6Background LD and other miscellania176.1Sequence data,tightly linked SNPs and haplotype data (17)6.2Multimodality (18)6.3Estimating admixture proportions when most individuals are admixed (18)7Running structure from the command line197.1Program parameters (19)7.2Parameters infile mainparams (19)7.3Parameters infile extraparams (21)7.4Command-line changes to parameter values (25)8Front End268.1Download and installation (26)8.2Overview (27)8.3Building a project (27)8.4Configuring a parameter set (28)8.5Running simulations (30)8.6Batch runs (30)8.7Exporting parameterfiles from the front end (30)8.8Importing results from the command-line program (31)8.9Analyzing the results (32)9Interpreting the text output339.1Output to screen during run (34)9.2Printout of Q (34)9.3Printout of Q when using prior population information (35)9.4Printout of allele-frequency divergence (35)9.5Printout of estimated allele frequencies(P) (35)9.6Site by site output for linkage model (36)10Other resources for use with structure3710.1Plotting structure results (37)10.2Importing bacterial MLST data into structure format (37)11How to cite this program37 12Bibliography371IntroductionThe program structure implements a model-based clustering method for inferring population struc-ture using genotype data consisting of unlinked markers.The method was introduced in a paper by Pritchard,Stephens and Donnelly(2000a)and extended in sequels by Falush,Stephens and Pritchard(2003a,2007).Applications of our method include demonstrating the presence of popu-lation structure,identifying distinct genetic populations,assigning individuals to populations,and identifying migrants and admixed individuals.Briefly,we assume a model in which there are K populations(where K may be unknown), each of which is characterized by a set of allele frequencies at each locus.Individuals in the sample are assigned(probabilistically)to populations,or jointly to two or more populations if their genotypes indicate that they are admixed.It is assumed that within populations,the loci are at Hardy-Weinberg equilibrium,and linkage equilibrium.Loosely speaking,individuals are assigned to populations in such a way as to achieve this.Our model does not assume a particular mutation process,and it can be applied to most of the commonly used genetic markers including microsatellites,SNPs and RFLPs.The model assumes that markers are not in linkage disequilibrium(LD)within subpopulations,so we can’t handle markers that are extremely close together.Starting with version2.0,we can now deal with weakly linked markers.While the computational approaches implemented here are fairly powerful,some care is needed in running the program in order to ensure sensible answers.For example,it is not possible to determine suitable run-lengths theoretically,and this requires some experimentation on the part of the user.This document describes the use and interpretation of the software and supplements the published papers,which provide more formal descriptions and evaluations of the methods.1.1OverviewThe software package structure consists of several parts.The computational part of the program was written in C.We distribute source code as well as executables for various platforms(currently Mac,Windows,Linux,Sun).The C executable reads a datafile supplied by the user.There is also a Java front end that provides various helpful features for the user including simple processing of the output.You can also invoke structure from the command line instead of using the front end.This document includes information about how to format the datafile,how to choose appropriate models,and how to interpret the results.It also has details on using the two interfaces(command line and front end)and a summary of the various user-defined parameters.1.2What’s new in Version2.3?The2.3release(April2009)introduces new models for improving structure inference for data sets where(1)the data are not informative enough for the usual structure models to provide accurate in-ference,but(2)the sampling locations are correlated with population membership.In this situation, by making explicit use of sampling location information,we give structure a boost,often allowing much improved performance(Hubisz et al.,2009).We hope to release further improvements in the coming months.loc a loc b loc c loc d loc eGeorge1-914566092George1-9-964094Paula110614268192Paula110614864094Matthew2110145-9092Matthew2110148661-9Bob210814264194Bob2-9142-9094Anja1112142-91-9Anja111414266194Peter1-9145660-9Peter1110145-91-9Carsten2108145620-9Carsten211014564192Table1:Sample datafile.Here MARKERNAMES=1,LABEL=1,POPDATA=1,NUMINDS=7, NUMLOCI=5,and MISSING=-9.Also,POPFLAG=0,LOCDATA=0,PHENOTYPE=0,EX-TRACOLS=0.The second column shows the geographic sampling location of individuals.We can also store the data with one row per individual(ONEROWPERIND=1),in which case thefirst row would read“George1-9-9145-96664009294”.2Format for the datafileThe format for the genotype data is shown in Table2(and Table1shows an example).Essentially, the entire data set is arranged as a matrix in a singlefile,in which the data for individuals are in rows,and the loci are in columns.The user can make several choices about format,and most of these data(apart from the genotypes!)are optional.For a diploid organism,data for each individual can be stored either as2consecutive rows, where each locus is in one column,or in one row,where each locus is in two consecutive columns. Unless you plan to use the linkage model(see below)the order of the alleles for a single individual does not matter.The pre-genotype data columns(see below)are recorded twice for each individual. (More generally,for n-ploid organisms,data for each individual are stored in n consecutive rows unless the ONEROWPERIND option is used.)2.1Components of the datafile:The elements of the inputfile are as listed below.If present,they must be in the following order, however most are optional(as indicated)and may be deleted completely.The user specifies which data are present,either in the front end,or(when running structure from the command line),in a separatefile,mainparams.At the same time,the user also specifies the number of individuals and the number of loci.2.2Rows1.Marker Names(Optional;string)Thefirst row in thefile can contain a list of identifiersfor each of the markers in the data set.This row contains L strings of integers or characters, where L is the number of loci.2.Recessive Alleles(Data with dominant markers only;integer)Data sets of SNPs or mi-crosatellites would generally not include this line.However if the option RECESSIVEALLE-LES is set to1,then the program requires this row to indicate which allele(if any)is recessive at each marker.See Section4.1for more information.The option is used for data such as AFLPs and for polyploids where genotypes may be ambiguous.3.Inter-Marker Distances(Optional;real)the next row in thefile is a set of inter-markerdistances,for use with linked loci.These should be genetic distances(e.g.,centiMorgans),or some proxy for this based,for example,on physical distances.The actual units of distance do not matter too much,provided that the marker distances are(roughly)proportional to recombination rate.The front end estimates an appropriate scaling from the data,but users of the command line version must set LOG10RMIN,LOG10RMAX and LOG10RSTART in thefile extraparams.The markers must be in map order within linkage groups.When consecutive markers are from different linkage groups(e.g.,different chromosomes),this should be indicated by the value-1.Thefirst marker is also assigned the value-1.All other distances are non-negative.This row contains L real numbers.4.Phase Information(Optional;diploid data only;real number in the range[0,1]).This isfor use with the linkage model