The global spread of antibiotic resistance genes (ARGs) and their acquisition by clini-cally relevant microorganisms is associated with the increased hospitalization and mor-tality rates of patients that are infected with such microorganisms, which constitutes a serious problem for the health and welfare of both humans and animals1. The effect of clinically relevant ARGs and antibiotic resistant bacteria (ARB) that are released from anthropogenic sources, together with the excessive use of antibiotics in both human and veterinary settings, is currently considered to be a serious environmental problem1–5. However, current risk assess-ment models are inadequate to evaluate the effect of antibiotics and ARGs on resistance emergence and selection, especially in non-clinical environments.
In contrast to many chemical contami-nants — the concentration of which typically diminishes owing to degradation, dilution or sorption — bacterial contaminants (and their ARGs, which are present both within bacterial genomes and in free DNA) are capable of persisting and even spreading in the environment. The ARGs carried by these bacterial contaminants can multiply in their
hosts, be passed on to other bacterial popu-
lations and be subject to further evolution.
As such, ARB that occur in the environment
represent potentially serious risks for
human health.
The increased dissemination of ARB in
the environment is probably caused by three
principle mechanisms, which can occur in
combination: horizontal gene transfer of
ARGs; genetic mutation and recombination
(which can be favoured by the existence of
hypermutator bacterial strains); and the
proliferation of ARB owing to selective
pressures that are imposed by antimicrobial
compounds or other contaminants, such as
biocides or heavy metals6–10. As bacterial
communities are shaped by a complex
array of evolutionary, ecological and envi-
ronmental factors, it is difficult to predict
the fate of ARGs and ARB that are released
into the environment, or to obtain a clear
understanding of the evolution and ecology
of antibiotic resistance in this setting. For
example, although the excessive use of anti-
biotics may select for resistant populations
in the environment, other biotic and abiotic
factors (such as physicochemical conditions,
environmental contaminants, induction of
stress responses, bacterial adaptation and
phenotypic heterogeneity) have the potential
to enhance the effect of selective pressures
and promote bacterial evolution towards
antibiotic resistance. Conversely, there is still
a poor understanding of the environmental
factors that may alleviate the spread of
antibiotic resistance.
Antibiotic resistance hotspots are found
not only in medical settings but also in envi-
ronmental compartments that are subjected
to anthropogenic pressure, such as munici-
pal wastewater systems, pharmaceutical
manufacturing effluents, aquaculture facili-
ties and animal husbandry facilities. These
sites are characterized by extremely high
bacterial loads coupled with subtherapeutic
concentrations of antibiotics, and they con-
tribute to the discharge of ARB and ARGs
into the environment. At present, it is not
clear to what extent environmental ARB and
ARGs promote the acquisition and spread
of antibiotic resistance among clinically
relevant bacteria, or whether ARGs that are
acquired by both clinically relevant bacteria
and strictly environmental bacteria origi-
nate from the same reservoirs11–13. However,
for these issues to be properly addressed,
global efforts are required to characterize
and quantify antibiotic resistance in the
environment. In particular, it is necessary
to create an adequate support for advanced
biological risk assessment evaluations, which
are needed to determine how contaminated
environments affect the proliferation of
antibiotic resistance. In parallel, the imple-
mentation of technological solutions that
can reduce the contamination of natural eco-
systems by clinically relevant and potentially
evolving ARB and ARGs is also a priority.
These topics were the focus of the
European COST (Cooperation in Science
and Technology) Action DARE (Detecting
Evolutionary Hotspots of Antibiotic
Resistance in Europe, TD 0803), which was
launched in 2009 and completed in 2013.
This interdisciplinary project involved 20
European countries and 123 scientists with
a wide range of backgrounds (for example,
engineers, microbiologists, chemists,
veterinarians and physicians, working at
O P I N I O N
T ackling antibiotic resistance: the
environmental framework
Thomas U. Berendonk, Célia M. Manaia, Christophe Merlin,
Despo Fatta?Kassinos, Eddie Cytryn, Fiona Walsh, Helmut Bürgmann,
Henning S?rum, Madelaine Norstr?m, Marie?No?lle Pons, Norbert Kreuzinger,
Pentti Huovinen, Stefania Stefani, Thomas Schwartz, Veljo Kisand,
Fernando Baquero and José Luis Martinez
Abstract | Antibiotic resistance is a threat to human and animal health worldwide,
and key measures are required to reduce the risks posed by antibiotic resistance
genes that occur in the environment. These measures include the identification
of critical points of control, the development of reliable surveillance and risk
assessment procedures, and the implementation of technological solutions that
can prevent environmental contamination with antibiotic resistant bacteria and
genes. In this Opinion article, we discuss the main knowledge gaps, the future
research needs and the policy and management options that should be prioritized
to tackle antibiotic resistance in the environment. PERSPECTIVES
310 | APRIL 2015 | VOLUME 13 https://www.doczj.com/doc/f46319258.html,/reviews/micro
? 2015 Macmillan Publishers Limited. All rights reserved
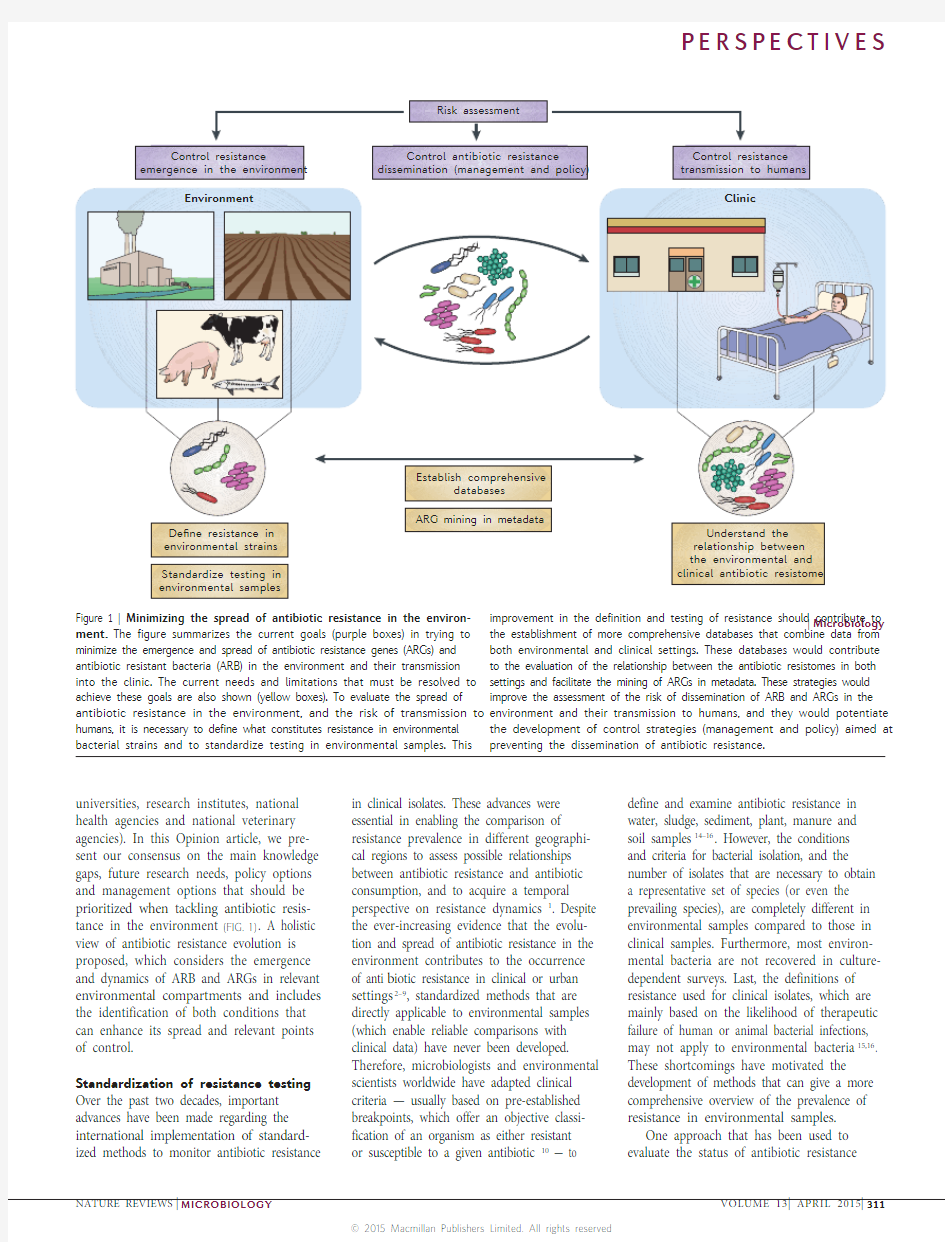
universities, research institutes, national health agencies and national veterinary agencies). In this Opinion article, we pre-sent our consensus on the main knowledge gaps, future research needs, policy options and management options that should be prioritized when tackling antibiotic resis-tance in the environment (FIG. 1). A holistic view of antibiotic resistance evolution is proposed, which considers the emergence and dynamics of ARB and ARGs in relevant environmental compartments and includes the identification of both conditions that can enhance its spread and relevant points of control.
Standardization of resistance testing Over the past two decades, important advances have been made regarding the international implementation of standard-ized methods to monitor antibiotic resistance in clinical isolates. These advances were
essential in enabling the comparison of
resistance prevalence in different geographi-
cal regions to assess possible relationships
between antibiotic resistance and antibiotic
consumption, and to acquire a temporal
perspective on resistance dynamics1. Despite
the ever-increasing evidence that the evolu-
tion and spread of antibiotic resistance in the
environment contributes to the occurrence
of anti b iotic resistance in clinical or urban
settings2–9, standardized methods that are
directly applicable to environmental samples
(which enable reliable comparisons with
clinical data) have never been developed.
Therefore, microbiologists and environmental
scientists worldwide have adapted clinical
criteria — usually based on pre-established
breakpoints, which offer an objective classi-
fication of an organism as either resistant
or susceptible to a given antibiotic10 — to
define and examine antibiotic resistance in
water, sludge, sediment, plant, manure and
soil samples14–16. However, the conditions
and criteria for bacterial isolation, and the
number of isolates that are necessary to obtain
a representative set of species (or even the
prevailing species), are completely different in
environmental samples compared to those in
clinical samples. Furthermore, most environ-
mental bacteria are not recovered in culture-
dependent surveys. Last, the definitions of
resistance used for clinical isolates, which are
mainly based on the likelihood of therapeutic
failure of human or animal bacterial infections,
may not apply to environmental bacteria15,16.
