crisprcas9技术-完整版
- 格式:pdf
- 大小:1.03 MB
- 文档页数:8

crisprcas9基因编辑技术流程下载温馨提示:该文档是我店铺精心编制而成,希望大家下载以后,能够帮助大家解决实际的问题。
文档下载后可定制随意修改,请根据实际需要进行相应的调整和使用,谢谢!并且,本店铺为大家提供各种各样类型的实用资料,如教育随笔、日记赏析、句子摘抄、古诗大全、经典美文、话题作文、工作总结、词语解析、文案摘录、其他资料等等,如想了解不同资料格式和写法,敬请关注!Download tips: This document is carefully compiled by theeditor. I hope that after you download them,they can help yousolve practical problems. The document can be customized andmodified after downloading,please adjust and use it according toactual needs, thank you!In addition, our shop provides you with various types ofpractical materials,such as educational essays, diaryappreciation,sentence excerpts,ancient poems,classic articles,topic composition,work summary,word parsing,copy excerpts,other materials and so on,want to know different data formats andwriting methods,please pay attention!CRISPR-Cas9 基因编辑技术流程一、准备工作阶段。

基因编辑技术CRISPRCas9的使用教程与最佳实践分享在现代生物学研究中,基因编辑技术的出现为研究人员提供了一种高效、精确、低成本的方式来研究基因功能和调控机制。
CRISPR-Cas9系统作为一种革命性的基因编辑工具被广泛应用于基因组编辑、疾病治疗和农业改良等领域。
本文将为您介绍CRISPR-Cas9基因编辑技术的使用教程,并分享一些最佳实践。
CRISPR-Cas9基因编辑技术概述CRISPR-Cas9是一种依靠细菌天然的免疫机制发展而来的基因编辑技术。
CRISPR是一种特殊的DNA序列,可与Cas9酶一起,通过识别和切割DNA序列来精确编辑基因组。
CRISPR-Cas9系统的主要组成部分包括CRISPR RNA (crRNA)、转录单元结构化RNA(tracrRNA)和Cas9酶。
crRNA负责识别目标DNA序列,而tracrRNA将crRNA与Cas9酶结合起来形成活跃的CRISPR-Cas9复合物。
CRISPR-Cas9基因编辑技术的使用教程1. 设计并合成RNA引导序列在使用CRISPR-Cas9进行基因编辑之前,首先需要设计并合成RNA引导序列。
该序列用于指导Cas9酶精确识别和切割目标基因组DNA。
合成的RNA引导序列通常由crRNA和tracrRNA合成而成,也可以合成一个融合的single-guide RNA (sgRNA)。
2. 构建CRISPR-Cas9载体CRISPR-Cas9基因编辑需要将Cas9酶和RNA引导序列导入目标细胞内。
可使用载体如质粒或病毒进行基因编辑构建。
选择合适的载体需考虑目标细胞类型、转染效率和所需编辑范围等因素。
将Cas9基因和RNA引导序列克隆至载体后,可通过转染或病毒介导转染等方法将其导入目标细胞。
3. 确定编辑效果在导入CRISPR-Cas9系统后,使用分子生物学方法来验证编辑效果。
例如,PCR、测序、Western blot或免疫组化等技术可以用于检测目标基因的突变、修复或敲除效果。

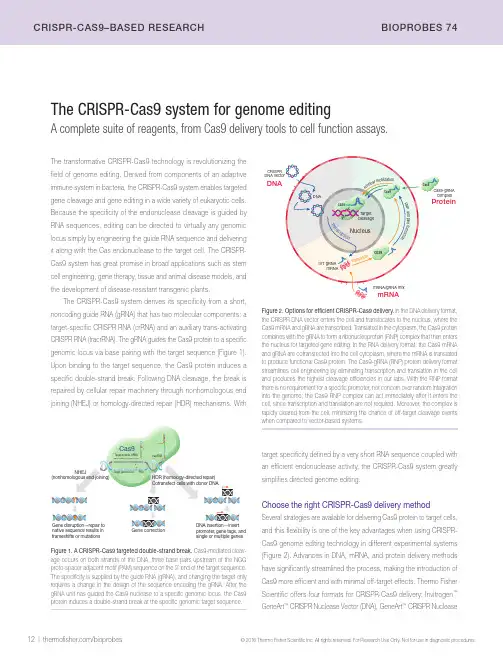
12 | /bioprobes© 2016 Thermo Fisher Scientific Inc. All rights reserved. For Research Use Only. Not for use in diagnostic procedures.CrIsPr-CAs9–BAsED rEsEArCH BIOPrOBEs 74The CRISPR-Cas9 system for genome editingA complete suite of reagents, from Cas9 delivery tools to cell function assays.The transformative CRISPR-Cas9 technology is revolutionizing the field of genome editing. Derived from components of an adaptive immune system in bacteria, the CRISPR-Cas9 system enables targeted gene cleavage and gene editing in a wide variety of eukaryotic cells. Because the specificity of the endonuclease cleavage is guided by RNA sequences, editing can be directed to virtually any genomic locus simply by engineering the guide RNA sequence and delivering it along with the Cas endonuclease to the target cell. The CRISPR-Cas9 system has great promise in broad applications such as stem cell engineering, gene therapy, tissue and animal disease models, and the development of disease-resistant transgenic plants.The CRISPR-Cas9 system derives its specificity from a short, noncoding guide RNA (gRNA) that has two molecular components: a target-specific CRISPR RNA (crRNA) and an auxiliary trans-activating CRISPR RNA (tracrRNA). The gRNA guides the Cas9 protein to a specific genomic locus via base pairing with the target sequence (Figure 1). Upon binding to the target sequence, the Cas9 protein induces a specific double-strand break. Following DNA cleavage, the break is repaired by cellular repair machinery through nonhomologous end joining (NHEJ) or homology-directed repair (HDR) mechanisms. Withtarget specificity defined by a very short RNA sequence coupled with an efficient endonuclease activity, the CRISPR-Cas9 system greatly simplifies directed genome editing.Choose the right CRISPR-Cas9 delivery methodSeveral strategies are available for delivering Cas9 protein to target cells,and this flexibility is one of the key advantages when using CRISPR-Cas9 genome editing technology in different experimental systems (Figure 2). Advances in DNA, mRNA, and protein delivery methods have significantly streamlined the process, making the introduction of Cas9 more efficient and with minimal off-target effects. Thermo Fisher Scientific offers four formats for CRISPR-Cas9 delivery: Invitrogen ™ GeneArt ™ CRISPR Nuclease Vector (DNA), GeneArt ™ CRISPR NucleaseFigure 1. A CRISPR-Cas9 targeted double-strand break. Cas9-mediated cleav-age occurs on both strands of the DNA, three base pairs upstream of the NGG proto-spacer adjacent motif (PAM) sequence on the 3’ end of the target sequence. The specificity is supplied by the guide RNA (gRNA), and changing the target only requires a change in the design of