only.This is a single row of L probabilities that appears after the genotype data for each individual.If phase is known completely,or no phase information is available,these rows are unnecessary.They may be useful when there is partial phase information from family data or when haploid X chromosome data from males and diploid autosomal data are input together.There are two alternative representations for the phase information:(1)the two rows of data for an individual are assumed to correspond to the paternal and maternal contributions,respectively.The phase line indicates the probability that the ordering is correct at the current marker(set MARKOVPHASE=0);(2)the phase line indicates the probability that the phase of one allele relative to the previous allele is correct(set MARKOVPHASE=1).Thefirst entry should befilled in with0.5tofill out the line to L entries.For example the following data input would represent the information from an male with5unphased autosomal microsatellite loci followed by three X chromosome loci, using the maternal/paternal phase model:102156165101143105104101100148163101143-9-9-90.50.50.50.50.5 1.0 1.0 1.0where-9indicates”missing data”,here missing due to the absence of a second X chromo-some,the0.5indicates that the autosomal loci are unphased,and the1.0s indicate that the X chromosome loci are have been maternally inherited with probability1.0,and hence are phased.The same information can be represented with the markovphase model.In this case the inputfile would read:102156165101143105104101100148163101143-9-9-90.50.50.50.50.50.5 1.0 1.0Here,the two1.0s indicate that thefirst and second,and second and third X chromosome loci are perfectly in phase with each other.Note that the site by site output under these two models will be different.In thefirst case,structure would output the assignment probabilities for maternal and paternal chromosomes.In the second case,it would output the probabilities for each allele listed in the inputfile.5.Individual/Genotype data(Required)Data for each sampled individual are arranged intoone or more rows as described below.2.3Individual/genotype dataEach row of individual data contains the following elements.These form columns in the datafile.bel(Optional;string)A string of integers or characters used to designate each individualin the sample.2.PopData(Optional;integer)An integer designating a user-defined population from which theindividual was obtained(for instance these might designate the geographic sampling locations of individuals).In the default models,this information is not used by the clustering algorithm, but can be used to help organize the output(for example,plotting individuals from the same pre-defined population next to each other).3.PopFlag(Optional;0or1)A Booleanflag which indicates whether to use the PopDatawhen using learning samples(see USEPOPINFO,below).(Note:A Boolean variable(flag)isa variable which takes the values TRUE or FALSE,which are designated here by the integers1(use PopData)and0(don’t use PopData),respectively.)4.LocData(Optional;integer)An integer designating a user-defined sampling location(orother characteristic,such as a shared phenotype)for each individual.This information is used to assist the clustering when the LOCPRIOR model is turned on.If you simply wish to use the PopData for the LOCPRIOR model,then you can omit the LocData column and set LOCISPOP=1(this tells the program to use PopData to set the locations).5.Phenotype(Optional;integer)An integer designating the value of a phenotype of interest,foreach individual.(φ(i)in table.)(The phenotype information is not actually used in structure.It is here to permit a smooth interface with the program STRAT which is used for association mapping.)6.Extra Columns(Optional;string)It may be convenient for the user to include additionaldata in the inputfile which are ignored by the program.These go here,and may be strings of integers or characters.7.Genotype Data(Required;integer)Each allele at a given locus should be coded by a uniqueinteger(eg microsatellite repeat score).2.4Missing genotype dataMissing data should be indicated by a number that doesn’t occur elsewhere in the data(often-9 by convention).This number can also be used where there is a mixture of haploid and diploid data (eg X and autosomal loci in males).The missing-data value is set along with the other parameters describing the characteristics of the data set.2.5Formatting errors.We have implemented reasonably careful error checking to make sure that the data set is in the correct format,and the program will attempt to provide some indication about the nature of any problems that exist.The front end requires returns at the ends of each row,and does not allow returns within rows;the command-line version of structure treats returns in the same way as spaces or tabs.One problem that can arise is that editing programs used to assemble the data prior to importing them into structure can introduce hidden formatting characters,often at the ends of lines,or at the end of thefile.The front end can remove many of these automatically,but this type of problem may be responsible for errors when the datafile seems to be in the right format.If you are importing data to a UNIX system,the dos2unix function can be helpful for cleaning these up.3Modelling decisions for the user3.1Ancestry ModelsThere are four main models for the ancestry of individuals:(1)no admixture model(individuals are discretely from one population or another);(2)the admixture model(each individual draws some fraction of his/her genome from each of the K populations;(3)the linkage model(like the admixture model,but linked loci are more likely to come from the same population);(4)models with informative priors(allow structure to use information about sampling locations:either to assist clustering with weak data,to detect migrants,or to pre-define some populations).See Pritchard et al.(2000a)and(Hubisz et al.,2009)for more on models1,2,and4and Falush et al.(2003a)for model3.1.No admixture model.Each individual comes purely from one of the K populations.The output reports the posterior probability that individual i is from population k.The prior probability for each population is1/K.This model is appropriate for studying fully discrete populations and is often more powerful than the admixture model at detecting subtle structure.2.Admixture model.Individuals may have mixed ancestry.This is modelled by saying that individual i has inherited some fraction of his/her genome from ancestors in population k.The output records the posterior mean estimates of these proportions.Conditional on the ancestry vector,q(i),the origin of each allele is independent.We recommend this model as a starting point for most analyses.It is a reasonablyflexible model for dealing with many of the complexities of real populations.Admixture is a common feature of real data,and you probably won’tfind it if you use the no-admixture model.The admixture model can also deal with hybrid zones in a natural way.Label Pop Flag Location Phen ExtraCols Loc1Loc2Loc3....Loc LM1M2M3....M Lr1r2r3....r L-1D1,2D2,3....D L−1,LID(1)g(1)f(1)l(1)φ(1)y(1)1,...,y(1)n x(1,1)1x(1,1)2x(1,1)3....x(1,1)LID(1)g(1)f(1)l(1)φ(1)y(1)1,...,y(1)n x(1,2)1x(1,2)2x(1,2)3....x(1,2)Lp(1)1p(1)2p(1)3....p(1)LID(2)g(2)f(2)l(2)φ(2)y(2)1,...,y(2)n x(2,1)1x(2,1)2x(2,1)3....x(2,1)LID(2)g(2)f(2)l(2)φ(2)y(2)1,...,y(2)n x(2,2)1x(2,2)2x(2,2)3....x(2,2)Lp(2)1p(2)2p(2)3....p(2)L ....ID(i)g(i)f(i)l(i)φ(i)y(i)1,...,y(i)n x(i,1)1x(i,1)2x(i,1)3....x(i,1)LID(i)g(i)f(i)l(i)φ(i)y(i)1,...,y(i)n x(i,2)1x(i,2)2x(i,2)3....x(i,2)Lp(3)1p(3)2p(3)3....p(3)L ....ID(N)g(N)f(N)l(N)φ(N)y(N)1,...,y(N)n x(N,1)1x(N,1)2x(N,1)3....x(N,1)LID(N)g(N)f(N)l(N)φ(N)y(N)1,...,y(N)n x(N,2)1x(N,2)2x(N,2)3....x(N,2)Lp(L)1p(L)2p(L)3....p(1)LTable2:Format of the datafile,in two-row format.Most of these components are optional(see text for details).M l is an identifier for marker l.r l indicates which allele,if any,is recessive at each marker(dominant genotype data only).D i,i+1is the distance between markers i and i+1.ID(i) is the label for individual i,g(i)is a predefined population index for individual i(PopData);f(i)is aflag used to incorporate learning samples(PopFlag);l(i)is the sampling location of individual i (LocData);φ(i)can store a phenotype for individual i;y(i)1,...,y(i)n are for storing extra data(ignoredby the program);(x i,1l ,x i,2l)stores the genotype of individual i at locus l.p(l)i is the phase informationfor marker l in individual i.3.Linkage model.This is essentially a generalization of the admixture model to deal with“ad-mixture linkage disequilibrium”–i.e.,the correlations that arise between linked markers in recently admixed populations.Falush et al.