These shortcomings have motivated the
development of methods that can give a more
comprehensive overview of the prevalence of
resistance in environmental samples.
One approach that has been used to
evaluate the status of antibiotic resistance
|Microbiology
ment. The figure summarizes the current goals (purple boxes) in trying to minimize the emergence and spread of antibiotic resistance genes (ARGs) and antibiotic resistant bacteria (ARB) in the environment and their transmission into the clinic. The current needs and limitations that must be resolved to achieve these goals are also shown (yellow boxes). To evaluate the spread of antibiotic resistance in the environment, and the risk of transmission to humans, it is necessary to define what constitutes resistance in environmental bacterial strains and to standardize testing in environmental samples. This the establishment of more comprehensive databases that combine data from both environmental and clinical settings. These databases would contribute to the evaluation of the relationship between the antibiotic resistomes in both settings and facilitate the mining of ARGs in metadata. These strategies would improve the assessment of the risk of dissemination of ARB and ARGs in the environment and their transmission to humans, and they would potentiate the development of control strategies (management and policy) aimed at preventing the dissemination of antibiotic resistance.
P E R S P E C T I V E S
NATURE REVIEWS |MICROBIOLOGY VOLUME 13 | APRIL 2015 |311
? 2015 Macmillan Publishers Limited. All rights reserved
in the environment involves the calculation of resistance percentage, which is based on the ratio between the number of bacteria that are able to grow on culture media that are supplemented with antibiotics at doses close to the minimal inhibitory concentra-tion (MIC) and the number of bacteria that grow on antibiotic-free media16. Culture-independent approaches, which are mainly based on information about ARGs that was previously acquired from the study of clinical isolates, have also been successfully developed17–21. In particular, quantitative PCR (qPCR) can give an approximation of the prevalence of known ARGs in environ-mental samples, which can be a good estima-tion of the level of contamination by ARGs, although careful standardization of gene copy numbers is needed. However, as dif-ferent methodologies are regularly used, the results obtained by these approaches cannot be compared with those commonly used
in surveillance reports on human medicine and veterinary medicine1,22. Therefore, har-monized guidelines are needed regarding the number of isolates and diversity of species or strains to be tested; the cultivation conditions or the DNA extraction methods; and the tar-geted resistance phenotypes and genotypes or primer sets. Such guidelines would enable direct comparisons between different envi-ronmental compartments, thus establishing bridges with clinical data. Standardization of resistance testing should further focus on bacterial indicators that are already in use and also on a subset of resistance determinants, preferably with both analysed concurrently. Primary bacte-rial indicators should be members of the class Gammaproteobacteria or the phylum Firmicutes, as these are the most frequent carriers of acquired ARGs (BOX 1). In partic-ular, we propose Escherichia coli and faecal enterococci — which are currently used to monitor microbiological water quality and are well characterized in terms of acquired antibiotic resistance — as indicator organ-isms16. In addition, Pseudomonas aeruginosa, which is also used as an indicator of water quality, and Aeromonas spp., which are typi-cal water inhabitants, should be used for the examination of environmental samples in which faecal contamination is not expected. Klebsiella spp., in particular Klebsiella pneumoniae, are also possible indicators
to consider, as members of this group are present in both the environment and the animal gut and have frequently been found to be pioneers in the emergence of antibiotic resistance23. Although other indicator bacte-ria would be eligible, the bacteria mentioned above have the advantage of being ubiqui-
tous as well as being important carriers of
antibiotic resistance and being responsible
for the transfer of ARGs between different
environmental compartments13,23–26.
For the selection of resistance determi-
nants, criteria such as the clinical relevance,
the prevalence in the environment, the
association with mobile genetic elements
(MGEs) and/or the potential to be acquired
by any mode of horizontal gene transfer
(such as conjugation, transformation or
transduction) are important. Possible candi-
date genes, frequently occurring in environ-
mental settings that are subjected to human
activities9,16,27,28, are shown in BOX 1.
These recommendations should not be
regarded as attempts to narrow the focus
of researchers; however, there is an urgent
need for standardization of a number of
core parameters to improve the comparability
between studies worldwide. This is a pre-
requisite for obtaining a global perspective
of the environmental antibiotic resistome
irrespective of the geographical region,
the time frame or the environmental
compartment being analysed.
The importance of global databases
The current state of knowledge is insufficient
to assess the distribution and abundance
of ARB and ARGs in the environment at
national, regional or global levels, which is a
major limitation in determining the current
risk of transmission of antibiotic resistance
from the environment to human-associated
bacteria. There are three main reasons for
this limitation: the existing data are not
comparable, as there are no guidelines for
data collection; there is no formal system of
data collation and curation for publication;
and surveillance in environmental compart-
ments or ecosystems (such as soil and water)
has not been encouraged. Therefore, most
of the available data come from sporadic
research studies rather than long-term
monitoring efforts.
Current limitations. Phenotypic resistance
is often interpreted based on clinical stand-
ards and recommended breakpoints, for
example, from the European Committee
on Antimicrobial Susceptibility Testing
(EUCAST) or the Clinical and Laboratory
Standards Institute (CLSI). However, the use
of clinical breakpoints to assess the antibiotic
susceptibility of environmental bacteria is
inadequate, as clinical breakpoints are based
on parameters that are only relevant for
therapeutic success. A more reliable alterna-
tive for the interpretation of the antibiotic
resistance of environmental bacteria may be
the epidemiological cut-off (ECOFF) value
developed by EUCAST, which, in a given
P E R S P E C T I V E S
312 | APRIL 2015 | VOLUME 13 https://www.doczj.com/doc/f46319258.html,/reviews/micro
? 2015 Macmillan Publishers Limited. All rights reserved
taxonomic group, separates the popula-tions with acquired resistance mechanisms (non-wild-type) from the wild-type popula-tions that have no resistance. In contrast to clinical breakpoints, the ECOFF values are epidemiologically based, do not relate to the therapeutic efficiency and do not differ among different committees (for example, EUCAST and CLSI)10. However, current ECOFF estimations use databases in which, for a given species, the number of isolates with a clinical origin is several orders of magnitude higher than that of isolates with an environmental origin. In addition, most ECOFF studies comprise microorganisms of relevance to human health (pathogens and commensals), whereas information regard-ing non-pathogenic environmental species, which can be important carriers of ARGs, is scarce. Hence, it is questionable whether the current ECOFF values correctly reflect the distribution of resistant and wild-type bac-teria in the environment. To improve the reliability of these data, and therefore their usefulness in the classification of bacteria of different origins, it is essential to supplement ECOFF databases with additional data from environmental species and isolates.
There are several public databases and global surveillance projects, such as the Antimicrobial Resistance Global Report
on Surveillance from the World Health Organization (WHO)1; the European Centre for Disease Prevention and Control (ECDC)-based European Antimicrobial Resistance Interactive Database (EARS-Net), EUCAST, and the European Antimicrobial Susceptibility Surveillance in Animals (EASSA)29 in Europe; the Surveillance Network Database (TSN) in the USA and Australia; and the Study for Monitoring Antimicrobial Resistance Trends (SMART) task force30 in the Asia–Pacific region. In addition, there are two centralized databases on ARGs: the Comprehensive Antibiotic Resistance Database (CARD)31 and the Antibiotic Resistance Genes Database (ARDB)32. However, the number of ARGs described in human and animal opportu-nistic pathogens in these databases is much higher than the number of ARGs described in environmental bacteria. Additionally, public databases that incorporate data from environmental bacteria or metagenomes contain genes that have been putatively annotated as ARGs based on the similarity of their nucleotide or amino acid sequences to those of previously described ARGs. However, a functional demonstration of the role of many sequences described as
‘resistance genes’ in specialized databases is lacking, which creates a background of
potentially misleading information for
researchers and clinicians. The lack of a
proper definition of antibiotic resistance
for environmental strains of bacteria, the
numerous databases with scant information
on antibiotic resistance in the environment
and the lack of functional demonstrations
for ARGs in environmental metagenomes
are considerable limitations for the charac-
terization of the environmental resistome
and the assessment of its clinical relevance8.
Perspectives. The consolidation of specialized
databases will certainly contribute to a stan-
dardized definition of ARGs and elucidate
which genes contribute to the future acquisi-
tion of resistance by human pathogens. In
parallel, the continuous improvement of
techniques for the cultivation of bacteria
using multiple culture media and conditions
(known as culturomics) is of vital impor-
tance for the identification of ARGs and
for understanding the cellular mechanisms
linking ARGs and resistance phenotypes in
different bacteria33. One of the greatest chal-
lenges in creating a consolidated database lies
in establishing a standardized methodology
and transforming it into a routine activity of
environmental-quality monitoring. However,
this is necessary if we are to acquire a coherent
picture of environmental resistance.
Conceptually, two types of data may be
integrated in public databases: data from
routine monitoring (that is, from surveil-
lance databases), which will give insights
into the distribution, prevalence, temporal
trends and geographical trends; and data
gathered by research studies (that is, from
emergence and evolution databases), which
are seeking to understand the acquisition
and molecular evolution processes. As one of
the most important issues concerning envi-
ronmental antibiotic resistance is its possible
implications in the health of humans and
animals, databases focusing on environmen-
tal bacteria should also be linked to existing
databases on ARB and ARGs in clinical,
veterinary and food-associated products.
The core entries of these integrative data-
bases should comprise the ARB and ARGs
described in BOX 1 and adopt the format of
existing databases (for example, EARS-Net
and REFS 31,32).
The information provided by surveillance
databases would be substantially improved
by the inclusion of other, systematically cho-
sen, meta-parameters. Examples include the
occurrence of heavy metals, antibiotic resi-
due concentrations and, whenever possible,
the composition of the microbial community,
which would be relevant for medium- to
long-term evaluations of resistance evolu-
tion34–36. This information could be linked to
routine monitoring assays of environmental
samples, particularly wastewater and sur-
face water samples37,38. Whenever possible,
links to existing databases on MGEs (for
example, A Classification of Mobile Genetic
Elements (ACLAME), Insertion Sequence
finder (ISfinder) or INTEGRALL) and
eco-toxicological information on mole c ules
with antibacterial effects (for example, from
the European Chemicals Agency (ECHA))
should be implemented to provide a compre-
hensive overview of the risk factors associ-
ated with specific antibiotic resistance (for
example, selection, mutation and
horizontal gene transfer).