the sequence encoding the gRNA. After the gRNA unit has guided the Cas9 nuclease to a specific genomic locus, the Cas9 protein induces a double-strand break at the specific genomic target sequence.Figure 2. Options for efficient CRISPR-Cas9 delivery. In the DNA delivery format, the CRISPR DNA vector enters the cell and translocates to the nucleus, where the Cas9 mRNA and gRNA are transcribed. Translated in the cytoplasm, the Cas9 protein combines with the gRNA to form a ribonucleoprotein (RNP) complex that then enters the nucleus for targeted gene editing. In the RNA delivery format, the Cas9 mRNA and gRNA are cotransfected into the cell cytoplasm, where the mRNA is translated to produce functional Cas9 protein. The Cas9-gRNA (RNP) protein delivery format streamlines cell engineering by eliminating transcription and translation in the cell and produces the highest cleavage efficiencies in our labs. With the RNP format there is no requirement for a specific promoter, nor concern over random integration into the genome; the Cas9 RNP complex can act immediately after it enters the cell, since transcription and translation are not required. Moreover, the complex is rapidly cleared from the cell, minimizing the chance of off-target cleavage events when compared to vector-based systems.Cas9native sequence results in frameshifts or mutations, gene tags, and single or multiple genesCotransfect cells with donor DNA/bioprobes |13 © 2016 Thermo Fisher Scientific Inc. All rights reserved. For Research Use Only. Not for use in diagnostic procedures.BIOPrOBEs 74 CrIsPr-CAs9–BAsED rEsEArCHdirectly, and when antibiotic selection is used to identify transfected cells, viability assays can be used to monitor the selection process.Monitoring the efficiency of genome editing. When using genome editing tools—such as CRISPR-Cas9, TAL effectors, or zinc finger nucleases—to obtain targeted mutations, you need to determine the efficiency with which these nucleases cleave the target sequence, prior to continuing with labor-intensive and expensive experiments. The Invitrogen ™ GeneArt ™ Genomic Cleavage Detection Kit provides a simple and reliable assay for the cleavage efficiency of genome editing tools at a given locus. In this assay, a sample of the edited cell population is used as a direct PCR template for amplification with primers specific to the targeted region. The PCR product is then denatured and reannealed to produce heteroduplex mismatches where double-strand breaks have occurred, resulting in insertion/deletion (indel) introduction. These mismatches are recognized and cleaved by the detection enzyme, and the cleavage is easily detectable and quantifiable by gel analysis.Cell phenotyping. The CRISPR-Cas9 system is routinely used for knockout, knock-in, or modulation of gene expression, and the primaryon-target effects can be measured using cell analysis techniques; west-ern blotting, flow cytometry, and fluorescence microscopy are often used to view changes to protein expression or structure in a cell population. Flow cytometry provides the throughput for multiparameter analysis on vast numbers of individual cells. Cell imaging (Figure 4) allows for direct analysis of changes in protein expression, compartmentalization, and cell morphology; high-content analysis (HCA) provides automation for the imaging process coupled with quantitative rigor.mRNA (mRNA), GeneArt ™ Platinum ™ Cas9 Nuclease (protein) (Figure 2), and CRISPR library services (see page 11). Based on the cell type and application, the most effective delivery format can be chosen and then paired with optimal cell culture reagents and analysis tools.Monitor the genome editing process from start to finishWhichever CRISPR-Cas9 delivery strategy you choose, it is important to carefully monitor the entire genome editing process to validate that Cas9 protein has been successfully incorporated into cells and that the target knockout or mutation has been accurately implemented. This monitoring can be broken down into four categories:Cell culture. The starting point for genome editing is healthy cells. Performing cell health assays prior to using the CRISPR-Cas9 system can serve as an important quality control step and help to avoid wasting time and reagents. Tests for viability, apoptosis, and stress responses should be a routine part of cell growth and can provide information to optimize experimental conditions to produce the most robust cells.Genome