(2003a)describes the model,and computations in more detail.The basic model is that,t generations in the past,there was an admixture event that mixed the K populations.If you consider an individual chromosome,it is composed of a series of“chunks”that are inherited as discrete units from ancestors at the time of the admixture.Admixture LD arises because linked alleles are often on the same chunk,and therefore come from the same ancestral population.The sizes of the chunks are assumed to be independent exponential random variables with mean length1/t(in Morgans).In practice we estimate a“recombination rate”r from the datathat corresponds to the rate of switching from the present chunk to a new chunk.1Each chunkin individual i is derived independently from population k with probability q(i)k ,where q(i)kis theproportion of that individual’s ancestry from population k.Overall,the new model retains the main elements of the admixture model,but all the alleles that are on a single chunk have to come from the same population.The new MCMC algorithm integrates over the possible chunk sizes and break points.It reports the overall ancestry for each individual,taking account of the linkage,and can also report the probability of origin of each bit of chromosome,if desired by the user.This new model performs better than the original admixture model when using linked loci to study admixed populations.It achieves more accurate estimates of the ancestry vector,and can extract more information from the data.It should be useful for admixture mapping.The model is not designed to deal with background LD between very tightly linked markers.Clearly,this model is a big simplification of the complex realities of most real admixed popu-lations.However,the major effect of admixture is to create long-range correlation among linked markers,and so our aim here is to encapsulate that feature within a fairly simple model.The computations are a bit slower than for the admixture model,especially with large K and unphased data.Nonetheless,they are practical for thousands of sites and individuals and multiple populations.The model can only be used if there is information about the relative positions of the markers(usually a genetic map).ing prior population information.The default mode for structure uses only genetic information to learn about population structure.However,there is often additional information that might be relevant to the clustering(e.g.,physical characteristics of sampled individuals or geographic sampling locations).At present,structure can use this information in three ways:•LOCPRIOR models:use sampling locations as prior information to assist the clustering–for use with data sets where the signal of structure is relatively weak2.There are some data sets where there is genuine population structure(e.g.,significant F ST between sampling locations),but the signal is too weak for the standard structure models to detect.This is often the case for data sets with few markers,few individuals,or very weak structure.To improve performance in this situation,Hubisz et al.(2009)developed new models that make use of the location information to assist clustering.The new models can often provide accurate inference of population structure and individual ancestry in data sets where the signal of structure is too weak to be found using the standard structure models.Briefly,the rationale for the LOCPRIOR models is as ually,structure assumes that all partitions of individuals are approximately equally likely a priori.Since there is an immense number of possible partitions,it takes highly informative data for structure to 1Because of the way that this is parameterized,the map distances in the inputfile can be in arbitrary units–e.g.,genetic distances,or physical distances(under the assumption that these are roughly proportional to genetic distances).Then the estimated value of r represents the rate of switching from one chunks to the next,per unit of whatever distance was assumed in the inputfile.E.g.,if an admixture event took place ten generations ago,then r should be estimated as0.1when the map distances are measured in cM(this is10∗0.01,where0.01is the probability of recombination per centiMorgan),or as10−4=10∗10−5when the map distances are measured in KB(assuming a constant crossing-over rate of1cM/MB).The prior for r is log-uniform.The front end tries to make some guesses about sensible upper and lower bounds for r,but the user should adjust these to match the biology of the situation.2Daniel refers to this as“Better priors for worse data.”conclude that any particular partition of individuals into clusters has compelling statistical support.In contrast,the LOCPRIOR models take the view that in practice,individuals from the same sampling location often come from the same population.Therefore,the LOCPRIOR models are set up to expect that the sampling locations may be informative about ancestry. If the data suggest that the locations are informative,then the LOCPRIOR models allow structure to use this information.Hubisz et al.(2009)developed a pair of LOCPRIOR models:for no-admixture and for admix-ture.In both cases,the underlying model(and the likelihood)is the same as for the standard versions.The key difference is that structure is allowed to use the location information to assist the clustering(i.e.,by modifying the prior to prefer clustering solutions that correlate with the locations).The LOCPRIOR models have the desirable properties that(i)they do not tend tofind struc-ture when none is present;(ii)they are able to ignore the sampling information when the ancestry of individuals is uncorrelated with sampling locations;and(iii)the old and new models give essentially the same answers when the signal of population structure is very strong.Hence,we recommend using the new models in most situations where the amount of available data is very limited,especially when the standard structure models do not provide a clear signal of structure.However,since there is now a great deal of accumulated experience with the standard structure models,we recommend that the basic models remain the default for highly informative data sets(Hubisz et al.,2009).To run the LOCPRIOR model,the user mustfirst specify a“sampling location”for each individual,coded as an integer.That is,we assume the samples were collected at a set of discrete locations,and we do not use any spatial information about the locations.(We recognize that in some studies,every individual may be collected at a different location,and so clumping individuals into a smaller set of discrete locations may not be an ideal representation of the data.)The“locations”could also represent a phenotype,ecotype,or ethnic group. The locations are entered into the inputfile either in the PopData column(set LOCISPOP=1), or as a separate LocData column(see Section2.3).To use the LOCPRIOR model you must first specify either the admixture or no-admixture models.If you are using the Graphical User Interface version,tick the“use sampling locations as prior”box.If you are using the command-line version,set LOCPRIOR=1.