In addition to the creation of integrative
databases, additional data on ARGs in the
environment are also necessary. Research on
uncharacterized ARGs and associated MGEs,
which are not included in the routine moni-
toring of environmental samples, is essential
to have a thorough understanding of the
environmental resistome. Whole-genome and
transcriptome analyses — including those
that apply next-generation sequencing (NGS)
technology — and functional metagenomic
studies present new possibilities for decipher-
ing resistance in environmental compart-
ments. Studies on the expression and function
of resistance genes will be fundamental in
understanding the interplay between the
environmental conditions and the genomic
context, and in understanding how this rela-
tionship will influence the selection of specific
ARGs39,40. These insights will offer the pos-
sibility of assessing the effect of external con-
ditions, such as the presence of subinhibitory
concentrations of antibiotics, on the presence,
expression and functionality of new and clin-
ically-associated ARGs in the environment.
Last, these strategies may unveil unforeseen
hotspots of antibiotic resistance. Continuous
improvements on annotation procedures,
such as those recently reported31,41–45, will not
only contribute to the annotation of ARGs in
metadata, but will also provide valuable infor-
mation about the enzymes that are respon-
sible for each resistance mechanism, mainly
concerning new genes. The association of
these data to both generic (for example, the
European Molecular Biology Laboratory
(EMBL) or GenBank) and metagenomic
databases (for example, Metagenomic Rapid
Annotations using Subsystems Technology
(MG-RAST)) would further provide access
to related sequence data and metadata
from gene fragments to metagenomes,
transcriptomes and proteomes.
P E R S P E C T I V E S
NATURE REVIEWS |MICROBIOLOGY VOLUME 13 | APRIL 2015 |313
? 2015 Macmillan Publishers Limited. All rights reserved
However, the most important associa-tions to achieve would be those between data from environmental and clinical settings, as well as the systematic combination of information obtained through research and routine monitoring practices. This bridging of information will be a step forwards in elucidating the role of environmental ARGs in the emergence and evolution of clinically-relevant antibiotic resistance.
Risk assessment
Integrated risk assessment of the evolution and emergence of antibiotic resistance in the environment addresses two main issues: first, the potential of subinhibitory con-centrations of antibiotics to promote the development of ARB in complex bacterial communities; and second, the capacity of resistance determinants to transfer from anthropogenic sources (such as treated wastewater, manure or others) to human commensal or pathogenic bacteria.
Potential of subinhibitory antibiotic concentrations. The Guideline on the envi-ronmental risk assessment of medicinal products for human use, produced by the European Medicines Agency46, does not recognize that the emergence and prolifera-tion of antibiotic resistance may be the most important risk associated with environmental contamination by antibiotics. Indeed, the endpoints for no-effect concentrations (NOECs; which correspond to the highest concentration at which a substance has no significant effect on the organisms exposed to it) are different from traditional environ-mental risk assessment, as the effects of antibiotics in promoting antibiotic resist-ance can go far beyond the toxicological implications. For example, even at levels that are considered safe according to the currently accepted Environmental Quality Standards47, antibiotics can still select for ARB48–50. Furthermore, these subinhibitory concentrations of antibiotics and antibiotic combinations may even induce the propa-gation of unforeseen multidrug-resistant opportunistic pathogens51–54. Moreover, the effects of antibiotics may be potentiated or extended by cofactors (general stress situ-ations and micro-contaminants, such as heavy metals and biocides), which possibly enhance the spread and evolution of anti-biotic resistance34,54–59. Therefore, combined molecular- and culture-based methods are necessary to determine the concentrations at which resistance acquisition and selec-tion is likely to occur in environmental compartments.
An addendum emphasizing the need
to assess the risks posed by antimicro-
bial agents of inducing ARB selection or
ARG emergence should be included in the
Guideline on the environmental risk assess-
ment of medicinal products for human use.
Although challenging, given the scarcity
of knowledge regarding the mechanisms
involved at the genetic, cellular and popu-
lation levels, this addendum is urgently
needed. Within this addendum the gold
standard of a reliable risk assessment should
determine the range of concentrations at
which, under defined conditions, an anti b iotic
can promote selection and the acquisition of
resistance.
Transmission of resistance determinants
from anthropogenic sources. Another
important aspect of antibiotic-resistance
risk assessment refers to the spread and
transmission of resistance determinants
from hotspots to downstream environ-
ments. Mathematical models capable of
predicting the influence of potential selec-
tive pressures, or the occurrence and the
evolutionary success of genetic recombina-
tion events, have proven to be promising
tools in predicting the spread of antibiotic-
resistance determinants54,60,61. As they are
specifically developed for environmental
niches and environment–human interfaces,
these mathematical models should rely on
parameters such as population size; bacte-
rial population growth rate and survival;
occurrence and frequency of horizontal
gene transfer and its implications on the
population fitness; and the influence of
other biotic and abiotic factors54,60–62. Such
models would allow predictions to be made
regarding the dynamics of ARB hosting
ARGs and the possible localization of ARGs
on MGEs, thus supporting the assessment
of their fate from anthropogenic sources to
downstream environments. A quantitative
risk-assessment framework should be devel-
oped by coupling data and analyses, such
as those outlined above, with a stochastic
assessment of exposure to clinically relevant
bacteria in the environment. Such a model
should then be used to predict the environ-
mental conditions that are associated with
the evolution of antibiotic resistance and
infer the probability of antibiotic resistance
determinants spreading. However, owing
to the scarcity of data on the occurrence of
antibiotic resistance and horizontal gene
transfer in the environment, it is currently
difficult to develop validated models that
can be applied in the framework of
environmental risk-assessment guidelines.
Management and policy options
Management and policy options aimed at
preventing and controlling antibiotic resist-
ance in the environment comprise several
different aspects, including the choice of
ARB and ARGs to be listed as contaminants
of emerging concern; the determination of
differentiated maximum admissible levels
of an antibiotic, ARB or ARG; and the
identification of critical points of control at
which prevention and remediation measures
should be implemented.
ARB and ARGs as contaminants of emerging
concern. The European Water Framework
Directive establishes the requirements for
determining the biological and chemical
quality standards of water bodies in Europe63.
Annex I of this directive sets obligations
for environmental quality standards (EQS)
for priority substances and certain other
pollu t ants, and it even identifies priority
hazardous substances. The inclusion of
ARB and ARGs as priority contaminants
would be justified based on the results of
numerous scientific studies, which show
that the occurrence of antibiotic resistance
increases in bodies of water (such as inland
surface waters, transitional waters, coastal
waters and groundwater) when they are
subjected to anthropogenic impacts, such
as wastewater effluents, animal manure,
agricultural runoff and wildlife living in
urban areas21,34,64–67. We suggest the inclu-
sion of a supplement to the European Water
Framework Directive for ARB hosting
clinically relevant ARGs. In this context,
it is important to establish differentiated
guidance levels for the abundance of these
biological contaminants in the environment.
The application of these guidance levels may
be especially important for the regulation of
specific practices, such as reuse of wastewa-
ter or soil fertilization with manure.
The inclusion of ARB and ARGs in the
list of contaminants of emerging concern
would require clear definitions on the nec-
essary monitoring methods. Although the
environmental survival of ARGs primarily
depends on the host and the type of MGEs,
the estimation of the levels of ARGs seems
a reliable and feasible method to monitor
antibiotic resistance. However, a crucial
issue that needs additional investigation is
the selection of target ARGs to be moni-
tored as indicators of resistance and the
determination of safe concentrations of
these genes in water. Ideally, such indicator
genes should be abundant in anthropogenic
sources and rare in native aquatic and ter-
restrial ecosystems to facilitate spatial and
P E R S P E C T I V E S
314 | APRIL 2015 | VOLUME 13 https://www.doczj.com/doc/f46319258.html,/reviews/micro
? 2015 Macmillan Publishers Limited. All rights reserved
temporal source tracking in the environ-ment. ARGs that are associated with MGEs or that present the highest environmental fitness (that is, long lasting survival and the capacity to proliferate) are good candi-dates — for example, sul 1 (which encodes for sulfonamide-resistant dihydropteroate synthase); bla TEM , bla CTX-M , bla VIM and bla NDM-1 (which encode β-lactamases); tet M (which encodes for tetracycline resistance); and van A (which encodes for vancomycin resist-ance) (BOX 1). However, determination of the maximum acceptable levels of these genes in the environment seems, at the current state of knowledge, a challenging objective. The establishment of a comprehensive database and the use of modelling approaches would be valuable contributions to estimate such limits. Despite these challenges, this know-ledge is an essential prerequisite, not only for establishing a strategy of direct action against antibiotic resistance in the environ-ment, but also for the application of drugs and interventions directed at preventing the emergence and evolution of ARB and ARGs (eco–evo drugs)68.
Critical control points. Environmental hotspots, where ARB are abundant or the transfer of ARGs is promoted, are critical points for resistance control. Good examples of such critical points are characterized by a high prevalence of resistance or by the occurrence of resistance determinants of emerging concern. These locations comprise habitats that are influenced by human activi-ties, such as wastewater (that is, hospital, urban and specific industrial wastewater) and waste, and wastewater from animal husbandry and intensive food-production facilities 16,69,70. Moreover, sites subjected to the frequent discharge of antibiotic residues have been shown to be potential hotspots for the selection, proliferation and spread of new resistance determinants to human com-mensal and pathogenic bacteria, and these sites should likewise be considered as critical control points 71,72.
Although some of the antimicrobials administered to animals are used exclusively in veterinary applications, most belong to the same structural families that are used in human medicine. As they share the same basic chemical molecular structures and mechanisms of action, these antibiotics are assumed to put selective pressures on human commensal and pathogenic bacteria. Large quantities of antibiotics that are adminis-tered to animals in intensive production sites are discharged, often un-metabolized, with manure and slurry when applied as fertilizer
(often in a raw and unstabilized state) and thus contaminate soils as well as surface water and groundwater. At present, it is diffi-cult to ascertain whether antibiotics reaching the environment at low concentrations exert a substantial selective pressure on ARGs or ARB 50,51. However, there is increasing evi-dence showing that repeated exposure of the environment to anthropogenically gener-ated ARGs (for example, soil manure) cor-relate with the emergence and proliferation of ARGs in indigenous microbiota 66,67,73,74. However, the impact of animal production on the propagation of antibiotic resistance is demonstrated by some zoonotic species of the genera Salmonella , Campylobacter , Listeria , Staphylococcus , Enterococcus and Escherichia , which are known to exhibit high levels of acquired antibiotic resistance 29,75–78. Although some animal production facili-ties implement systems that decontaminate liquid and solid wastes, such treatments are not intended to remove ARB and might even promote resistance 79,80. The same might be true for biogas reactors, which use manure as a substrate, because the residues of these reactors are used as fertilizers on agricul-tural fields 81. The confinement and treat-ment of effluents and (processed) manure from intensive animal production is thus a priority.