editing. Immunochemical assays such as western blots can effectively measure the presence of Cas9 in cells. Figure 3 shows that the accumulation of Cas9 protein varies considerably depending on the choice of delivery method (plasmid, mRNA, or protein). Together with immunocytochemistry, antibiotic selection and gene expression are frequently used to monitor the assembly of CRISPR components for gene editing in the cell. Fluorescent protein expression can be measuredFigure 3. Western blot detection of Cas9 accumulation over time in cells transfected with Cas9-expressing plasmid DNA, Cas9 mRNA, or Cas9 protein. HEK293FT cells were transfected with Cas9 plasmid DNA, mRNA, or protein and then harvested at indicated times for western blot analysis. Proteins in the cell lysates were separated on an Invitrogen ™ NuPAGE ™ Novex ™ 4–12% Bis-Tris Protein Gel, transferred to a PVDF membrane using the Invitrogen ™ iBlot ™ 2 Gel Transfer Device, and incubated with an anti-Cas9 mouse monoclonal antibody at 1:3,000 dilution and an HRP-conjugated rabbit anti–mouse IgG antibody at 1:2,000. The membrane was developed using Thermo Scientific ™ Pierce ™ ECL Western Blotting Substrate (Cat. No. 32106). Reprinted with permission from Liang X, Potter J, Kumar S et al. (2015) Rapid and highly efficient mammalian cell engineering via Cas9 protein transfection. J Biotechnol 208:44–53.DNA 72ProteinmRNA 4824840(hr)Time Figure 4. Absence of LC3B in CRISPR-Cas9–edited HAP1 cells after chloro-quine treatment. HAP1 cells were modified using CRISPR-Cas9 gene editing to knock out the ATG5 gene. After chloroquine treatment, which normally causes LC3-containing autophagosomes to accumulate, edited cells (right panel) show the expected absence of LC3B-positive puncta, whereas wild-type cells (left panel) show an increase in LC3B accumulation. Cells were labeled with rabbit anti-LC3B antibody (Cat. No. L10382) followed by Invitrogen ™ Alexa Fluor ™ 647 goat anti–rabbit IgG antibody (red, Cat. No. A21245) and counterstained with Hoechst ™ 33342 (blue, Cat. No. H3570). Images were acquired on a Thermo Scientific ™ CellInsight ™ CX5High-Content Screening (HCS) Platform.14 | /bioprobes© 2016 Thermo Fisher Scientific Inc. All rights reserved. For Research Use Only. Not for use in diagnostic procedures.CrIsPr-CAs9–BAsED rEsEArCH BIOPrOBEs 74Product Quantity Cat. No.CRISPR proteinGeneArt ™ Platinum Cas9 Nuclease (1 µg/µL)GeneArt ™ Platinum Cas9 Nuclease (1 µg/µL)GeneArt ™ Platinum Cas9 Nuclease (3 µg/µL)10 µg 25 µg 75 µg B25642B25640B25641CRISPR mRNAGeneArt ™ CRISPR Nuclease mRNA 15 µg A29378GeneArt ™ Strings U6 DNA >200 ng **************************************GeneArt ™ Strings T7 DNA >200 ng **************************************Custom in vitro –transcribed gRNA 250 nmol ***********************************CRISPR plasmidGeneArt ™ CRISPR Nuclease Vector with OFP Reporter Kit10 reactions A21174GeneArt ™ CRISPR Nuclease Vector with OFP Reporter, with competent cells 10 reactions A21178GeneArt ™ CRISPR Nuclease Vector with CD4 Enrichment Kit10 reactions A21175GeneArt ™ CRISPR Nuclease Vector with CD4 Enrichment Kit, with competent cells 10 reactionsA21177CRISPR-Cas9 gRNAGeneArt ™ Precision gRNA Synthesis KitA29377CRISPR libraries: see page 11 and go to /crisprlibraries .Forcustom(arrayedorpooled)CRISPRlibraries,***********************************.CRISPR engineered cell lines: go to /engineeredcells .Forcustomstablecelllinegenerationservices,***********************************.Detection and analysis reagents GeneArt ™ Genomic Cleavage Detection Kit GeneArt ™ Genomic Cleavage Selection Kit20 reactions 10 reactionsA24372A27663Figure 5. Rapid analysis of various cell health parameters using a high-content analysis (HCA) platform. Wild-type and CRISPR-edited HAP1 cells were analyzed using the Thermo Scientific ™ CellInsight ™ CX5 High-Content Screening Platform for (A) apoptosis, using CellEvent ™ Caspase-3/7 Green Detection Reagent (Cat. No. R37111), (B) oxidative stress, using CellROX ™ Green Reagent (Cat. No. C10444), (C) protein degradation, with the Click-iT ™ HPG Alexa Fluor ™ 488 Protein Synthesis Assay Kit (Cat. No. C10428), and (D) protein synthesis, using the Click-iT ™ Plus OPP Alexa Fluor ™ 488 Protein Synthesis Assay Kit (Cat. No. C10456).ABCChloroquine concentration (μM)M e a n c i r c a v g i n t e n s i t yMenadione concentration (μM)M e a n c i r c a v g i n t e n s it yStaurosporine concentration (μM)M e a n c i r c a v g i n t