(Note that LOCPRIOR is incompatible with the linkage model.)Our experience so far is that the LOCPRIOR model does not bias towards detecting structure spuriously when none is present.You can use the same diagnostics for whether there is genuine structure as when you are not using a LOCPRIOR.Additionally it may be helpful to look at the value of r,which parameterizes the amount of information carried by the locations. Values of r near1,or<1indicate that the locations are rger values of r indicate that either there is no population structure,or that the structure is independent of the locations.•USEPOPINFO model:use sampling locations to test for migrants or hybrids–for use with data sets where the data are very informative.In some data sets,the user mightfind that pre-defined groups(eg sampling locations)correspond almost exactly to structure clusters,except for a handful of individuals who seem to be misclassified.Pritchard et al.(2000a)developed a formal Bayesian test for evaluating whether any individuals in the sample are immigrants to their supposed populations,or have recent immigrant ancestors.Note that this model assumes that the predefined populations are usually correct.It takes quite strong data to overcome the prior against misclassification.Before using the USEPOPINFO model,you should also run the program without population information to ensure that the pre-defined populations are in rough agreement with the genetic information.To use this model set USEPOPINFO to1,and choose a value of MIGRPRIOR(which isνin Pritchard et al.(2000a)).You might choose something in the range0.001to0.1forν.The pre-defined population for each individual is set in the input datafile(see PopData).In this mode,individuals assigned to population k in the inputfile will be assigned to cluster k in the structure algorithm.Therefore,the predefined populations should be integers between 1and MAXPOPS(K),inclusive.If PopData for any individual is outside this range,their q will be updated in the normal way(ie without prior population information,according to the model that would be used if USEPOPINFO was turned off.3).•USEPOPINFO model:pre-specify the population of origin of some individuals to assist ancestry estimation for individuals of unknown origin.A second way to use the USEPOPINFO model is to define“learning samples”that are pre-defined as coming from particular clusters.structure is then used to cluster the remaining individuals.Note:In the Front End,this option is switched on using the option“Update allele frequencies using only individuals with POPFLAG=1”,located under the“Advanced Tab”.Learning samples are implemented using the PopFlag column in the datafile.The pre-defined population is used for those individuals for whom PopFlag=1(and whose PopData is in(1...K)).The PopData value is ignored for individuals for whom PopFlag=0.If there is no PopFlag column in the datafile,then when USEPOPINFO is turned on,PopFlag is set to1 for all individuals.Ancestry of individuals with PopFlag=0,or with PopData not in(1...K) are updated according to the admixture or no-admixture model,as specified by the user.As noted above,it may be helpful to setαto a sensible value if there are few individuals without predefined populations.This application of USEPOPINFO can be helpful in several contexts.For example,there may be some individuals of known origin,and the goal is to classify additional individuals of unknown origin.For example,we might collect data from a set of dogs of known breeds (numbered1...K),and then use structure to estimate the ancestry for additional dogs of unknown(possibly hybrid)origin.By pre-setting the population numbers,we can ensure that the structure clusters correspond to pre-defined breeds,which makes the output more interpretable,and can improve the accuracy of the inference.(Of course,if two pre-defined breeds are genetically identical,then the dogs of unknown origin may be inferred to have mixed ancestry.Another use of USEPOPINFO is for cases where the user wants to update allele frequen-cies using only a subset of the individuals.Ordinarily,structure analyses update the allele frequency estimates using all available individuals.However there are some settings where you might want to estimate ancestry for some individuals,without those individuals affecting the allele frequency estimates.For example you may have a standard collection of learning samples,and then periodically you want to estimate ancestry for new batches of genotyped 3If the admixture model is used to estimate q for those individuals without prior population information,αis updated on the basis of those individuals only.If there are very few such individuals,you may need tofixαat a sensible value.。

利用 RACE 技术获得甜菜 M14 品系特异表达基因 M14-86 的 5′末端陈速黑龙江大学生命科学学院,黑龙江哈尔滨(150080)E-mail:chensu208@摘 要:本文利用 RACE 技术对从 M14 品系花期特异表达基因 cDNA 文库中筛选获得的 cDNA Me-c86(Me-c86 所在基因命名为 M14-86)进行 5′-RACE 扩增,并将所获得的 5′末 端 cDNA 与其 3′端 cDNA 序列进行拼接,获得的基因 M14-86 的 cDNA 全长为 1 330bp。
将 M14-86 全长 cDNA 序列提交到 GenBank 数据库进行 BLASTn 分析,结果表明, 该基因与大 花马齿苋(Portulaca grandiflora)等植物的 26S rRNA 基因序列相似性高达 97%,该基因全 长 cDNA 序列的获得,为进一步研究甜菜 M14 品系特异表达基因生物学特性提供了有用的 信息。
关键词:甜菜 M14 品系; M14-86;5′-RACE;BLAST 中图分类号:Q 71. 引言自 1998 年开始,郭德栋教授等以二倍体栽培甜菜(B.vulgaris L.)与四倍体野生种白花 甜菜(B.corolliflora Zoss.)进行种间杂交,获得了真实杂种 F1 VC88-1(VVCC,2n=36) , 然后通过与栽培甜菜回交,首次合成了异源三倍体甜菜(VVC,2n=27) ;获得了带有白花 甜菜染色体的完整栽培甜菜单体附加系,共 9 种类型(VV+1C1-9,2n=19) ,其中带有白花 甜菜 9 号染色体的单体附加系 M14 品系(VV+1C9,2n=19)其传递率在 96.5%以上。
通过 专家鉴定,认为带有白花甜菜第 9 号染色体的栽培甜菜单体附加系 M14 品系是克隆无融合 生殖基因极其难得的材料[1-5]。
cDNA 末端快速扩增技术[6](rapid amplification of cDNA ends, RACE) ,是一种从低丰度 转 录 本 中 快 速 扩 增 cDNA 5′ 和 3′ 末 端 简 单 而 有 效 的 方 法 , 也 被 称 为 锚 定 PCR (Anchored-PCR)或单边 PCR (one- sided PCR) 。
基因家族分析套路近年来,测序价格的下降,导致越来越多的基因组完成了测序,在数据库中形成了大量的可用资源。
如何利用这些资源呢?今天小编带你认识一下不测序也能发文章的思路--全基因组基因家族成员鉴定与分析(现在这一领域可是很热奥);一、基本分析内容数据库检索与成员鉴定进化树构建保守domain和motif分析.基因结构分析.转录组或荧光定量表达分析.二、数据库检索与成员鉴定1、数据库检索1)首先了解数据库用法,学会下载你要分析物种的基因组相关数据。
一般也就是下面这些数据库了Brachypodiumdb Genome Annotation Project :NCBI基因组数据库:)已鉴定的家族成员获取。
如何获得其他物种已发表某个基因家族的所有成员呢,最简单的就是下载该物种蛋白序列文件(可以从上述数据库中下载),然后按照文章中的ID,找到对应成员。
对于没有全基因组鉴定的,可以下列数据库中找:a. NCBI: nucleotide and protein db.b. EBI:c. UniProtKB、比对工具。
一般使用blast 和hmmer,具体使用命令如下:Local BLASTformatdb–i –p F/T;blastall–p blastp(orelse) –i –d –m 8 –b 2(or else) e 1 e-5 –o .-b:output two different members in subject sequences (db).Hmmer (hidden Markov Model) search. Thesame as PSI-BLAST in function. It has a higher sensitivity, but the speed islower.Command:、过滤。
Identity: 至少50%.Cover region: 也要超过50%或者蛋白结构域的长度.domain: 必须要有完整的该蛋白家族的。
2021,43(2)DOI :10.13836/j.jjau.2021049江西农业大学学报Acta Agriculturae Universitatis Jiangxiensishttp :// 平榛PYL 基因家族全基因组鉴定及果实发育表达分析张兴政,赵豫川,陈玥,孙博,刘剑锋*(吉林师范大学生命科学学院/吉林省植物资源科学与绿色生产重点实验室,吉林四平136000)摘要:【目的】植物激素脱落酸(Abscisic acid ,ABA )在植物生长发育、种子成熟及非生物胁迫响应中发挥重要作用。
PYR/PYL/RCAR (Pyrabactin Resistance1/PYR1-like/Regulatory Component of ABA Receptor ,PYLs )作为ABA 直接受体,在ABA 信号转导通路中起关键作用。
平榛(Corylus heteropylla )是东北地区重要的特色经济林木,其果实发育分子机制研究对于榛子实际生产具有重要意义。
【方法】为明确平榛ChPYLs 基因家族的进化关系,研究借助生物信息学,以课题组测序组装获得的平榛全基因组数据为基础,从中筛选获得8个ChPYLs 基因,且进一步明确了各基因的基因结构及其编码蛋白的理化性质、亚细胞定位、保守结构域、模体,并以现有高通量转录组数据为基础,明确了各成员在平榛子房及胚珠发育不同时期的表达水平,并采用qRT-PCR 对后者进行了验证。
【结果】ChPYLs 在进化上可分为3大群组,且同一物种不同成员在进化过程中可能由多次复制事件产生;基因家族各成员大多不含内含子,ChPYL4、ChPYL5和ChPYL8则含有1~2个内含子;各成员编码蛋白均含有典型的PYR_PYL_RCAR_like 结构域,且ChPYL1、ChPYL7、ChPYL3和ChPYL5还含有1个保守的Bet_v_1结构域;蛋白保守模体分析结果显示,在该基因家族编码蛋白中预测得到10种保守模体,且成员间所含的模体数量存在一定差异,所含的保守模体数量在4~6个,其中所有蛋白均含有Motif 1、Motif 2和Motif 3;基于子房和胚珠不同发育时期转录组数据,结果表明基因家族成员间表达模式存在显著差异,且大部分成员在子房和胚珠发育各时期表达水平较低;在子房发育不同时期,ChPYL4和ChPYL8在不同时期均高表达,后者表达水平更高;在胚珠发育不同时期,只有ChPYL4、ChPYL6和ChPYL8有表达,且只有ChPYL8在4个时期均高表达,且呈现双峰表达模式;结合qRT-PCR 结果分析发现,ChPYL4在OV1时期表达量最高,其他3个时期均低表达;ChPYL8表达呈现双峰模式,即OV2和OV4时期表达量更高。