Urban, hospital and pharmaceutical industry wastewater is among the main sources of antibiotic and ARB contamina-tion in soil and water ecosystems 16,21,82. In the environment, these contaminants can reach water resources for drinking water produc-tion, enter the food chain or reach clini-cally relevant niches 9,16,17,76,82. These effects can be potentially even more pronounced when irrigation with wastewater effluents (wastewater reuse schemes) is applied. Water reuse is already a common practice in many regions of the world owing to increased water scarcity, mainly in arid and semi-arid regions 20. Most of the wastewater treatment plants worldwide, in particular those using mechanical and biological treatments, are primarily designed to remove organic com-pounds, nutrients (for example, nitrogen and phosphorous) and suspended solids. However, the currently available wastewater treatment processes have limited capability to efficiently remove organic micropollutants, including antibiotics and other antimicro-bial agents 82. Similarly, certain ARB and ARGs can survive the wastewater treatment processes with a maintenance (or even an increase) of resistance prevalence compared to the pretreatment levels 16,17,36,64. These fea-tures require the immediate attention of the
scientific community and the development and implementation of technological solu-tions capable of mitigating ARB and ARGs in wastewater to safe levels. Although the defini-tion of a ‘safe level’ may be difficult to achieve, it is at least necessary to find an agreement on the threshold values below which the prob-ability of significant proliferation of an ARG is severely impaired.
Technologies for the removal of micro-pollutants, including antibiotics, and microorganisms from wastewater are
becoming increasingly available (for exam-ple, membrane filtration, activated carbon, photo-driven technologies and ozonation)82. However, additional research is needed to determine the effectiveness of these processes for the elimination of ARB and ARGs and to characterize the associated microbiological risks 16,82. Recommendations of effective and economically sustainable interventions at critical points within the wastewater stream are urgently needed.Concluding remarks
Given the public health threat posed by antibiotic resistance, the development and implementation of national and interna-tional guidelines for the biological risk assessment of the emergence and propaga-tion of ARB in the environment is a strategic priority. The generation of reliable compari-sons and evaluation of temporal trends in antibiotic resistance in the environment are currently seriously limited owing to the dis-parity of surveillance strategies. To address this issue, the interdisciplinary scientific community involved in the DARE Action has proposed specific priority measures.It is necessary to improve the com-parability between studies worldwide to provide the basis for a global perspective on the antibiotic resistome irrespective of geographical, temporal or environmental constraints. A formal system for the col-lation and curation of data for publication must be implemented, and surveillance of environmental samples must be encouraged, to comprehensively assess antibiotic resist-ance in environmental bacteria. Databases with multiple levels of information and metadata, supporting the understanding of antibiotic resistance dynamics on a global scale, must be established. An integrated risk-assessment platform must include the potential of subinhibitory concentrations of antibiotics to promote the development of ARB in complex environmental communi-ties, and mathematical models are required to devise reliable risk-assessment guidelines on the spread of antibiotic resistance from
P E R S P E C T I V E S
NATURE REVIEWS | MICROBIOLOGY
VOLUME 13 | APRIL 2015 | 315
? 2015 Macmillan Publishers Limited. All rights reserved
the environment to human commensal or pathogenic bacteria. A selection of sentinel ARB and ARGs should be included in the list of contaminants of emerging concern and maximum admissible levels of these contaminants should be defined, as well as the critical points of control at which pre-vention and remediation measures can be implemented. Other pressing management options include the reduction of antibiotic usage; the confinement and treatment of problematic reservoirs, such as effluents and manure from intensive animal production or hospital effluents; and the improvement of current wastewater treatment technologies. Finally, it is important to acquire a more comprehensive understanding of the mole-cular, evolutionary and ecological mecha-nisms associated with the acquisition and spread of antibiotic resistance. In parallel, the implementation of effective management options, mainly seeking to impose barriers against the dissemination of resistance from well-established resistance reservoirs, is urgently needed to combat the evolution and spread of antibiotic resistance, while protect-ing human health and the environment. Better control of the emergence and dissemination of ARB and ARGs does not replace the need to seek new therapeutic drugs. On the contrary, by minimising the evolution of ARB and ARGs, it also prepares the ground for new drugs to be effective for a longer period of time.
Thomas U. Berendonk is at the Institute for Hydrobiology, T echnische Universit?t Dresden,
Dresden 01062, Germany.
Célia M. Manaia is at the Centro de Biotecnologia e
Química Fina – Laboratório Associado, Escola Superior de Biotecnologia, Universidade Católica Portuguesa/Porto, Rua Arquiteto Lob?o Vital, Apartado 2511, Porto 4202–401, Portugal. Christophe Merlin is at the Université de Lorraine and le Centre National de la Recherche Scientifique, Laboratoire de Chimie Physique et Microbiologie pour l’Environnement, Unite Mixte de Recherche 7564,
Vandoeuvre?lès?Nancy F?54500, France. Despo Fatta?Kassinos is at the Civil and Environmental Engineering Department and Nireas, International Water Research Center, University of Cyprus,
PO Box 20537, Nicosia 1678, Cyprus.
Eddie Cytryn is at the Institute of Soil, Water and Environmental Sciences, Volcani Center,
Agricultural Research Organization,
PO Box 6, Bet Dagan 50250, Israel.
Fiona Walsh is at the Department of Biology, Biosciences and Electronic Engineering Building,
National University of Ireland Maynooth,
Maynooth,County Kildare, Ireland.
Helmut Bürgmann is at the Eawag: Swiss Federal Institute of Aquatic Science and T echnology, Department of Surface Waters – Research and Management, Kastanienbaum (LU) 6047, Switzerland.
Henning S?rum is at the Norwegian University of
Life Sciences, Faculty of Veterinary Medicine and Life
Sciences, Department of Food Safety and Infection
Biology, PO Box 8146 Dep, Oslo N?0033, Norway.
Madelaine Norstr?m is at the Norwegian
Veterinary Institute, Department of Epidemiology,
Oslo N?0106, Norway.
Marie?No?lle Pons is at the Laboratoire Reactions et
Génie des Procédés (UMR CNRS 7274), Université de
Lorraine, Nancy54000, France.
Norbert Kreuzinger is at the Vienna University of
T echnology, Institute of Water Quality, Resources
and Waste Management, Karlsplatz 13/226,
Vienna 1040, Austria.
Pentti Huovinen is at the Department of Medical
Microbiology and Immunology, University of T urku,
Kiinamyllynkatu 10, 20520 T urku, Finland
Stefania Stefani is at the Laboratory of Molecular
Microbiology and Antibiotic Resistance, Department
of Biomedical Sciences, University of Catania,
Via Androne 81, Catania 95124, Italy.
Thomas Schwartz is at the Karlsruhe Institute of
T echnology Campus North, Institute of Functional
Interfaces, Microbiology of Natural and T echnical
Interfaces Department, Hermann?von?Helmholtz?Platz 1,
Eggenstein?Leopoldshafen 76344, Germany.
Veljo Kisand is at the Institute of T echnology,
T artu University, Nooruse 1, T artu 50411, Estonia.
Fernando Baquero is at the Ramón y Cajal Institute for
Health Research, Ramón y Cajal University Hospital,
Department of Microbiology, Madrid 28034, Spain.
José Luis Martinez is at the Departamento
de Biotecnologia Microbiana, Centro Nacional de
Biotecnologia, Consejo Superior de Investigaciones,
Darwin 3, Cantoblanco, Madrid 28049, Spain.
T.U.B. and C.M.M. contributed equally to this work.
Correspondence to C.M.M.
e?mail: cmanaia@porto.ucp.pt
doi:10.1038/nrmicro3439
Published online 30 March 2015
1. World Health Organization. Antimicrobial resistance:
global report on surveillance (WHO, 2014).
2. Cantón, R. Antibiotic resistance genes from the
environment: a perspective through newly identified
antibiotic resistance mechanisms in the clinical setting.
Clin. Microbiol. Infect. 15 (Suppl. 1), 20–25 (2009).
3. Allen, H. K.
et al. Call of the wild: antibiotic resistance
genes in natural environments. Nature Rev. Microbiol.
8, 251–259 (2010).
4. Bush, K.
et al. T ackling antibiotic resistance. Nature
Rev. Microbiol. 9, 894–896 (2011).
5. D’Costa, V. M.
et al. Antibiotic resistance is ancient.
Nature 477, 457–461 (2011).
6. Baquero, F., Martinez, J. L. & Canton, R. Antibiotics
and antibiotic resistance in water environments. Curr.
Opin. Biotechnol. 19, 260–265 (2008).
7. Martinez, J. L. Environmental pollution by antibiotics
and by antibiotic resistance determinants. Environ.
Pollut. 157, 2893–2902 (2009).
8. Wright, G. D. Antibiotic resistance in the environment:
a link to the clinic? Curr. Opin. Microbiol. 13, 589–594
(2010).
9. Vaz-Moreira, I., Nunes, O. C. & Manaia, C. M.
Bacterial diversity and antibiotic resistance in water
habitats: searching the links with the human
microbiome. FEMS Microbiol. Rev. 38, 761–778
(2014).
10. Kahlmeter, G. Defining antibiotic resistance — towards
international harmonization. Ups. J. Med. Sci. 119,
78–86 (2014).
11. De Gelder, L., Williams, J. J., Ponciano, J. M., Sota, M.
& T op, E. M. Adaptive plasmid evolution results in
host-range expansion of a broad-host-range plasmid.
Genetics 178, 2179–2190 (2008).
12. Carattoli, A., Villa, L., Poirel, L., Bonnin, R. A. &
Nordmann, P. Evolution of IncA/C bla
CMY-2
-carrying
plasmids by acquisition of the bla
NDM-1
carbapenemase
gene. Antimicrob. Agents Chemother. 56, 783–786
(2012).
13. Walsh, T. R., Weeks, J., Livermore, D. M. &
T oleman, M. A. Dissemination of NDM-1 positive
bacteria in the New Delhi environment and its
implications for human health: an environmental point
prevalence study. Lancet Infect. Dis. 11, 355–362
(2011).
14. Dantas, G., Sommer, M. O., Oluwasegun, R. D. &
Church, G. M. Bacteria subsisting on antibiotics.
Science 320, 100–103 (2008).
15. Popowska, M. et al. Influence of soil use on prevalence
of tetracycline, streptomycin, and erythromycin
resistance and associated resistance genes.