e n s i t yDCycloheximide concentration (μM)M e a n c i r c a v g i n t e n s i t yWith modulation of any cellular signaling pathway comes the risk of proximal and distal consequences. It is important to track your targeted protein and also monitor the impact on other aspects of cell health and behavior (off-target phenotyping). HCA is particularly suited to this type of multiparameter investigation (Figure 5).Resources to help you get startedThermo Fisher Scientific offers a wide range of reagents, kits, and tools to support your genome editing experiments (Table 1). In addi-tion to our state-of-the-art online Invitrogen ™ CRISPR Search and Design Tool, we offer several different Cas9 delivery systems as well as cell culture reagents and cell analysis tools that can be matched to your experimental system. Our suite of genome editing products isTable 1. Online CRISPR-Cas9 resources from Thermo Fisher Scientific.selection guide Delivery formatproduct selection guide /genomeedit101gRNA design tool /crisprdesign Products for monitoring genome editing/detectcrisprcontinually expanding to include the entire cell engineering workflow, from reagents for cell culture, transfection, and sample preparation to kits for genome modification and for detection and analysis of known genetic variants. Go to /detectcrisprbp74 for an up-to-date view of our products and technologies. ■。

CRISPRCas9基因编辑操作步骤及详细说明实验材料与方法一、细胞培养人宫颈癌细胞 HeLa,常规培养使用含 10% FBS 的 DMEM 培养基 ( 含 1.5 mg/L-Glutamine,100 U/mL Penicillin,100 μg/mL Streptomycin) 中,37ºC 5% CO2 饱和湿度培养箱中培养。
二、基因信息及双 gRNA 设计基因信息及分析1.hsa-mir-152 基因信息:pubmed2.hsa-mir-152 基因位于蛋白编码基因 COPZ2 内含子内,敲除hsa-mir-152 基因不会影响该蛋白编码3.hsa-mir-152 precursor 序列(87 bp):TGTCCCCCCCGGCCCAGGTTCTGTGATACACTCCGACTCGGGCTCTGGAGCAGTCAGTGCATGACAGAACTTGGGCCCGGAAGGACC双 gRNA 设计使用在线 gRNA 设计软件在 hsa-mir-152 precursor 基因组序列两侧设计双 gRNA注:dgRNA 即为双 gRNA.三、慢病毒侵染实验材料及试剂DMEM 培养基 + 10% FBSD-Hank’s SolutionTrypsin-EDTA Solution96 孔板24 孔板Lentivirus- 病毒液(GenePharma)步骤靶细胞侵染实验1.靶细胞铺板:24-well,加入2.5×105 cells/well(根据细胞种类调整),0.5 mL 完全培养基,37℃,5% CO2 过夜;2.稀释病毒:稀释液(靶细胞维持液培养基)400 μL + 终浓度 5 μg/mL Polybrene,将慢病毒原液按 1:9 加入到稀释液中;3.移去 Step1 中细胞培养液,加入 Step2 稀释后的病毒液,同时建立对照(blank、negative),37℃,5% CO2 过夜;4.12~24 小时移去细胞侵染后的病毒液,加入 0.5 mL 完全培养基,37℃,5% CO2 过夜;5.根据细胞状态和类型,如果必要分出 1/3~1/5,加入0.5 mL 完全培养基,继续培养 24~48 小时,荧光倒置显微镜下观察结果。
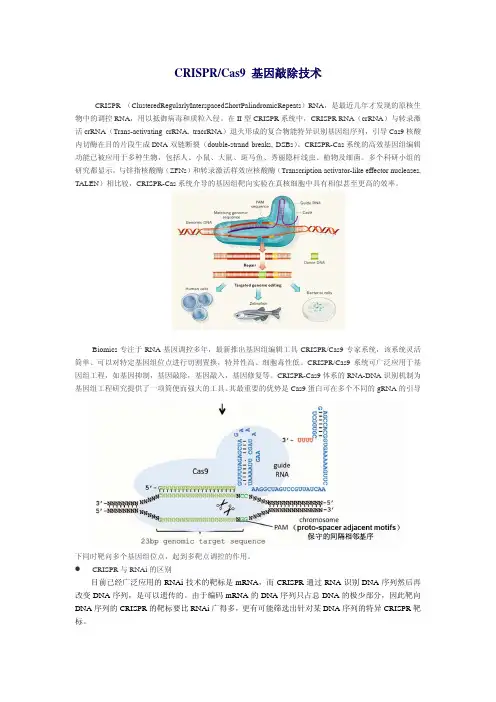
CRISPR/Cas9 基因敲除技术CRISPR (ClusteredRegularlyInterspacedShortPalindromicRepeats)RNA,是最近几年才发现的原核生物中的调控RNA,用以抵御病毒和质粒入侵。
在II型CRISPR系统中,CRISPR RNA(crRNA)与转录激活crRNA(Trans-activating crRNA, tracrRNA)退火形成的复合物能特异识别基因组序列,引导Cas9核酸内切酶在目的片段生成DNA双链断裂(double-strand breaks, DSBs)。
CRISPR-Cas系统的高效基因组编辑功能已被应用于多种生物,包括人、小鼠、大鼠、斑马鱼、秀丽隐杆线虫、植物及细菌。
多个科研小组的研究都显示,与锌指核酸酶(ZFNs)和转录激活样效应核酸酶(Transcription activator-like effector nucleases, TALEN)相比较,CRISPR-Cas系统介导的基因组靶向实验在真核细胞中具有相似甚至更高的效率。
Biomics专注于RNA基因调控多年,最新推出基因组编辑工具CRISPR/Cas9专家系统,该系统灵活简单、可以对特定基因组位点进行切割置换,特异性高、细胞毒性低。
CRISPR/Cas9系统可广泛应用于基因组工程,如基因抑制,基因敲除,基因敲入,基因修复等。
CRISPR-Cas9体系的RNA-DNA识别机制为基因组工程研究提供了一项简便而强大的工具。
其最重要的优势是Cas9蛋白可在多个不同的gRNA的引导下同时靶向多个基因组位点,起到多靶点调控的作用。
z CRISPR与RNAi的区别目前已经广泛应用的RNAi技术的靶标是mRNA,而CRISPR通过RNA识别DNA序列然后再改变DNA序列,是可以遗传的。
由于编码mRNA的DNA序列只占总DNA的极少部分,因此靶向DNA序列的CRISPR的靶标要比RNAi广得多,更有可能筛选出针对某DNA序列的特异CRISPR靶标。

crispr-cas9技术操作流程下载温馨提示:该文档是我店铺精心编制而成,希望大家下载以后,能够帮助大家解决实际的问题。
文档下载后可定制随意修改,请根据实际需要进行相应的调整和使用,谢谢!并且,本店铺为大家提供各种各样类型的实用资料,如教育随笔、日记赏析、句子摘抄、古诗大全、经典美文、话题作文、工作总结、词语解析、文案摘录、其他资料等等,如想了解不同资料格式和写法,敬请关注!Download tips: This document is carefully compiled by theeditor.I hope that after you download them,they can help yousolve practical problems. The document can be customized andmodified after downloading,please adjust and use it according toactual needs, thank you!In addition, our shop provides you with various types ofpractical materials,such as educational essays, diaryappreciation,sentence excerpts,ancient poems,classic articles,topic composition,work summary,word parsing,copy excerpts,other materials and so on,want to know different data formats andwriting methods,please pay attention!CRISPR-Cas9技术的操作流程详解CRISPR-Cas9,全称为“簇状规律间隔短回文重复序列-CRISPR相关蛋白9”,是一种革命性的基因编辑工具,它使得科学家能够精确地添加、删除或修改DNA序列。


CRISPR/Cas9基因编辑技术具体步骤及方法CRISPR/Cas9 是一种能够对基因组的特定位点进行精确编辑的技术。
其原理是核酸内切酶Cas9蛋白通过导向性RNA(guide RNA, gRNA)识别特定基因组位点并对双链DNA进行切割,细胞随之利用非同源末端连接(Non homologous End Joining,NHEJ)或者同源重组(Homologous Recombination, HR)方式对切割位点进行修复,实现DNA水平基因敲除或精确编辑。
CRISPR基因敲除利用CRISPR / Cas9 进行单基因敲除目前研究最透彻、应用最广泛的II 型-CRISPR/Cas9 系统由两部分组成:1. 单链的guide RNA(single-guide RNA,sgRNA)2. 有核酸内切酶活性的Cas9 蛋白CRISPR/Cas9 系统利用sgRNA 来识别靶基因DNA,并引导Cas9 核酸内切酶剪切DNA(图1)。