收稿日期:2023-05-05作者简介:韩君(1988—),男,黑龙江哈尔滨人,硕士,工程师,研究方向为生物信息学与多组学数据分析。
刺猬线粒体基因组密码子偏好性分析韩君(北京康仁堂药业有限公司,北京101301)摘要:为利用分子技术探究刺猬皮等组织作为中药使用的机制,促进远东刺猬分子进化研究。
以远东刺猬线粒体全基因组序列为材料,从中筛选出长度大于300bp 的非重复编码序列(CDS )12条,利用CodonW1.4.2、SPSS 25.0和Excel 2007等软件分析其密码子偏好性。
结果显示:密码子第3位的碱基平均GC 含量为24.30%;有效密码子数目(ENC )分布范围为31.83~50.67,平均值为43.37;相对同义密码子使用度(RSCU )值>1.00的密码子共有32个,偏好以碱基A 或U (T )结尾。
中性绘图分析结果显示,GC 1和GC 2的平均值(GC 12)与GC 3之间的相关系数为0.443;ENC-plot 分析结果显示,多数基因在标准曲线附近聚集;对应性分析结果表明,第1~4个向量轴的贡献率分别为35.64%、16.22%、10.26%和9.13%,同义密码子第3位的GC 含量(GC3s )、ENC 与第1向量轴(Axis1)呈显著正相关;密码子适应指数(CAI )与Axis1呈负相关,最终确定CUA 、AUA 、GUU 、UCU 、CCC 、ACA 、GCU 、CAU 、AAA 、GAA 、UGA 、CGC 、GGC 和GGA 为最优密码子。
通过优化远东刺猬线粒体基因组密码子以及应用分子手段进行深入研究,有助于探究远东刺猬组织入药机制。
关键词:远东刺猬;线粒体;密码子;偏好性;中药中图分类号:S862;R282文献标志码:A文章编号:1001-0084(2023)04-0031-07Codon Preference Analysis on MitochondrialGenome of Erinaceus amurensisHAN Jun(Beijing Tcmages Pharmaceutical Co.,Ltd.,Beijing 101301,China )Abstract:Using molecular technique to explore the mechanism of hedgehog hide and other tissues applied astraditional Chinese medicine,and promote molecular evolution of Erinaceus amurensis in the Far East,taking the complete mitochondrial genome sequence of Erinaceus amurensis as the material,12non-repeating coding sequences (CDS)with a length greater than 300bp were selected as the research objects in this study,and their codon preference was analyzed by using CodonW1.4.2,SPSS 25.0,Excel 2007and other software.The average GC content of the third codon was 24.30%;the number of effective codons (ENC)ranged from 31.83to 50.67,with an average value of 43.37;and the relative synonymous codon usage (RSCU)value of 32codons was greater than 1.00,and the preference ends with either A or U (T).According to neutral plot analysis,the correlation coefficientbetween the average value (GC 12)of GC 1,and GC 2and GC 3was 0.443.In addition,ENC-plot analysis also revealed that most genes cluster near the standard curve;besides,the corresponding analysis showed that the contribution rates of the 1-4vector axes were 35.64%,16.22%,10.26%and 9.13%,respectively;and the GC content of the third synonymous codon (GC3s )and ENC were significantly positively correlated with the first vector axis (Axis1).In addition,the codon adaptation index (CAI)was negatively correlated with Axis1.Hence,it could finally beDOI:10.20041/ki.slbl.2023.04.006猬科在我国共有5个属7个种。
Effect of genetic divergence in identifying ancestral origin using HAPAAAndreas Sundquist,1Eugene Fratkin,1Chuong B.Do,Serafim Batzoglou2Department of Computer Science,Stanford University,Stanford,California94305,USAThe genome of an admixed individual with ancestors from isolated populations is a mosaic of chromosomal blocks, each following the statistical properties of variation seen in those populations.By analyzing polymorphisms in the admixed individual against those seen in representatives from the populations,we can infer the ancestral source of the individual’s haploblocks.In this paper we describe a novel approach for ancestry inference,HAPAA (HMM-based analysis of polymorphisms in admixed ancestries),that models the allelic and haplotypic variation in the populations and captures the signal of correlation due to linkage disequilibrium,resulting in greatly improved accuracy.We also introduce a methodology for evaluating the effect of genetic divergence between ancestral populations and time-to-admixture on inference ing HAPAA,we explore the limits of ancestry inference in closely related populations.[HAPAA is available at .]Human population migration,adaptation,and admixture have a chaotic and mostly undocumented history.However,nature has auspiciously recorded its account of events within our genomes, and we are at the cusp of an era where we will be able to unlock these records.An individual’s genome is a mosaic of ancestral haploblocks whose sizes depend on how far back in the ancestry we compare them.Because recombination can occur essentially anywhere in the genome,the precise boundaries and sources of these haploblocks cannot be easily inferred.However,if the hap-loblocks are derived from isolated human subpopulations,they will tend to follow the patterns of variation seen in those ing these patterns,we can partition an admixed indi-vidual’s genome into a mosaic of blocks derived from different populations.The inference of admixed ancestries is intriguing from a personal perspective because it speaks to an individual’s origins.In addition,it can be used in association mapping studies to identify loci relevant in genetic disease(McKeigue1998;Hog-gart et al.2004;Montana and Pritchard2004;Patterson et al. 2004;Zhu et al.2004,2005)and will help unravel some of the complexities in the history of human evolution.Although recent work suggests that human genomes differ significantly in many ways(Redon et al.2006),single nucleotide polymorphisms(SNPs)are ubiquitous and can serve as markers for the variation.Recent advances in genotyping technology al-low us to genotype hundreds of thousands of SNPs in a single experiment,making them a convenient vehicle for studying ge-nome-wide variation.For example,the Illumina HumanHap550 genotyping chip can assay over550,000tag-SNP loci for a few hundred dollars(/pages.ilmn?ID=154).Be-cause linkage disequilibrium(LD)has a strong effect at short genetic distances,the high-density coverage of such genotyping chips makes it possible to infer much of the intervening genomic variation(Carlson et al.2004).Using SNPs as a basis for variation, methods have been described recently that infer the ancestral population composition of admixed individuals,known as the ancestral haploblock reconstruction or inference problem.These methods are often probabilistic models that use the statistical properties of alleles seen in different populations to derive the most likely ancestral origin of each locus.For example,some methods use a first-order hidden Markov model(HMM)whose hidden states each correspond to an ancestral population(Falush et al.2003;Hoggart et al.2004;Patterson et al.2004;Zhu et al. 2004).Other methods use more complex models that account for some amount of LD between loci(Tang et al.2006).Here,we present two main contributions:(1)HAPAA(HMM-based analy-sis of polymorphisms in admixed ancestries),a novel approach for ancestral haploblock inference that is more accurate than previous methods();and(2)a method-ology that studies the limitations of inference as a function of both the genetic similarity between ancestral populations and the number of generations since first admixture between those populations.Unlike other methods,our inference methodology models long-range allelic correlations due to LD via a represen-tation that makes explicit the haplotypes seen in different popu-lations.By conducting large simulations of population evolu-tion,we are able to test the dependence of population divergence on ancestry inference.In contrast,tests done in the past have relied on a few specific populations with fixed divergence,for example the four in the HapMap data set(International HapMap Consortium2005).Together,our study allows us to better un-derstand the limitations of genomic analysis in decoding an in-dividual’s history of admixture.In Methods,we summarize the ancestral haploblock infer-ence problem in technical detail,review some previous inference methodologies,and finally describe the HAPAA method.In Re-sults,we first compare the performance of HAPAA to the best previous method,and then study the effect of population genetic divergence on ancestry inference.Finally,we describe our experi-ments in varying the input to our methodology and show that it is robust to changes in representing the populations. MethodsProblem