Antimicrob. Agents Chemother. 56, 1434–1443
(2012).
16. Rizzo, L. et al. Urban wastewater treatment plants as
hotspots for antibiotic resistant bacteria and genes
spread into the environment: a review. Sci. T otal
Environ. 447, 345–360 (2013).
17. Schwartz, T., Kohnen, W., Jansen, B. & Obst, U.
Detection of antibiotic-resistant bacteria and their
resistance genes in wastewater, surface water, and
drinking water biofilms. FEMS Microbiol. Ecol. 43,
325–335 (2003).
18. Volkmann, H., Schwartz, T., Bischoff, P., Kirchen, S. &
Obst, U. Detection of clinically relevant antibiotic-
resistance genes in municipal wastewater using
real-time PCR (T aqMan). J. Microbiol. Methods 56,
277–286 (2004).
19. Szczepanowski, R. et al. Detection of 140 clinically
relevant antibiotic-resistance genes in the plasmid
metagenome of wastewater treatment plant bacteria
showing reduced susceptibility to selected antibiotics.
Microbiology 155, 2306–2319 (2009).
20. Negreanu, Y., Pasternak, Z., Jurkevitch, E. & Cytryn, E.
Impact of treated wastewater irrigation on antibiotic
resistance in agricultural soils. Environ. Sci. T echnol.
46, 4800–4808 (2012).
21. Czekalski, N., Gascon Diez, E. & Burgmann, H.
Wastewater as a point source of antibiotic-resistance
genes in the sediment of a freshwater lake. ISME J. 8,
1381–1390 (2014).
22. European Centre for Disease Prevention and Control.
Antimicrobial resistance surveillance in Europe 2011
report (ECDC, 2012).
23. Tzouvelekis, L. S., Markogiannakis, A., Psichogiou, M.,
T assios, P. T. & Daikos, G. L. Carbapenemases in
Klebsiella pneumoniae and other Enterobacteriaceae:
an evolving crisis of global dimensions. Clin. Microbiol.
Rev. 25, 682–707 (2012).
24. Livermore, D. M. & Woodford, N. The β-lactamase
threat in Enterobacteriaceae, Pseudomonas and
Acinetobacter. T rends Microbiol. 14, 413–420
(2006).
25. Jacobs, L. & Chenia, H. Y. Characterization of
integrons and tetracycline resistance determinants in
Aeromonas spp. isolated from South African
aquaculture systems. Int. J. Food Microbiol. 114,
295–306 (2007).
26. Cytryn, E. The soil resistome: the anthropogenic, the
native, and the unknown. Soil Biol. Biochem. 63,
18–23 (2013).
27. Allen, H. K. Antibiotic resistance gene discovery in
food-producing animals. Curr. Opin. Microbiol. 19,
25–29 (2014).
28. Wang, F. H. et al. High throughput profiling of
antibiotic resistance genes in urban park soils with
reclaimed water irrigation. Environ. Sci. T echnol. 48,
9079–9085 (2014).
29. de Jong, A. et al. Pan-European resistance monitoring
programmes encompassing food-borne bacteria and
target pathogens of food-producing and companion
animals. Int. J. Antimicrob. Agents 41, 403–409
(2013).
30. Morrissey, I. et al. A review of ten years of the study
for monitoring antimicrobial resistance trends
(SMART) from 2002 to 2011. Pharmaceuticals (Basel)
6, 1335–1346 (2013).
31. McArthur, A. G. et al. The comprehensive antibiotic
resistance database. Antimicrob. Agents Chemother.
57, 3348–3357 (2013).
32. Liu, B. & Pop, M. ARDB — antibiotic resistance genes
database. Nucleic Acids Res. 37, D443–D447
(2009).
33. Lagier, J. C. et al. Microbial culturomics: paradigm
shift in the human gut microbiome study. Clin.
Microbiol. Infect. 18, 1185–1193 (2012).
P E R S P E C T I V E S
316 | APRIL 2015 | VOLUME 13 https://www.doczj.com/doc/f46319258.html,/reviews/micro
? 2015 Macmillan Publishers Limited. All rights reserved
34. Graham, D. W. et al. Antibiotic resistance gene
abundances associated with waste discharges to the
Almendares River near Havana, Cuba. Environ. Sci.
T echnol. 45, 418–424 (2011).
35. Huerta, B. et al. Exploring the links between antibiotic
occurrence, antibiotic resistance, and bacterial
communities in water supply reservoirs. Sci. T otal
Environ.456–457, 161–170 (2013).
36. Novo, A., Andre, S., Viana, P., Nunes, O. C. &
Manaia, C. M. Antibiotic resistance, antimicrobial
residues and bacterial community composition in
urban wastewater. Water Res. 47, 1875–1887 (2013).
37. American Public Health Association. Standard
Methods for the Examination of Water and
Wastewater 21st edn (APHA, 2005).
38. Council of the European Union. Council Directive
98/83/EC. of 3 November 1998 on the quality of
water intended for human consumption as amended
by regulation 1882/2003/EC. FAOLEX [online],
https://www.doczj.com/doc/f46319258.html,/docs/pdf/eur18700.pdf (1998). 39. Fajardo, A. & Martinez, J. L. Antibiotics as signals that
trigger specific bacterial responses. Curr. Opin.
Microbiol. 11, 161–167 (2008).
40. Baquero, F., T edim, A. P. & Coque, T. M. Antibiotic
resistance shaping multi-level population biology of
bacteria. Front. Microbiol. 4, 15 (2013).
41. Gibson, M. K., Forsberg, K. J. & Dantas, G. Improved
annotation of antibiotic resistance determinants
reveals microbial resistomes cluster by ecology.
ISME J.9, 207–216 (2014).
42. Forsberg, K. J. et al. Bacterial phylogeny structures
soil resistomes across habitats. Nature 509, 612–616 (2014).
43. Port, J. A., Cullen, A. C., Wallace, J. C., Smith, M. N. &
Faustman, E. M. Metagenomic frameworks for
monitoring antibiotic resistance in aquatic
environments. Environ. Health Perspect. 122,
222–228 (2014).
44. Donato, J. J. et al. Metagenomic analysis of apple
orchard soil reveals antibiotic resistance genes
encoding predicted bifunctional proteins. Appl.
Environ. Microbiol. 76, 4396–4401 (2010).
45. Nesme, J. et al. Large-scale metagenomic-based study
of antibiotic resistance in the environment. Curr. Biol.
24, 1096–1100 (2014).
46. European Medicines Agency. Guideline on the
environmental risk assessment of medicinal products
for human use. Pre?authorisation evaluation of
medicines for human use (EMA,2006).
47. European Commission. Common Implementation
Strategy for the EU Water Framework Directive
(2000/60/EC). The guidance document No. 27:
technical guidance for deriving environmental quality
standards (European Communities, 2011).
48. Negri, M. C., Lipsitch, M., Blazquez, J., Levin, B. R. &
Baquero, F. Concentration-dependent selection of
small phenotypic differences in TEM β-lactamase-
mediated antibiotic resistance. Antimicrob. Agents
Chemother. 44, 2485–2491 (2000).
49. Andersson, D. I. & Hughes, D. Evolution of antibiotic
resistance at non-lethal drug concentrations. Drug
Resist. Updat. 15, 162–172 (2012).
50. Gullberg, E. et al. Selection of resistant bacteria at
very low antibiotic concentrations. PLoS Pathog. 7,
e1002158 (2011).
51. Goh, E. B. et al. T ranscriptional modulation of bacterial
gene expression by subinhibitory concentrations of
antibiotics. Proc. Natl Acad. Sci. USA 99,
17025–17030 (2002).
52. Davies, J., Spiegelman, G. B. & Yim, G. The world of
subinhibitory antibiotic concentrations. Curr. Opin.
Microbiol. 9, 445–453 (2006).
53. Bruchmann, J., Kirchen, S. & Schwartz, T. Sub-
inhibitory concentrations of antibiotics and
wastewater influencing biofilm formation and gene
expression of multi-resistant Pseudomonas
aeruginosa wastewater isolates. Environ. Sci. Pollut.
Res. Int. 20, 3539–3549 (2013).
54. T ello, A., Austin, B. & T elfer, T. C. Selective pressure of
antibiotic pollution on bacteria of importance to public
health. Environ. Health Perspect. 120, 1100–1106
(2012).
55. Ciusa, M. L. et al. A novel resistance mechanism to
triclosan that suggests horizontal gene transfer and
demonstrates a potential selective pressure for
reduced biocide susceptibility in clinical strains of
Staphylococcus aureus. Int. J. Antimicrob. Agents 40,
210–220 (2012).
56. Davies, J. Everything depends on everything else.
Clin. Microbiol. Infect. 15 (Suppl. 1), 1–4 (2009).
57. Seiler, C. & Berendonk, T. U. Heavy metal driven
co-selection of antibiotic resistance in soil and water
bodies impacted by agriculture and aquaculture.
Front. Microbiol. 3, 399 (2012).
58. Baquero, F. Environmental stress and evolvability in
microbial systems. Clin. Microbiol. Infect. 15
(Suppl. 1), 5–10 (2009).
59. Martinez, J. L. et al. A global view of antibiotic
resistance. FEMS Microbiol. Rev. 33, 44–65 (2009).
60. Nielsen, K. M., Bohn, T. & T ownsend, J. P. Detecting
rare gene transfer events in bacterial populations.
Front. Microbiol. 4, 415 (2014).
61. Nguyen, T. T. et al. Mathematical modeling of bacterial
kinetics to predict the impact of antibiotic colonic
exposure and treatment duration on the amount of
resistant enterobacteria excreted. PLoS Comput. Biol.
10, e1003840 (2014).
62. Kohanski, M. A., DePristo, M. A. & Collins, J. J.
Sublethal antibiotic treatment leads to multidrug
resistance via radical-induced mutagenesis. Mol. Cell
37, 311–320 (2010).
63. European Parliament and the Council of the European
Union. Directive 2000/60/EC of the European
Parliament and of the Council of 23 October 2000
establishing a framework for community action in the
field of water policy. FAOLEX [online], http://faolex.fao.
org/docs/pdf/eur23005.pdf (2000).
64. Czekalski, N., Berthold, T., Caucci, S., Egli, A. &
Burgmann, H. Increased levels of multiresistant
bacteria and resistance genes after wastewater
treatment and their dissemination into Lake Geneva,
Switzerland. Front. Microbiol. 3, 106 (2012).