当基因组发生双链DNA 断裂后,细胞通过非同源性末端接合(Non-homologous end joining, NHEJ) 将断裂接合,在此过程中,将随机引入N 个碱基的缺失或增加,若N 非3 的倍数,则目的基因发生移码突变,实表1 CRISPR/Cas9 基因敲除与RNAi 比较CRISPR过表达利用CRISPR / Cas9 进行单基因过表达通过修饰CRISPR/Cas9 系统中的一些元件,形成一种蛋白复合物-协同激活介质(SAM),可实现对多数细胞内源基因的特异性激活。
该系统灵活方便,为研究基因功能提供了极为便利的工具。
CRISPR-SAM 系统由三部分组成:1. 失去核酸酶活性的dCas9(deactivated Cas9)-VP64 融合蛋白2. 含2 个MS2 RNA adapter 的sgRNA3. MS2-P65-HSF1 激活辅助蛋白CRISPR-SAM 系统中的MS2-P65-HSF1 激活辅助蛋白就是SAM,全称为SynergisticActivation Mediator( 协同激活调节器),这也就是CRISPR-SAM 的命名由来。
CRISPR-Cas9基因编辑技术简介中学生物科学登录CRISPR-Cas9基因编辑技术简介一林黄叶731 人赞同了该文章来源|公众号:中学生物科学CRISPR-Cas9基因编辑技术简介/s?__biz=MzUzMjE2ODkxOA==&mid=224749 0541&idx=1&sn=1db981f771aab1bf6ff57f070d35f468&chksm =fab63254cdc1bb4243bd7e781027a571dd3a76f2355e38eff36b 5bd2635ad5636e8ea9fe914c&token=1724268749&lang=zh_CN #rdCRISPR-Cas9是继ZFN、TALENs等基因编辑技术推出后的第三代基因编辑技术,短短几年内,CRISPR-Cas9技术风靡全球, 成为现有基因编辑和基因修饰里面效率最高、最简便、成本最低、最容易上手的技术之一,成为当今最主流的基因编辑系统。
一、什么是CRISPR-Cas系统CRISPR-Cas系统是原核生物的一种天然免疫系统。
某些细菌在遭到病毒入侵后,能够把病毒基因的一小段存储到自身的 DNA 里一个称为CRISPR 的存储空间。
当再次遇到病毒入侵时,细菌能够根据存写的片段识别病毒,将病毒的DNA切断而使之失效。
C RISPR-Cas系统包含CRISPR基因座和Cas基因(CRISPR关联基因)两部分。
1、CRISPR(/'krɪspər/)是原核生物基因组内的一段重复序列。
CRISPR全称Clustered Regularly Interspersed Short Palindromic Repeats(成簇的规律性间隔的短回文重复序列)。
分布在40%的已测序细菌和90%的已测序古细菌当中。
(注:生活在深海的火山口、陆地的热泉以及盐碱湖等极端环境中,有一些独特结构的细菌,称为古细菌)CRISPR基因序列主要由前导序列(leader)、重复序列(repeat)和间隔序列(spacer)构成。

CRISPR/Cas9技术介绍一、CRISPR/Cas9系统的构成CRISPR(clustered,regularly interspaced,short palindromic repeats)是一种来自细菌降解入侵的病毒DNA或其他外源DNA的免疫机制。
在细菌及古细菌中,CRISPR系统共分成3类,其中Ⅰ类和Ⅲ类需要多种CRISPR相关蛋白(Cas蛋白)共同发挥作用,而Ⅱ类系统只需要一种Cas蛋白即可,这为其能够广泛应用提供了便利条件。
目前,来自Streptococcus pyogenes 的CRISPR/Cas9系统应用最为广泛。
Cas9蛋白(含有两个核酸酶结构域,可以分别切割DNA两条单链。
Cas9首先与crRNA及tracrRNA结合成复合物,然后通过PAM序列结合并侵入DNA,形成RNA-DNA复合结构,进而对目的DNA双链进行切割,使DNA双链断裂。
研究人员为了将CRISPR/Cas9技术发展为高效的基因打靶工具,又进行了优化和改造。
Cong, L.等人[1]在不影响系统效率的情况下,将crRNA和tracrRNA融合为一条RNA。
通过这种简化,CRISPR/Cas9系统现仅包括两个元素:Cas9蛋白和sgRNA(single guide RNA)。
因此现在人们将CRISPR/Cas9技术也称为Cas9/sgRNA技术。
二、CRISPR/Cas9技术的基因编辑机制CRISPR/Cas9通过对预设的DNA位点进行切割,造成DNA双链断裂(DSB, double strand break)。
这种DNA的损伤可以启动细胞内的修复机制,主要包括两种途径:一是低保真性的非同源末端连接途径(NHEJ,Non-homologous end joining),此修复机制非常容易发生错误,导致修复后发生碱基的缺失或插入(Indel),从而造成移码突变,最终达到基因敲除的目的。
NHEJ是细胞内主要的DNA断裂损伤修复机制。
CRISPR-Cas9文库技术原理及应用CRISPR-Cas9技术原理CRISPR-Cas9技术凭借着成本低廉,操作方便,效率高等优点,CRISPR-Cas9技术迅速风靡全球的实验室,成为了生物科研的有力帮手,是继“锌指核酸内切酶(ZFN)”、“类转录激活因子效应物核酸酶(TALEN)”之后出现的第三代“基因组定点编辑技术”。
CRISPR-Cas9系统最初在大肠杆菌基因组中被发现,是细菌中抵抗外源病毒的免疫系统。
CRISPR-Cas9系统由两部分组成,一部分是用来识别靶基因组的,长度为20bp左右的sgRNA 序列,另外一部分是存在于CRISPR位点附近的双链DNA核酸酶——Cas9,能在sgRNA的引导下对靶位点进行切割,最终通过细胞内的非同源性末端连接机制(NHEJ)和同源重组修复机制(HDR)对形成断裂的DNA进行修复,从而形成基因的敲除和插入,最终实现基因的(定向)编辑。
与前两代技术相比,CRISPR-Cas9技术最大的突破是不仅可以对单个基因进行编辑,更重要的是可以同时对多个基因进行编辑,这也为全基因组筛选提供了有效的方法。
目前比较常见的文库类型包括:●CRISPR-Cas9 knock out文库●CRISPR panal文库●CRISPRa/i文库●psgRNA文库CRISPR-Cas9文库建库流程●靶位点确认及sgRNA文库设计●sgRNA文库芯片合成●sgRNA文库构建●QC验证文库质量●sgRNA文库慢病毒包装●感染稳定细胞株●药物筛选实验●细胞表型筛选●NGS测序验证功能基因CRISPR-Cas9文库应用方向1、药物靶点确定与验证CRISPR-Cas9筛选技术可以应用于药物靶点筛选中,通过大规模筛选技术,可以系统的分析、验证一些与抗药性相关的基因,从而为疾病治疗提供相关数据。
SCIENCE发表文章[1],研究人员利用CRISPR-Cas9文库筛选人类黑色素瘤A375细胞中的18,080个基因进行筛选,最终发现NF2、CUL3等4个基因参与了黑色素瘤A375细胞中的耐药调节过程。
CRISPR/Cas9 基因敲除技术CRISPR (ClusteredRegularlyInterspacedShortPalindromicRepeats)RNA,是最近几年才发现的原核生物中的调控RNA,用以抵御病毒和质粒入侵。
在II型CRISPR系统中,CRISPR RNA(crRNA)与转录激活crRNA(Trans-activating crRNA, tracrRNA)退火形成的复合物能特异识别基因组序列,引导Cas9核酸内切酶在目的片段生成DNA双链断裂(double-strand breaks, DSBs)。
CRISPR-Cas系统的高效基因组编辑功能已被应用于多种生物,包括人、小鼠、大鼠、斑马鱼、秀丽隐杆线虫、植物及细菌。
多个科研小组的研究都显示,与锌指核酸酶(ZFNs)和转录激活样效应核酸酶(Transcription activator-like effector nucleases, TALEN)相比较,CRISPR-Cas系统介导的基因组靶向实验在真核细胞中具有相似甚至更高的效率。
Biomics专注于RNA基因调控多年,最新推出基因组编辑工具CRISPR/Cas9专家系统,该系统灵活简单、可以对特定基因组位点进行切割置换,特异性高、细胞毒性低。
CRISPR/Cas9系统可广泛应用于基因组工程,如基因抑制,基因敲除,基因敲入,基因修复等。
CRISPR-Cas9体系的RNA-DNA识别机制为基因组工程研究提供了一项简便而强大的工具。
其最重要的优势是Cas9蛋白可在多个不同的gRNA的引导下同时靶向多个基因组位点,起到多靶点调控的作用。
z CRISPR与RNAi的区别目前已经广泛应用的RNAi技术的靶标是mRNA,而CRISPR通过RNA识别DNA序列然后再改变DNA序列,是可以遗传的。
由于编码mRNA的DNA序列只占总DNA的极少部分,因此靶向DNA序列的CRISPR的靶标要比RNAi广得多,更有可能筛选出针对某DNA序列的特异CRISPR靶标。