formulationSuppose we have N populations P={P1,P2,...,P N},each repre-sented by a set of n p model individuals P p={a p1,a p2,...,a pnp}.For1These authors contributed equally to the work.2Corresponding author.E-mail serafim@;fax(650)725-1449.Article published online before print.Article and publication date are at http:///cgi/doi/10.1101/gr.072850.107.Freely available onlinethrough the Genome Research Open Access option.RECOMB Special/Methods676Genome Research18:676–682©2008by Cold Spring Harbor Laboratory Press;ISSN1088-9051/08;each individual a pk we have SNP genotypes sampled at L loci spaced across the genome,phased into two putative haplotypes a pk 0=〈a pk 01,a pk 02,...,a pk 0L 〉and a pk 1,where at each locus we have a pkhi ∈{A ,C ,G ,T ,מ}.We assume that the per-generation probability of recombina-tion (the genetic distance)between any two adjacent loci i and (i +1)is known to be R i for all populations.Given a new,potentially ad-mixed individual genotyped at thesame loci a g =〈a 1g ,a 2g ,...,a L g〉,we would like to determine the unob-served,true ancestral origin of each locus in the two haplotypes z m =〈z 1m ,z 2m ,...,z L m〉(maternally derived)andz f (paternal),where the ancestral ori-gin is confined to one of the givenpopulations z i m ,z i f ∈{1,...,N }.Thus,the problem of ancestral hap-loblock reconstruction can be seenas using a set of model individualsrepresenting the populations P and observed SNP genotypes a g to infer the “most likely ”ancestral assignment z i m∈{1,...,N }and z i f.For simplicity,let us begin by assuming that we know the truephasing of the individual,so that we can do inference on each haplotype independently.The problem thus reduces to assigning an ancestral origin to each SNP locus z i ∈{1,...,N }from a haplotype of alleles a i ∈{A ,C ,G ,T ,מ}.After we have solved thisproblem,we will extend our solution to unphased genotypes.Previous work Existing approaches vary considerably;our work follows meth-ods that model SNPs as the successive emissions of a probabilistic graphical model (Falush et al.2003;Hoggart et al.2004;Patter-son et al.2004;Zhu et al.2004).The model allows us to perform inference on a set of hidden states {S 1,S 2,...,S N },each corre-sponding to one of the N ancestral populations.Transitions be-tween the populations as we move along the genome are gov-erned by a Markov process.In a population state S p ,the modelprobabilistically emits alleles based on the frequencies seen in the model individuals in P p .An example of emission probabilities for a first-order HMM is P (a i =x |z i =S p )=(1/2n p )∑n pk =1∑1h =01[a pkhi =x ]where 1[condition ]∈{0,1}is the indicator function and x ∈{A ,C ,G ,T ,מ}.The method used in SABER (Tang et al.2006)improved on this by emitting alleles according to pair-allele frequencies P (a i =x |z ˆi =S p ,a i מ1).The probability of transi-tioning states P (z i +1=S p Ј|z i =S p )between two loci i and (i +1)depends on the genetic distance between the loci R i and genome-wide model parameters p ,the time since admixing for chromo-some blocks derived from population p ,which are learned from examples.The state diagram is depicted in Figure 1B.Although SABER attempts to address the problem via a sec-ond-order model,fixed-order models do not fully exploit the information available by examining the full haplotypes in the model individuals.Even though it is possible to further expand on SABER by devising a third-order or fourth-order model,the size of these models grows exponentially and becomes intrac-table to learn.HAPAA methodologyThe model To capture the effects of linkage disequilibrium at larger dis-tances,our methodology uses a representation of possible emis-sions that models long-range correlations between alleles in hap-lotypes.The HMM,depicted in Figure 1A,has an emitting state S pkh for the two haplotypes h ∈{0,1}of each model individual k in population p .In addition,there are non-emitting states {In p }and {Out p }for each population p ,that serve as the primary means of transitioning between haplotypes {S pkh }.If the hidden statevariable is denoted y i ,the probability of emission is given by the 5ן5matrix P (a i =x |y i =S pkh )=M (a pkhi ,x ).Here,M (x ,x )is typi-cally very likely,while M (x Ј,x x Ј)provides a small allowance for haplotypes not seen in the representative individuals,muta-tions,and genotyping error.Our HMM starts in an emitting state with equal probability for each population given by P (y 1=S pkh )=1/2Nn p .Each state S pkh can transition to three places:back to itself with probability (1מw pki )e מp R i ,to the other putative haplotype within the samemodel individual S pk (1מh )with probability w pki иe מp R i ,or to the exit state Out p with probability 1מe מp R i .The recombination rate parameters p are learned from training examples and can be interpreted as the reciprocal of the expected genetic length of ahaploblock inherited from population p .The constants w pki rep-resent the probability of a phasing switch error between loci i and (i +1)for model individual k in population p .In the ideal situa-tion with no phasing errors,we set w pki =0,in which case we will never transition directly between the two putative haplotypes ofan individual.The other way of transitioning between haplo-blocks is from S pkh to an Out p state,then to an In p Јstate with probability specified by the N ןN admixture matrix P (Out p →In p Ј)=A (p ,p Ј),and finally back to an emitting haplo-type state S p Јk Јh Јwith uniform probability 1/2n p Ј.Note that,in order to switch between haploblocks within the same population p ,we still transition to Out p and then In p with probability A (p ,p ).This hierarchical structure of our HMM is depicted in Figure1A.Figure 1.(A )Hierarchical HMM state diagram for HAPAA.On the left ,inter-and intra-populationtransitions occur with probabilities governed by matrix A (p ,p Ј).In the middle ,each population P p has a similar structure:entry state In p transitions with uniform probability to a diploid model individual a pk ,then to exit state Out p .On the right ,in a pk we transition into one of two states representing the haplotypes S pk 0and S pk 1of model individual a pk with equal probability.Each haplotype emits its alleles a pkhi via a mutation/error probability distribution M (a pkhi ,a i ).Haplotypes transition to each other with probability proportional to the phase switch error W pki ,and transition out of the diploid sample with probability governed by genetic distance to the next locus R i and the population-specific recombination rate parameter p .(B )HMM state diagram for previous methods.Each state represents a population and emits alleles accordingto frequency estimates for the populations,and admixture transition probabilities depend on the degree of admixture expected and other learned parameters.By construction,these methods assume a greaterdegree of independence between adjacent loci.Identifying ancestral origin using HAPAAGenome Research677Inference and testingWe infer the ancestral origins z i by first computing the standardforward␣pkhi,backwardpkhi,and posterior probability matrices ␥pkhi(Durbin et al.1998).We then compute the population-total posterior probability⌫pi=∑n p k=1∑1h=0␥pkhi and finally set zi=argmax p⌫pi,the population with maximal total posterior probability.In order to reduce the occurrence of false positives,we then apply a filtering procedure with a single parameter,the genetic length of the minimum acceptable block size.We partition z i into the largest consecutive blocks{j}of equal ancestry assign-ments.Every block that is larger thanis marked“solid”,and for each remaining smaller blockj we find the population of the last preceding solid block pop L(j)(if it exists)and the population of the first subsequent solid