65. Vredenburg, J. et al. Quinolone-resistant Escherichia
coli isolated from birds of prey in Portugal are
genetically distinct from those isolated from water
environments and gulls in Portugal, Spain and
Sweden. Environ. Microbiol. 16, 995–1004 (2014).
66. Jechalke, S. et al. Increased abundance and
transferability of resistance genes after field
application of manure from sulfadiazine-treated pigs.
Appl. Environ. Microbiol. 79, 1704–1711 (2013).
67. Udikovic-Kolic, N., Wichmann, F., Broderick, N. A. &
Handelsman, J. Bloom of resident antibiotic-resistant
bacteria in soil following manure fertilization. Proc.
Natl Acad. Sci. USA 111, 15202–15207 (2014).
68. Baquero, F., Coque, T. M. & de la Cruz, F. Ecology and
evolution as targets: the need for novel eco–evo drugs
and strategies to fight antibiotic resistance. Antimicrob.
Agents Chemother. 55, 3649–3660 (2011).
69. Soonthornchaikul, N. et al. Resistance to three
antimicrobial agents of Campylobacter isolated from
organically- and intensively-reared chickens purchased
from retail outlets. Int. J. Antimicrob. Agents 27,
125–130 (2006).
70. Silbergeld, E. K., Graham, J. & Price, L. B. Industrial
food animal production, antimicrobial resistance, and
human health. Annu. Rev. Publ. Health 29, 151–169
(2008).
71. Li, D. et al. Antibiotic-resistance profile in
environmental bacteria isolated from penicillin
production wastewater treatment plant and the
receiving river. Environ. Microbiol. 11, 1506–1517
(2009).
72. Kristiansson, E. et al. Pyrosequencing of antibiotic-
contaminated river sediments reveals high levels of
resistance and gene transfer elements. PLoS ONE 6,
e17038 (2011).
73. Jechalke, S., Heuer, H., Siemens, J., Amelung, W. &
Smalla, K. Fate and effects of veterinary antibiotics in
soil. T rends Microbiol. 22, 536–545 (2014).
74. Shade, A. et al. Streptomycin application has no
detectable effect on bacterial community structure in
apple orchard soil. Appl. Environ. Microbiol. 79,
6617–6625 (2013).
75. Kühn, I. et al. Occurrence and relatedness of
vancomycin-resistant enterococci in animals, humans,
and the environment in different European regions.
Appl. Environ. Microbiol. 71, 5383–5390 (2005).
76. World Health Organization. T ackling antibiotic
resistance from a food safety perspective in Europe
(WHO, 2011).
77. Brooks, J. P., Adeli, A. & McLaughlin, M. R. Microbial
ecology, bacterial pathogens, and antibiotic resistant
genes in swine manure wastewater as influenced by
three swine management systems. Water Res. 57,
96–103 (2014).
78. Garcia-Migura, L., Hendriksen, R. S., Fraile, L. &
Aarestrup, F. M. Antimicrobial resistance of zoonotic
and commensal bacteria in Europe: the missing link
between consumption and resistance in veterinary
medicine. Vet. Microbiol. 170, 1–9 (2014).
79. Moura, A., Henriques, I., Ribeiro, R. & Correia, A.
Prevalence and characterization of integrons from
bacteria isolated from a slaughterhouse wastewater
treatment plant. J. Antimicrob. Chemother. 60,
1243–1250 (2007).
80. Pei, R., Cha, J., Carlson, K. H. & Pruden, A. Response
of antibiotic resistance genes (ARG) to biological
treatment in dairy lagoon water. Environ. Sci. T echnol.
41, 5108–5113 (2007).
81. Resende, J. A. et al. Prevalence and persistence of
potentially pathogenic and antibiotic resistant
bacteria during anaerobic digestion treatment of
cattle manure. Bioresour. T echnol. 153, 284–291
(2014).
82. Michael, I. et al. Urban wastewater treatment plants
as hotspots for the release of antibiotics in the
environment: a review. Water Res. 47, 957–995
(2013).
Acknowledgements
The authors thank the EU for support provided through COST
Action TD0803: DARE, and the contributions of A. T ello,
A. von Haeseler, H.-P. Grossart, H. Garelick, L. Rizzo,
I. Henriques, M. Cani?a, M. Coci, M. W?gerbauer and
T. T enson. C.M.M., T.U.B., T.S. and D.F.K. thank the National
Funding Agencies through the WATER JPI project StARE
(WaterJPI/0001/2013). Acknowledged funding: T.U.B. is
funded by ANTI-Resist (02WRS1272A; Bundesministerium für
Bildung und Forschung (BMBF)) and the ‘support the best’
programme of the T echnische Universit?t Dresden and Deutsch
Forschung Gründung; T.S. is funded by T ransRisk (BMBF); C.M.
is funded by the Agence Nationale de la Recherche,
Programmes ECOTECH and BIOADAPT, and the Zone Atelier
Moselle (ZAM); E.C. is funded by grant #821014213 from the
Israeli Ministry of Agriculture; V.K. is funded by a European
Regional Development Fund through the Centre of Excellence
in Chemical Biology, Estonia; and C.M.M. is funded by the
Funda??o para a Ciência e a T ecnologia, Portugal, projects
PEst-OE/EQB/LA0016/2013.
Competing interests statement
The authors declare no competing interests.
P E R S P E C T I V E S
NATURE REVIEWS |MICROBIOLOGY VOLUME 13 | APRIL 2015 |317
? 2015 Macmillan Publishers Limited. All rights reserved
英语广告语集锦 本文是关于经典句子的,仅供参考,如果觉得很不错,欢迎点评和分享。 英语广告语集锦 1.Time is what you make of it.(Swatch) 天长地久。(斯沃奇手表) 2.Make yourself heard.(Ericsson) 理解就是沟通。(爱立信) 3.Engineered to move the human spirit.(Mercedes-Benz) 人类精神的动力。(梅塞德斯-奔驰) 4.Start Ahead.(Rejoice) 成功之路,从头开始。(飘柔) 5.A diamond lasts forever.(De Bierres) 钻石恒久远,一颗永流传。(第比尔斯) 6.Fresh-up with Seven-up.(Seven-up) 提神醒脑,喝七喜。(七喜) 7.Intel Inside.(Intel Pentium) 给电脑一颗奔腾的“芯”。(英特尔奔腾) 8.Connecting People.(Nokia) 科技以人为本。(诺基亚) 9.For the Road Ahead.(Honda)
康庄大道。(本田) 10.Let us make things better.(Philips) 让我们做的更好。(飞利浦) 11.Enjoy Coca-Cola.(Coca-Cola) 请喝可口可乐。(可口可乐) 12.Generation Next.(Pepsi) 新的一代。(百事) 13.The Relentless Pursuit of Perfection.(Lexus) 追求完美永无止境。(凌志汽车) https://www.doczj.com/doc/f46319258.html,munication unlimited.(Motorola) 沟通无极限。(摩托罗拉) 15.Feast your eyes.(Pond’s Cucumber Eye Treatment) 滋润心灵的窗户。(庞氏眼贴片) 16.Focus on life.(Olympus) 瞄准生活。(奥林巴斯) 17.Behind that healthy smile,t here ’s a Crest kid.(Crest toothpaste) 健康笑容来自佳洁士。(佳洁士牙膏) 关于英语的广告词英语培训学校的广告语经典广告词(英语)感谢阅读,希望能帮助您!
英语口语8000句:喜欢上某人时常用口语句子 恋爱和结婚 ●喜欢、爱上…… ◎汤姆是个美男子。 Tom is a lady-killer. *lady-killer直译是“少女杀手”,其实不是杀手,而是指一下子就能迷住女人的男子。 = Tom dates around a lot. (汤姆和好多女人来往。) = Tom is a real playboy. (汤姆真是个花花公子。) ◎汤姆真让我神魂颠倒。 Tom really turns me on. *turn...on“有性方面的吸引力”、“使人着迷”。 I didn't know you felt that way. (我一点儿都不知道你的感觉。) = I'm crazy about Tom. = I have strong feelings for Tom. = I love Tom. = I have the hots for Tom. *俚语。 ◎克里斯长得真帅。 Chris is really a heartbreaker. *用heartbreaker表示“长得很帅,对异性有吸引力的人”。进 一步讲,“heartbreaker”是指给异性带来撕心裂肺的痛苦和失望的人,魅惑他人,让人沉醉的人。Elvis Presley (爱尔维斯·普里斯利)有一首成名曲“Heartbreak Hotel”,指的就是“(因恋爱而)绝望的 人住的饭店”。
Chris breaks a lot of hearts. (克里斯使很多女人尝到了失恋的痛苦。) Chris dates a lot of women. (克里斯和很多女人有来往。) ◎珍妮特真迷人。Janet is a knockout. *如同拳击中的“knock out”一样,表示极具魅力、使对方晕头转向的人,多指女性。 You can say that again! (颇有同感。) = Janet is sexy. = Janet is beautiful. ◎他好像看上你了。I think he has a crush on you. *have a crush on...“看上……”。 Give me a break. (别随便瞎说。) = I think he is infatuated with you. * be infatuated with...“被……迷住,为……神魂颠倒”。 = I think he likes you. ◎简好像喜欢上我了。Jane seems to like me. *seem“好像,看上去像……”。 = I've got the feeling that Jane likes me. = I think Jane likes me. = I have a hunch (that) Jane likes me. ◎戴安娜对杰克有意思。Diana's been coming on to Jack.