What is CRISPR/Cas9?Melody Redman,1Andrew King,2Caroline Watson,3David King41HYMS Centre for Education Development(CED),Hull York Medical School,University of York,York,UK2Molecular Haematology Unit, MRC Weatherall Institute of Molecular Medicine,University of Oxford,John Radcliffe Hospital,Oxford,UK3Department of Haematology, Oxford University NHS Foundation Trust,Churchill Hospital,Oxford,UK4Academic Unit of Child Health, Sheffield Children’s Hospital, Sheffield,UK Correspondence toDr David King,Academic Unit of Child Health,Sheffield Children’s Hospital,Western Bank, Sheffield S102TH,UK;*********************.uk Received5January2016 Revised18February2016 Accepted19February2016INTRODUCTIONClustered regularly interspaced palin-dromic repeats(CRISPR)/Cas9is agene-editing technology causing a majorupheaval in biomedical research.It makesit possible to correct errors in the genomeand turn on or off genes in cells and organ-isms quickly,cheaply and with relativeease.It has a number of laboratory applica-tions including rapid generation of cellularand animal models,functional genomicscreens and live imaging of the cellulargenome.1It has already been demonstratedthat it can be used to repair defective DNAin mice curing them of genetic disorders,2and it has been reported that humanembryos can be similarly modified.3Otherpotential clinical applications include genetherapy,treating infectious diseases such asHIV and engineering autologous patientmaterial to treat cancer and other diseases.In this review we will give an overview ofCRISPR/Cas9with an emphasis on how itmay impact on the specialty of paediatrics.Although it is likely to have a significanteffect on paediatrics through its impact inthe laboratory,here we will concentrate onits potential clinical applications.W e willalso describe some of the difficulties andethical controversies associated with thisnovel technology.OVERVIEW OF CRISPR/CAS9CRISPR/Cas9is a gene-editing technologywhich involves two essential components:a guide RNA to match a desired targetgene,and Cas9(CRISPR-associatedprotein9)—an endonuclease which causesa double-stranded DNA break,allowingmodifications to the genome(seefigure1).POTENTIAL CLINICAL USESCorrection of genetic disordersOne of the most exciting applications ofCRISPR/Cas9is its potential use to treatgenetic disorders caused by single genemutations.Examples of such diseasesinclude cystic fibrosis(CF),Duchenne’smuscular dystrophy(DMD)and haemo-globinopathies.The approach so far hascurrently only been validated in preclin-ical models,but there is hope it can soonbe translated to clinical practice.Schwank et al used CRISPR/Cas9toinvestigate the treatment of ingadult intestinal stem cells obtained fromtwo patients with CF,they successfullycorrected the most common mutationcausing CF in intestinal organoids.Theydemonstrated that once the mutation hadbeen corrected,the function of the CFtransmembrane conductor receptor(CFTR)was restored.4Another disease in which CRISPR/Cas9has been investigated is DMD.T abebordbaret al recently used adeno-associated virus(AA V)delivery of CRISPR/Cas9endonu-cleases to recover dystrophin expression ina mouse model of DMD,by deletion of theexon containing the original mutation.Thisproduces a truncated,but still functionalprotein.T reated mice were shown to par-tially recover muscle functional deficien-cies.5Significantly,it was demonstrated thatthe dystrophin gene was edited in musclestem cells which replenish mature muscletissue.This is important to ensure anytherapeutic effects of CRISPR/Cas9do notfade over time.T wo similar studies havedescribed using the CRISPR/Cas9system invivo to increase expression of the dys-trophin gene and improve muscle functionin mouse models of DMD.67Other studieshave used CRISPR/Cas9to target duplica-tion of exons in the human dystrophin genein vitro and have shown that this approachcan lead to production of full-length dys-trophin in the myotubules of an individualwith DMD.8CRISPR/Cas9could also be used totreat haemoglobinopathies.Canver et al9recently showed BCL11A enhancer dis-ruption by CRISPR/Cas9could inducefetal haemoglobin in both mice andprimary human erythroblast cells.In thefuture such an approach could allow fetalhaemoglobin to be expressed in patientswith abnormal adult haemoglobin.Thiswould represent a novel therapeutic strat-egy in patients with diseases such assickle cell disease orthalassaemias.RESEARCH IN PRACTICE Redman M,et al.Arch Dis Child Educ Pract Ed2016;0:1–3.doi:10.1136/archdischild-2016-3104591 Education & Practice Online First, published on April 8, 2016 as 10.1136/archdischild-2016-310459 Copyright Article author (or their employer) 2016. Produced by BMJ Publishing Group Ltd under licence. copyright. on December 25, 2023 by guest. Protected by / Arch Dis Child Educ Pract Ed: first published as 10.1136/archdischild-2016-310459 on 8 April 2016. Downloaded fromKnock-in of a fully functional β-globin gene is much more