block pop R(j).Next,we recompute the forward,backward,and posterior matrices with additional con-straints:(1)for each solid blockj,we force the emitting states to be in population,pop(j),and(2)for each small blockj,we force the emitting states to be in either population pop L(j)or popula-tion pop R(j),and the only A(p,pЈ)transitions allowed are frompopL(j)or pop R(j)back to themselves,or one-way from pop L{j}topopR(j).Finally,we once again infer z i as described above.To test our model,ideally we would use real,labeled,ad-mixed individuals.Such data may become available in the future, but for now we synthesize test individuals using a model that we believe more closely reflects the properties of recombination.We construct a G th generation admixed individual by selecting2G (potentially redundant)ancestors from individuals left out for test set construction and simulating the mating process over G generations.For each chromosome,the number of recombina-tion points is chosen from a normal distribution with mean equal to the chromosome’s genetic length,with a minimum of one crossover per meiosis.The result is an admixed individual where each locus is annotated with its source population. TrainingFrom the above description,our model consists of the following parameters:the emission probability matrix M(xЈ,x),the recom-bination ratesp,and the admixture transition matrix A(p,pЈ). We perform supervised learning of these parameters using an EM algorithm on training examples(Durbin et al.1998).The ex-amples are labeled with their true ancestral origins z i,and we constrain the HMM so that if z i=p then y i=S pkh for some k and h,restricting ourselves to model haplotypes within the true population.Our filtering procedure adds an additional parameter ,which we train by maximizing one of our scoring metrics(de-scribed later in the paper)via a grid-search method.When real admixed training examples are not available,it is still possible to train using simulated admixed examples con-structed from the model individuals themselves,while at the same time avoiding overfitting.For all our experiments,we syn-thesize training examples from the model individuals using the same procedure described above for the generation of admixed test individuals.The result is a synthetic admixed haplotype a˜i, where,at each locus i,an allele can be annotated with the model haplotype from which it is derived:b˜i=〈p,k,h〉indicates that locus i is derived from model individual k haplotype h of popu-lation p.When training on a˜,we constrain the HMM so that at each locus i it is not allowed to be in the state corresponding toits source haplotype:y i S b˜i ,forcing it to model the trainingexample using the remaining model individuals.Extension to genotypesEarlier we assumed that we knew the true phasing of a g,but typically we would be presented with unphased genotypes a g=〈a1g,a2g,...,a L g〉.We extend our method to genotypes using the following iterative procedure.InitializationBased on the precomputed haplotypes of the model individuals apkh,construct an initial phasing of the genotypes of a g into two halotypes a m=〈a1m,a2m,...,a L m〉and a f using a program such as HAP(Halperin and Eskin2004),PHASE(Stephens et al.2001), fastPHASE(Scheet and Stephens2006),or an algorithm we de-veloped that is significantly faster at the expense of a marginal performance decrease(not described here).Between each pair of consecutive loci we describe the likelihood of a phase switch between the two haplotypes with a vector w i,the probability of a phase switch between loci i and(i+1).For phasing methods that do not estimate this directly,we set the vector to a uniform switch probability between heterozygous locations.Iterative stepCompute the forward and backward matrices using our HMM on each haplotype independently,producing a pkhim andpkhim for the putative maternal haplotype,and␣pkhif andpkhif for the paternal. Given the current phasing,we use our HMM model to compute the probability of witnessing these two haplotypes for any locus i asP͑a m,a f|C⌬i͒=͚ͩp=1N͚k=1n p͚h=01␣pkhi mиpkhi m͚ͪͩp=1N͚k=1n p͚h=01␣pkhi fиpkhi fͪwhere⌬i is the event that there is a phase switch error between locus i and(i+1).Suppose now that the haplotypes had exactly one phase switch error between locus i and(i+1).Then,we could compute the probability of witnessing the two haplotypes as:P͑a m,a f|⌬i͒=͚ͩp=1N͚k=1n p͚h=01␣pkhi mиpkhi f͚ͪͩp=1N͚k=1n p͚h=01␣pkhi fиpkhi mͪUsing the vector w i as a prior for the phase switch at i,we can use Bayes’rule to computeP͑⌬i|a m,a f͒=P͑a m,a f|⌬i͒иw iP͑a m,a f|⌬i͒иw i+P͑a m,a f|C⌬i͒и͑1−w i͒We compute this conditional probability for each locus and heuristically pick a set of loci H with the following procedure: 1.Find h=argmax i P(⌬i|a m,a f).If this probability is>1/2thenadd h to H,otherwise stop.2.Find maximum h L<h such that∑hמ1i=h Lwi>2and minimumhR>h such that∑h R i=h w i>2.Exclude the range[h L,h R]from further consideration and repeat step1.The limit of2was chosen to avoid selecting multiple nearby loci that stem from a single phase switch error.If the set H is empty,then we terminate the iterative procedure.Otherwise,we update the two haplotypes a m and a f by switching the phase at each locus in H and repeat the iterative step,not allowing the same loci in H to be picked again.Empirically,this procedure terminates after seven to20iterations.Sundquist et al.678Genome ResearchFinalizationCompute the posterior probabilities for the two haplotypes ␥pkhimand ␥pkhi f ,the population-total posteriors ⌫pi m and ⌫pi f,and finallydecode the inferred ancestries z i m and z i f.All tests in Results were conducted on unphased genotypes using this methodology.ResultsComparison to previous workWe benchmarked HAPAA against the current best-performing method,the Markov-HMM-based SABER (Tang et al.2006).We used the HapMap data set (International HapMap Consortium 2005),representing three populations:60unrelated North-Western Europeans (CEU),60Yoruban-Africans (YRI),and 90East Asians (ASN =CHB [Han Chinese]+JPT [Japanese]).We re-stricted the data set to the loci in the Illumina HumanHap550genotyping chip (/pages.ilmn?ID=154)within chromosome 22,spaced 4.5kb apart on average,and used a recombination rate map computed from HapMap (McVean et al.2004;Winckler et al.2005).We partitioned each population into two sets of individuals:5/6for the model individuals and for training,and 1/6for test set construction.Our test set comprised 400individuals,consisting of 20simulated diploid genotypes for each value of G ∈{1,2,...,20},which we phased using our own algorithm.Each test individual was derived by simulating the mating process over G generations,beginning with 2G ancestral individuals drawn with equal probability from each of the three populations.We constructed a training set in a similar fashion,picking ancestors from the model individuals instead,at the same time avoiding overfitting via the technique described in Methods for training HAPAA.We trained a single set of model parameters for all tests using our EM algorithm and optimized the filtering procedure by maximizing the accuracy of ancestry recall.To measure the performance of the two methods,we used the mean-square-error metric (Tang et al.2006),MSE =1L͚i =1L͚p =1N ͩ12͑⌫pi m+⌫pif͒−12͑1͓z i m =p ͔+1͓z i f =p ͔͒ͪ2where each of the maternal and paternal haplotypes contributes1/2to the measure.Figure 2is a demonstration of results pro-duced at different stages of inference by HAPAA compared to those by SABER.The performance comparison in Figure 3shows that HAPAA ’s inference is significantly more accurate,though there is a clear correlation between the methods.Because HAPAA relies on inferring a phasing of genotypes into two haplotypes,we found that for G =1,where entire chromosomes come from the same