英语语法大全 初中英语语法 学习提纲 一、词类、句子成分和构词法: 1、词类:英语词类分十种: 名词、形容词、代词、数词、冠词、动词、副词、介词、连词、感叹词。 1、名词(n.):表示人、事物、地点或抽象概念的名称。如:boy, morning, bag, ball, class, orange. 2、代词(pron.):主要用来代替名词。如:who, she, you, it . 3、形容词(adj..):表示人或事物的性质或特征。如:good, right, white, orange . 4、数词(num.):表示数目或事物的顺序。如:one, two, three, first, second, third, fourth. 5、动词(v.):表示动作或状态。如:am, is,are,have,see . 6、副词(adv.):修饰动词、形容词或其他副词,说明时间、地点、程度等。如:now, very, here, often, quietly, slowly. 7、冠词(art..):用在名词前,帮助说明名词。如:a, an, the. 8、介词(prep.):表示它后面的名词或代词与其他句子成分的关系。如in, on, from, above, behind. 9、连词(conj.):用来连接词、短语或句子。如and, but, before . 10、感叹词(interj..)表示喜、怒、哀、乐等感情。如:oh, well, hi, hello. 2、句子成分:英语句子成分分为七种:主语、谓语、宾语、定语、状语、表语、宾语补足语。 1、主语是句子所要说的人或事物,回答是“谁”或者“什么”。通常用名词或代词担任。如:I’m Miss Green.(我是格林小姐) 2、谓语动词说明主语的动作或状态,回答“做(什么)”。主要由动词担任。如:Jack cleans the room every day. (杰克每天打扫房间) 3、表语在系动词之后,说明主语的身份或特征,回答是“什么”或者“怎么样”。通常由名词、代词 或形容词担任。如:My name is Ping ping .(我的名字叫萍萍) 4、宾语表示及物动词的对象或结果,回答做的是“什么”。通常由名词或代词担任。如:He can spell the word.(他能拼这个词) 有些及物动词带有两个宾语,一个指物,一个指人。指物的叫直接宾语,指人的叫间接宾语。间 接宾语一般放在直接宾语的前面。如:He wrote me a letter . (他给我写了一封信) 有时可把介词to或for加在间接宾语前构成短语,放在直接宾语后面,来强调间接宾语。如: He wrote a letter to me . (他给我写了一封信) 5、定语修饰名词或代词,通常由形容词、代词、数词等担任。如:
经典英语广告语大全 our wheels are always turning. 我们的车轮常转不停。(五十铃汽车) the world smiles with reader’s digest. 《读者文摘》给全世界带来欢笑。(《读者文摘》) one should love animals. they are so tasty. 每个人都应该热爱动物,因为它们很好吃. love the neighbor. but don‘t get caught. 要用心去爱你的邻居,不过不要让她的老公知道. anything is possible. 没有不可能的事。(东芝电子) /take toshiba, take the world. 拥有东芝,拥有世界。(东芝电子) nobody is perfect. 没有一个人的身材是十全十美的。(苗条健身器材) behind every successful man, there is a woman. and behind every unsuccessful man, there are two. 每个成功男人的背后都有一个女人,每个不成功男人的背后都有两个女人。
every man should marry. after all, happiness is not the only thing in life. 再快乐的单身汉迟早也会结婚,幸 福不是永久的嘛. no business too small, no problem too big. 没有不做 的小生意,没有解决不了的大问题。(ibm公司) the wise never marry, and when they marry they become otherwise. 聪明人都是未婚的,结婚的人很难再聪明起来. we’re the dot. in. com. 我们就是网络。(太阳微系统 公司) children in backseats cause accidents. accidents in backseats cause children. 后排座位上的小孩会生出意外,后排 座位上的意外会生出小孩. the new digital era. 数码新时代。(索尼影碟机) love is photogenic. it needs darkness to develop. 爱情就象照片, 需要大量的暗房时间来培养. good to the last drop. 滴滴香浓,意犹未尽。(麦氏咖啡) a kodak moment. 就在柯达一刻。(柯达胶卷)/share moments. share life. (柯达胶卷)
1.在家中 ●从起床到出门 早晨好 Good morning 闹钟响了吗? *go off是闹钟“响”的意思。 Did the alarm clock go off? 该起床了 -It's time to get up! -I don't wanna get up. 快点儿起床! -Get up soon. -I don't want to. 你醒了吗? *get up是动词,表示“起床”、“起”的动作。awake是形容词,表示“醒了”、“没睡”-Are you awake? -I am now. (我刚醒。) 你不舒服吗? -Are you feeling sick? -No, I'm just tired. 睡得好吗? -Did you sleep well? -Yes, I slept very well. / -No, I couldn't fall asleep. ? Would you turn off the alarm clock? -You finally got up. -I'm still sleepy. (我还困着呢!) 今天是个好天! -It's a nice day! -It sure is. ? Did you stay up late last night? 把被子叠好。 Let's fold up the futon. *snore“打呼噜”。 -Did I keep you up? -You were snoring last night. 过去进行时 我做了个可怕的梦。 -I had a nightmare. -It's all right now. 你一直没关灯啊。 You left the light on. I have to go wash my face. 该吃早饭了。 It's time to eat/have breakfast. 我还困着呢。 I'm still sleepy. 我还打哈欠呢。 I'm still yawning. ['j?:ni?] I have a hangover. I'm a night person. 我是用咖啡来提神的。 Coffee wakes me up. 刷牙了吗? Did you brush your teeth? I have to comb my hair. [k?um] vt 梳头发 穿什么好呢 -What should I wear/put on? -The red one. (穿红的吧!) 快换衣服。 Hurry up and get dressed. 把睡衣收好。*put away 收拾,放好-Put those pajamas away! [p?'d?ɑ:m?z] 睡衣 啊,我正要洗呢。) 我走了。妈妈再见! -I'm leaving. Bye mom! -Study hard. 今天我们逃学吧。*play hooky为俚语“逃学”。 -Yeah, let's. 你毛衣穿反了。 You're wearing your sweater inside out. 上下颠倒了。 It's upside down. 可别忘了扔垃圾! ['ɡɑ:bid?] n垃圾,废物 -Don't forget to take out the garbage. -I won't. 今天该你扔垃圾了。 It's your turn to take out the garbage. 今天你们干嘛? -What are you doing today? -We're having a track and field meet. *运动会 你快点儿,我们该迟到了! If you don't hurry, we'll be late. 快点儿,上学该迟到了。 -Hurry or you'll be late for school. -What time is it? 你锁门了吗? Did you lock the door? 没忘了什么东西吧? -Aren't you forgetting something? 都已经8点了! It's already 8:00. 我晚了! I'm late! 我得赶紧走! I have to rush! 你今天会回来得晚吗? -Are you gonna be late today? -No, I'll be home at the usual time. 几点回来? -What time are you coming home? -Around seven o'clock. 饭盒带了吗? -Have you got your lunch box? -Yes, right here. 今天好像要下雨。 -It might rain today. -Take your umbrella with you. [?m'brel?] 出门的时候,可别忘了锁门。 Don't forget to lock the door when you leave. ●从回家到就寝 我回来了。 I'm home. 你回来了。 Welcome home! 今天过得愉快吗? Did you have a good time? 今天怎么样? How did it go today? 我可以出去玩儿会儿吗? -Can I go out to play? -After you finish your homework. 我饿了。 -I'm hungry. -We have some snacks. *[sn?k] 点心 点心在哪儿?
初中英语语法大全汇总 (一) 一.词类( ) 名词英文名称(缩写为n.) 表示人或事物的名称例词等 冠词英文名称(缩写为.) 用在名词前帮助说明名词所指的人和或事物。例词a() 代词英文名称(缩写为) 用来代替名词、形容词或是数词例词 形容词英文名称(缩写为.) 用以修饰名词,表示人或事物的特征 例词. 数词英文名称(缩写为.) 表示数量或是顺序。例词 动词英文名称(缩写为v.) 表示动作或状态。例词() 副词英文名称(缩写为.) 修饰动词、形容词或其他副词。例词 介词英文单词(缩写为.) 表示名词、代词等和句中其他词的关系。例词. 连词英文单词(缩写为.) 用来连接词与词、短语与短语或句与句。例词. 感叹词英文单词(缩写为.) 表示说话时的喜悦、惊讶等情感。例词. 二.名词() 1.总的说来,名词分专有名词和普通名词两类。 专有名词: 表示具体的人,事物,地点或机构的专有名称。 中国亚洲北京。 专有名词的第一个字母要大写。 普通名词: 表示某些人,某类事物,某种物质或抽象概念的名称。例如: 老师茶改革 普通名词又可进一步分为四类 1) 个体名称: 表示单个的人和事物。 马汽车房间苹果风扇照片 2) 集体名称: 表示一群人或一些事物的名称。 人们家庭军队政府集团 3) 物质名词:表示物质或不具备确定形状和大小的个体的物质。 火钢空气水牛奶 4)抽象名词:表示动作,状态,品质或其他抽象概念。 劳动健康生活友情耐力 2.名词按其所表现的事物的性质分为可数名词和不可数名词。 可数名词( )有复数形式,如: a 不可数名词( )一般没有复数形式. 抽象名词, 物质名词和专有名词一般是不可数名词。 沙糖 有少数名词即可作可数名词,也可作不可数名词,但含义不同。 玻璃玻璃杯纸报纸,文件 名词的功能 名词在句中作主语, 宾语,介词宾语,宾语补助语,表语以及名词短语作状语。
附录A Introduction to database information management system The database is stored together a collection of the relevant data, the data is structured, non-harmful or unnecessary redundancy, and for a variety of application services, data storage independent of the use of its procedures, insert new data on the database, revised, and the original data can be retrieved by a common and can be controlled manner. When a system in the structure of a number of entirely separate from the database, the system includes a "database collection". Database management system is a manipulation and large-scale database management software is being used to set up, use and maintenance of the database. Its unified database management and control so as to ensure database security and integrity. Database management system users access data in the database, the database administrator through Database management system database maintenance work. It provides a variety of functions, allows multiple applications and users use different methods at the same time or different time to build, modify, and asked whether the database. It allows users to easily manipulate data definition and maintenance of data security and integrity, as well as the multi-user concurrency control and the restoration of the database. Using the database can bring many benefits: such as reducing data redundancy, thus saving the data storage space; to achieve full sharing of data resources, and so on. In addition, the database technology also provides users with a very simple means to enable users to easily use the preparation of the database applications. Especially in recent years introduced micro-computer relational database management system , intuitive operation, the use of flexible, convenient programming environment to extensive (generally 16 machine, such as IBM / PC / XT, China Great Wall 0520, and other species can run software), data-processing capacity strong. Database in our country are being more and more widely used, will be a powerful tool of economic management. The database is through the database management system (DBMS-DATA BASE
英语广告语集锦 导读: 英语广告语集锦 1.Time is what you make of it.(Swatch) 天长地久。(斯沃奇手表) 2.Make yourself heard.(Ericsson) 理解就是沟通。(爱立信) 3.Engineered to move the human spirit.(Mercedes-Benz) 人类精神的动力。(梅塞德斯-奔驰) 4.Start Ahead.(Rejoice) 成功之路,从头开始。(飘柔) 5.A diamond lasts forever.(De Bierres) 钻石恒久远,一颗永流传。(第比尔斯) 6.Fresh-up with Seven-up.(Seven-up) 提神醒脑,喝七喜。(七喜) 7.Intel Inside.(Intel Pentium) 给电脑一颗奔腾的“芯”。(英特尔奔腾) 8.Connecting People.(Nokia) 科技以人为本。(诺基亚) 9.For the Road Ahead.(Honda) 康庄大道。(本田) 10.Let us make things better.(Philips)
让我们做的更好。(飞利浦) 11.Enjoy Coca-Cola.(Coca-Cola) 请喝可口可乐。(可口可乐) 12.Generation Next.(Pepsi) 新的一代。(百事) 13.The Relentless Pursuit of Perfection.(Lexus) 追求完美永无止境。(凌志汽车) https://www.doczj.com/doc/f46319258.html,munication unlimited.(Motorola) 沟通无极限。(摩托罗拉) 15.Feast your eyes.(Pond’s Cucumber Eye Treatment) 滋润心灵的窗户。(庞氏眼贴片) 16.Focus on life.(Olympus) 瞄准生活。(奥林巴斯) 17.Behind that healthy smile,there ’s a Crest kid.(Crest toothpaste) 健康笑容来自佳洁士。(佳洁士牙膏)
英语口语8000 句 ●从起床到出门 Good morning, mom. 早晨好,妈妈。 Did the alarm clock go off ? Did the alarm clock buzz? Did the alarm clock ring? It's time to get up! 该起床了! I don't wanna get up. 我真不想起。w a n n a=w a n t t o;g o n n a=b e g o i n g t o美语口语常用说法It's time to get ready. Get up soon. 快点儿起床! I don't want to. 我真不想起。 Are you awake? 你醒了吗? I am now. 我刚醒。 Are you feeling sick? 你不舒服吗? No, I'm just tired. 没有,只是有点儿累 Did you sleep well? 睡得好吗? Yes, I slept very well. 嗯,睡得挺好。 No, I couldn't fall asleep.哪儿啊几乎没睡着Would you turn off the alarm clock? Please turn off the alarm clock. You finally got up. 你终于起来了。 It's a nice day! 今天是个好天! It sure is. 是不错啊。 It's a beautiful day! It's a wonderful day! It's a great day! Did you stay up late last night? Did you go to bed late last night? Let's fold up the futon. 把被子叠好 Let's put the futon away. 把被子收起 You were snoring last night. 打呼噜 Did I keep you up? 影响你睡觉了吗? I had a nightmare. 我做了个可怕的梦 It's all right now. 现在没事了/挺好的 You left the light on. 你一直没关灯啊 You forgot to turn off the light.你忘关灯了 I have to go wash my face. 我得.... It's time to eat breakfast. It's time to have breakfast.