challenging,which is the reason for this some-what unusual approach.Treatment of HIVAnother potential clinical application of CRISPR/Cas9is to treat infectious diseases,such as HIV .Although antiretroviral therapy provides an effective treatment for HIV ,no cure currently exists due to permanent integration of the virus into the host genome.Hu et al showed the CRISPR/Cas9system could be used to target HIV-1genome activity .This inactivated HIV gene expression and replication in a variety of cells which can be latently infected with HIV ,without any toxic effects.Furthermore,cells could also be immu-nised against HIV-1infection.This is a potential thera-peutic advance in overcoming the current problem of how to eliminate HIV from infected individuals.After further refinement,the authors suggest their findings may enable gene therapies or transplantation of genet-ically altered bone marrow stem cells or inducible pluripotent stem cells to eradicate HIV infection.10Engineering somatic cells ex vivo to treat malignancy or other diseasesThere has been increasing interest in the possibility of using CRISPR/Cas9to modify patient-derived T-cells and stem/progenitor cells which can then be reintro-duced into patients to treat disease.This approach may overcome some of the issues associated with how to efficiently deliver gene editing to the right cells.T-cell genome engineering has already shown success in treating haematological malignancies and has the potential to treat solid cancers,primary immune deficiencies and autoimmune diseases.Genetic manipulation of T-cells has previously been inefficient.However,Schumann et al recently reported a more effective approach in human CD4+T-cells based on the CRISPR/Cas9system.Their tech-nique allowed experimental and therapeutic knock-out and knock-in editing of the genome in primary human T-cells.They demonstrated T-cells could be manipulated to prevent expression of the protein PD-1,which other work has shown may allow T-cells to target solid cancers.11There is also interest in using CRISPR/Cas9-mediated genome editing in pluripotent stem cells or primary somatic stem cells to treat disease.For example Xie et al 12showed the mutation causing β-thalassaemia could be corrected in human induced pluripotent stem cells ex vivo.They suggest that in the future such an approach could provide a source of cells for bone marrow transplantation to treat β-thalassaemia and other similar monogenic diseases.LIMITATIONSA number of challenges remain before the potential of CRISPR/Cas9can be translated to effective treatments at the bedside.A particular issue is how to deliver gene editing to the right cells,especially if the treat-ment is to be delivered in vivo.T o safely deliver Cas9-nuclease encoding genes and guide RNAs in vivo without any associated toxicity ,a suitable vector is needed.AA V has previously been a favoured option for gene delivery .1However,this delivery system may be too small to allow efficient transduction of the Cas9gene.1A smaller Cas9gene could be used,but this has additional implications on efficacy .1A number of other non-viral delivery systems are under investi-gation and this process requires further optimisation.Another significant concern is the possibility of off-target effects on parts of the genome separate from the region being targeted.Unintentional edits of the genome could have profound long-term complications for patients,including malignancy .The concentration of the Cas9nuclease enzyme and the length of time Cas9is expressed are both important when limiting off-target activity .1Although recent modifications in the nuclease have increased specificity ,further work is required to minimise off-target effects and to establish the long-term safety of any treatment.The therapeutic applications of CRISPR/Cas9con-sidered in this article have predominantly been direc-ted at somatic cells.A particularly controversial issue surrounding CRISPR/Cas9is that of gene editinginFigure 1The CRISPR/Cas9system.1Clustered regularlyinterspaced palindromic repeats (CRISPR)refers to sequences in the bacterial genome.They afford protection against invading viruses,when combined with a series of CRISPR-associated (Cas)proteins.Cas9,one of the associated proteins,is anendonuclease that cuts both strands of DNA.Cas9is directed to its target by a section of RNA.This can be synthesised as a single strand called a synthetic single guide RNA (sgRNA);the section of RNA which