ancestry,phasing errors impair our performance com-pared to SABER.As the number of generations G increases,the problem of inferring the recombinations between ancestries dominates the problem of determining phase.However,HAPAA manages to infer the ancestral origin with higher fidelity than SABER by better modeling the effects of linkage disequilibrium in each population.As G approaches 20,the errors appear to level off as the distribution of expected haploblock sizes remains rela-tively stable.Effect of genetic divergence on inferenceAlthough the HapMap data set is useful for some basic validation,it is somewhat limiting for the purpose of studying the problem of ancestral inference.The genetic divergences between the four populations exemplify two extremes of the problem:Distin-guishing between haploblocks derived from CEU and YRI is rela-tively straightforward,while haploblocks from CHB and JPT are virtually indistinguishable.To better assess the performance of ancestry inference we created a novel testing methodology that measures performance as a function of the genetic distance be-tween populations.First,we construct pairs of populations separated by D ∈{100,200,...,2000}generations via simulation:Starting with the whole-genome HapMap CEU population restricted to the Illumina 550K sites,we simulate the divergence of two popu-lations over the course of D generations of random mating with fixed population sizes of 5000.The results have a strong depen-dence on this parameter —we chose it to be between the effective population sizes of 3100and 7500estimated by Tenesa et al.(2007).Other numbers for the effective human population size exist,but we chose this estimate specifically because it was based on the HapMap data set.Although we simulate recombination and genetic drift,we do not model selection or novel mutations,which would tend to make the populations more divergent and the ancestry inference problem easier.Other models incorporat-ing effects such as continuous gene flow may also affect the di-vergence.However,since human population history issuffi-Figure 3.Performance comparison between HAPAA and SABER.We measured the mean-square-error of the inferred posterior probability of population ancestry on chromosome 22for a varying number of genera-tions of admixture.Tests were constructed by simulating admixture over G generations from 2G individuals selected randomly from three HapMappopulations.Figure 2.Example inference on chromosome 22of an individual ad-mixed between three HapMap populations.The top two tracks represent the true ancestries,followed by three stages of HAPAA processing,and finally posterior probabilities and Viterbi decoding by SABER.The gray bars highlight two locations with correctly inferred ancestry but with phase switching errors between the haplotypes.Identifying ancestral origin using HAPAAGenome Research679ciently complex that there is no consensus on the most accurate model,we have chosen to use a simple,reasonable one.We ran-domly divide each population into a model/training partition consisting of 60individuals and the remainder for test set con-struction.For this data set,we measure the ability of our inference algorithm to recall a trace amount of ancestry from one popula-tion in a background of the other population.For example,sup-pose an individual ’s ancestors G generations ago consisted of 2G מ1individuals from population A and only one individual from population B.This methodology is illustrated in Figure 4.Testing different values of G ∈{1,2,...,20}and conditioned on the event that there exists some remaining ancestry derived from the minor population B,we measured our ability to detect the signal from the minor population.We report on our recall =true positives/(true positives +false negatives)and precision =true positives/(true positives +false positives)for correctly assigningthe minor ancestry to each locus z i m and z i f.To train our parameters,we constructed 2000simulated training genomes from the model individuals for each pair of populations parameterized by the number of generations of di-vergence D .We trained our model using EM and optimized our filtering procedure by maximizing the product of the recall and precision measure.We benchmarked the performance on a test set that consisted of 100admixed individuals derived from the test partition for each D and G ,for a total of 40,000full-genome inferences,and plot the results in Figure 5.It is clear that both genetic distance between populations and generations of admixture significantly affect the accuracy of inference.For populations that are not very divergent (D =100),it is possible to infer the ancestry of very recent admixture (G Յ2).However,as we increase the number of generations of admixture,there is not enough divergence between the popula-tions to correctly classify the haploblocks.In the other extreme,for populations that have been reproductively isolated by many generations (D =2000),inference is possible with high recall andprecision.From our simulations,even with G =10generations of admixture,HAPAA is able to detect the presence of haploblocks inherited from one individual in the minor population among haploblocks derived from 2G מ1ancestors in the other popula-tion.As our genomes recombine over a large number of genera-tions,most ancestral haploblocks will disappear.However,we estimate that even a 10th-generation ancestor has a significant probability of 26%of having a remaining haploblock.Therefore,for many individuals with ancestry admixed within 10genera-tions,we anticipate being able to detect the presence of both populations.Varying the number of model individualsUnlike profile-HMM approaches or the Markov-HMM of SABER,in HAPAA each model individual haplotype is a separate state in the HMM.To understand how the number of model individuals affects performance,we performed the following two experi-ments.Uniform population sizeAs in our previous comparison between HAPAA and SABER,we partitioned the HapMap data set on chromosome 22into model individuals and individuals used for test set generation.We con-structed an equal number of simulated test individuals for each G ∈{1,2,...,20}by mating 2G individuals drawn from the three populations with equal probability over G generations.Then,for x ∈{4,8,...,48}we restricted HAPAA to x model individuals in each population.We constructed a training set in a similar fash-ion to the testing set from the reduced number of model indi-viduals,and trained different parameters for each x to maximize inference accuracy.The mean-square-error for each test is plotted in Figure 6.Performance improves monotonically as we increase the number of model individuals in the HMM.Although we quickly see diminishing returns,the size of the underlying Hap-Map data set makes it impossible to assess at what pointperfor-Figure 4.Methodology for studying the effect of genetic divergence on ancestry inference.We simulate pairs of randomly mating populations of fixed size 5000derived from the HapMap CEU population over D generations.We construct training and test individuals derived over G generations of admixture from 2G מ1ancestors from one population and one ancestor from the other (minor)population.Figure 5.Recall and precision of detecting minor population.We simu-lated 20pairs of populations separated by D ∈{100,200,...,2000}generations of drift on the whole genome of Illumina 550K loci.For each D we constructed test individuals that were derived over G ∈{1,2,...,20}generations of admixture from 2G מ1ancestors from one popula-tion and one ancestor from the other (minor)population.Conditioned on the existence of at least one haploblock derived from the minor popu-lation,we measure the ability of HAPAA to identify these loci.Sundquist et al.680Genome Research。