世界名牌英文广告语大全 世界名牌英文广告语举例 1. Everything from head to toe should be coordinated, Wee bok brand clothing and shoes to do this. Wee bok brand clothing 2.Luzon ------ more your style! Luzon brand clothing 3. Put on “Janssen”, you just like to put on a smile. Jansen garments 4. No matter how you look, “Lands'End” swimsuit always make you satisfied, perfect body shape. Lands'End swimsuit 5. Dancing graceful puter promotional advertising language, to develop Italian pride! Cool! Shuai! Italian modern clothing 6. Elegant chic Leiteng Meng! Reiter Mongolian clothing 7. Not seeking the position of the emperor, but the emperor demeanor! Emperor clothing 8. Silver are fashion, international brand! Silver are brand fashion 9. Li Ning slogan: everything is possible.
REVIEW ARTICLE published:28September2011 doi:10.3389/fmicb.2011 .00203 Acquired antibiotic resistance genes:an overview Angela H.A.M.van Hoek1,Dik Mevius2,3,Beatriz Guerra4,Peter Mullany5,Adam Paul Roberts5and Henk J.M.Aarts1* 1Laboratory for Zoonoses and Environmental Microbiology,Centre for Infectious Disease Control,National Institute of Public Health and the Environment,Utrecht, Netherlands 2Central Veterinary Institute of Wageningen UR,Lelystad,Netherlands 3Department of Infectious Diseases and Immunology,Utrecht University,Utrecht,Netherlands 4National Salmonella Reference Laboratory,Federal Institute for Risk Assessment,Berlin,Germany 5Department of Microbial Diseases,University College London Eastman Dental Institute,University College London,London,UK Edited by: Timothy Rutland Walsh,Cardiff University,UK Reviewed by: M.Pilar Francino,Center for Public Health Research,Spain Jun Liu,Mount Sinai School of Medicine,USA *Correspondence: Henk J.M.Aarts,National Institute of Public Health and the Environment, Antonie van Leeuwenhoekla9,3721 MA Bilthoven,Utrecht,Netherlands. e-mail:henk.aarts@rivm.nl In this review an overview is given on antibiotic resistance(AR)mechanisms with special attentions to the AR genes described so far preceded by a short introduction on the dis-covery and mode of action of the different classes of antibiotics.As this review is only dealing with acquired resistance,attention is also paid to mobile genetic elements such as plasmids,transposons,and integrons,which are associated with AR genes,and involved in the dispersal of antimicrobial determinants between different bacteria. Keywords:antimicrobial resistance mechanisms,acquired,antibiotics,mobile genetic elements INTRODUCTION The discovery and production of(synthetic)antibiotics in the?rst half of the previous century has been one of medicine’s greatest achievements.The use of antimicrobial agents has reduced mor-bidity and mortality of humans and contributed substantially to human’s increased life span.Antibiotics are,either as therapeutic or as prophylactic agents,also widely used in agricultural practices. The?rst discovered antimicrobial compound was penicillin (Flemming,1929)aβ-lactam antibiotic.Soon after this very important discovery,antibiotics were used to treat human infec-tions starting with sulfonamide and followed by the aminoglyco-side streptomycin and streptothricin(Domagk,1935;Schatz and Waksman,1944).Nowadays numerous different classes of antimi-crobial agents are known and they are classi?ed based on their mechanisms of action(Neu,1992).Antibiotics can for instance inhibit protein synthesis,like aminoglycoside,chloramphenicol, macrolide,streptothricin,and tetracycline or interact with the syn-thesis of DNA and RNA,such as quinolone and rifampin.Other groups inhibit the synthesis of,or damage the bacterial cell wall asβ-lactam and glycopeptide do or modify,like sulfonamide and trimethoprim,the energy metabolism of a microbial cell. Upon the introduction of antibiotics it was assumed that the evolution of antibiotic resistance(AR)was unlikely.This was based on the assumption that the frequency of mutations generating resistant bacteria was negligible(Davies,1994).Unfortunately, time has proven the opposite.Nobody initially anticipated that microbes would react to this assault of various chemical poi-sons by adapting themselves to the changed environment by developing resistance to antibiotics using such a wide variety of mechanisms.Moreover,their ability of interchanging genes, which is now well known as horizontal gene transfer(HGT)emergence of resistance actually began before the?rst antibiotic, penicillin,was characterized.The?rstβ-lactamase was identi-?ed in Escherichia coli prior to the release of penicillin for use in medical practice(Abraham and Chain,1940).Besidesβ-lactams, the aminoglycoside–aminocyclitol family was also one of the?rst groups of antibiotics to encounter the challenges of resistance (Wright,1999;Bradford,2001).Over the years it has been shown by numerous ecological studies that(increased)antibiotic con-sumption contributes to the emergence of AR in various bacterial genera(MARAN,2005,2007;NethMap,2008).Some examples of the link between antibiotic dosage and resistance development are the rise of methicillin-resistant Staphylococcus aureus(MRSA) and vancomycin-resistant enterococci(VRE).The initial appear-ance of MRSA was in1960(Jevons et al.,1963),whereas VRE were ?rst isolated about20years ago(Uttley et al.,1988).Over the last decades they have remained a reason for concern,but additional public health threats in relation to resistant microorganisms have also arisen(see for example Cantón et al.,2008;Goossens,2009; Allen et al.,2010). Bacteria have become resistant to antimicrobials through a number of mechanisms(Spratt,1994;McDermott et al.,2003; Magnet and Blanchard,2005;Wright,2005): I.Permeability changes in the bacterial cell wall which restricts antimicrobial access to target sites, II.Active ef?ux of the antibiotic from the microbial cell, III.Enzymatic modi?cation of the antibiotic, IV.Degradation of the antimicrobial agent, V.Acquisition of alternative metabolic pathways to those inhib-ited by the drug, VI.Modi?cation of antibiotic targets,
1 (see 、hear 、notice 、find 、feel 、listen to 、 look at (感官动词)+do eg:I like watching monkeys jump 2 (比较级 and 比较级)表示越来越怎么样 3 a piece of cake =easy 小菜一碟(容易) 4 agree with sb 赞成某人 5 all kinds of 各种各样 a kind of 一样 6 all over the world = the whole world 整个世界 7 along with同……一道,伴随…… eg : I will go along with you我将和你一起去 the students planted trees along with their teachers 学生同老师们一起种树 8 As soon as 一怎么样就怎么样 9 as you can see 你是知道的 10 ask for ……求助向…要…(直接接想要的东西) eg : ask you for my book 11 ask sb for sth 向某人什么 12 ask sb to do sth 询问某人某事 ask sb not to do 叫某人不要做某事 13 at the age of 在……岁时 eg:I am sixteen I am at the age of sixteen 14 at the beginning of …………的起初;……的开始 15 at the end of +地点/+时间最后;尽头;末尾 eg : At the end of the day 16 at this time of year 在每年的这个时候 17 be /feel confident of sth /that clause +从句感觉/对什么有信心,自信 eg : I am / feel confident of my spoken English I feel that I can pass the test 18 be + doing 表:1 现在进行时 2 将来时 19 be able to (+ v 原) = can (+ v 原)能够…… eg : She is able to sing She can sing 20 be able to do sth 能够干什么 eg :she is able to sing 21 be afraid to do (of sth 恐惧,害怕…… eg : I'm afraed to go out at night I'm afraid of dog 22 be allowed to do 被允许做什么 eg: I'm allowed to watch TV 我被允许看电视 I should be allowed to watch TV 我应该被允许看电视 23 be angry with sb 生某人的气 eg : Don't be angry with me 24 be angry with(at) sb for doing sth 为什么而生某人的气 25 be as…原级…as 和什么一样 eg : She is as tall as me 她和我一样高 26 be ashamed to 27 be away from 远离 28 be away from 从……离开 29 be bad for 对什么有害 eg : Reading books in the sun is bad for your eyes 在太阳下看书对你的眼睛不好 30 be born 出生于 31 be busy doing sth 忙于做什么事 be busy with sth 忙于…… 32 be careful 当心;小心 33 be different from……和什么不一样 34 be famous for 以……著名 35 be friendly to sb 对某人友好 36 be from = come from 来自 eg :He is from Bejing He comes from Bejing Is he from Bejing Does he come from Bejing