binds to the genomic DNA is 18–20nucleotides.In order to cut,a specific sequence of DNA of between 2and 5nucleotides (the exact sequence depends upon the bacteria which produces the Cas9)must lie at the 3’end of the guide RNA:this is called the protospacer adjacent motif (PAM).Repair after the DNA cut may occur via twopathways:non-homologous end joining,typically leading to a random insertion/deletion of DNA,or homology directed repair where a homologous piece of DNA is used as a repair template.It is the latter which allows precise genome editing:thehomologous section of DNA with the required sequence change may be delivered with the Cas9nuclease and sgRNA,theoretically allowing changes as precise as a single base-pair.Research in practice2Redman M,et al .Arch Dis Child Educ Pract Ed 2016;0:1–3.doi:10.1136/archdischild-2016-310459copyright.on December 25, 2023 by guest. Protected by /Arch Dis Child Educ Pract Ed: first published as 10.1136/archdischild-2016-310459 on 8 April 2016. Downloaded fromembryos.It has already been shown that CRISPR/ Cas9technology can alter the genome of human embryos3which theoretically could prove useful in the preimplantation treatment of genetic diseases. However,any genetic modification of the germline would be permanent and the long-term consequences are unclear.Many oppose germline modification under any circumstances,reasoning that an eventual consequence could be non-therapeutic genetic enhancement.13It is clear that the ethical boundaries, within which CRISPR/Cas9can be used,remain to be fully determined.▸CRISPR/Cas9technology has the potential to revolu-tionise the treatment of many paediatric conditions.▸A number of practical and ethical challenges must be overcome before this potential can be realised at thebedside.Contributors DK conceived the idea for this article.All authors were involved in writing and reviewing the final manuscript. Funding AK is supported by a W ellcome T rust Fellowship (108785/Z/15/Z).Competing interests None declared.Provenance and peer review Not commissioned;externally peer reviewed.Open Access This is an Open Access article distributed in accordance with the terms of the Creative Commons Attribution(CC BY4.0)license,which permits others to distribute,remix,adapt and build upon this work,for commercial use,provided the original work is properly cited. See:/licenses/by/4.0/REFERENCES1Hsu PD,Lander ES,Zhang F.Development and Applications of CRISPR-Cas9for Genome Engineering.Cell2014;157:1262–78.2Yin H,Xue W,Chen S,et al.Genome editing with Cas9in adult mice corrects a disease mutation and phenotype.NatBiotech2014;32:551–3.3Liang P,Xu Y,Zhang X,et al.CRISPR/Cas9-mediated gene editing in human tripronuclear zygotes.Protein Cell2015;6:363–72.4Schwank G,Koo BK,Sasselli V,et al.Functional repair of CFTR by CRISPR/Cas9in intestinal stem cell organoids ofcystic fibrosis patients.Cell Stem Cell2013;13:653–8.5T abebordbar M,Zhu K,Cheng JK,et al.In vivo gene editing in dystrophic mouse muscle and muscle stem cells.Science2016;351:407–11.6Nelson CE,Hakim CH,Ousterout DG,et al.In vivo genome editing improves muscle function in a mouse model ofDuchenne muscular dystrophy.Science2016;351:403–7.7Long C,McAnally JR,Shelton JM,et al.Prevention of muscular dystrophy in mice by CRISPR/Cas9–mediated editing of germline DNA.Science2014;345:1184–8.8W ojtal D,Kemaladewi Dwi U,Malam Z,et al.Spell checking nature:versatility of CRISPR/Cas9for developing treatmentsfor inherited disorders.Am J Hum Genet2016;98:90–101.9Canver MC,Smith EC,Sher F,et al.BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis.Nature2015;527:192–7.10Hu W,Kaminski R,Y ang F,et al.RNA-directed gene editing specifically eradicates latent and prevents new HIV-1infection.Proc Natl Acad Sci USA2014;111:11461–6.11Schumann K,Lin S,Boyer E,et al.Generation of knock-in primary human T cells using Cas9ribonucleoproteins.ProcNatl Acad Sci USA2015;112:10437–42.12Xie F,Y e L,Chang JC,et al.Seamless gene correction of β-thalassemia mutations in patient-specific iPSCs usingCRISPR/Cas9and piggyBac.Genome Res2014;24:1526–33. 13Lanphier E,Urnov F,Haecker SE,et al.Don’t edit the human germ line.Nature2015;519:410–1.Research in practiceRedman M,et al.Arch Dis Child Educ Pract Ed2016;0:1–3.doi:10.1136/archdischild-2016-3104593copyright. on December 25, 2023 by guest. Protected by / Arch Dis Child Educ Pract Ed: first published as 10.1136/archdischild-2016-310459 on 8 April 2016. Downloaded from。