
Jpn.J.Infect.Dis.,63,157-165,2010
Review
The Molecular Virology and Reverse Genetics of Influenza C Virus
Yasushi Muraki1,2*and Seiji Hongo2
1Department of Microbiology,Kanazawa Medical University School of Medicine,Ishikawa920-0293;
and2Department of Infectious Diseases,Yamagata University Faculty of Medicine,
Yamagata990-9585,Japan
(Received January18,2010.Accepted March29,2010)
CONTENTS
1.Introduction
2.Reverse genetics of influenza viruses
3.Molecular virology of influenza C virus
4.Epidemiology of influenza C virus
5.Generation of influenza C virus-like particles and a
recombinant influenza C virus 6.Analysis of the structure-function relationship of
the M1protein
https://www.doczj.com/doc/2a5145719.html,parison of the generation efficiencies of in-
fluenza C and A viruses
8.Prospects for research on influenza C virology
9.Conclusion
SUMMARY:Influenza C virus,an enveloped virus containing seven single-stranded RNA segments of negative polarity,belongs to the genus Influenza C Virus of the family Orthomyxoviridae.A number of questions remain to be resolved with regard to the molecular virology and epidemiology of the virus.To address them,we have established a virus-like particle(VLP)generation system and reverse genetics of the virus and succeeded in clarifying the structure-function relationship of the M1protein of the virus. Although the approach adopted was similar to that for influenza A virus reverse genetics,the number of infectious influenza C viruses generated was much lower than that for influenza A virus.Based on a comparison of the number of influenza C VLPs with that of influenza A VLPs generated using a similar system,we proposed a virion generation mechanism unique to influenza C virus.
1.Introduction
Reverse genetics,as the term is used in molecular virology,describes the generation of viruses possessing genome(s)derived from cloned cDNA(s).Of the viruses belonging to the family Orthomyxoviridae,reverse genetics has hitherto been reported for influenza A,in-fluenza B and Thogoto viruses.Recently,our research group,as well as Crescenzo-Chaigne and van der Werf, reported the successful reverse genetics of influenza C virus.
In this review,we will first provide an overview of research on reverse genetics of influenza A virus,and then summarize the molecular virology and epidemiolo-gy of influenza C virus,including a discussion of presently unresolved issues of the virus.In the latter part of this review,we will deal with a virus-like particle (VLP)generation and reverse genetics of influenza C vi-rus by our research group and provide a hypothesis to explain influenza C virion generation as observed in the established reverse-genetics system.
2.Reverse genetics of influenz aviruses
The genomes of negative-sense RNA viruses,includ-ing influenza viruses,are noninfectious.Therefore,for the generation of negative-sense RNA viruses,re-searchers faced the obstacle of providing the viral RNA(vRNA)with viral RNA polymerase and nucleo-protein.In the case of influenza A virus,the viral ribonucleoprotein(vRNP)complex,composed of three polymerase subunits(PB2,PB1,and PA), nucleoprotein(NP)and vRNA,is minimally required. Initially,by transfecting an artificially reconstituted vRNP complex into eukaryotic cells followed by infec-tion with an influenza helper virus,successful recovery of a recombinant influenza virus containing a viral gene segment derived from the cloned cDNA was reported (1,2).
In1996,Pleschka et al.reported the successful gener-ation of a transfectant influenza virus(3).To synthesize influenza vRNA in the nucleus,they made use of a nucleolar enzyme,RNA polymerase I,in a system that had been established by Zobel et al.(4).Cotransfection of the Pol Iplasmid encoding neuraminidase(N A) vRNA together with viral protein-expressing plasmids for PB2,PB1,PA,and NP,followed by infection with an influenza helper virus,resulted in the recovery of a recombinant containing the NA gene of interest.Even in using this method,however,the helper virus-depen-dent system remained an obstacle to the efficient recov-ery of the recombinant virus.
In1999,a recombinant influenza A virus was report-ed to be generated entirely from cloned cDNA for the first time(5).The cDNAs of the eight vRNA segments of A/WSN/33or A/PR/8/34virus were each cloned into the pHH21vector in negative-sense orientation be-tween the RN A polymerase Ipromoter and terminator sequences.The resulting eight plasmids were transfected
157
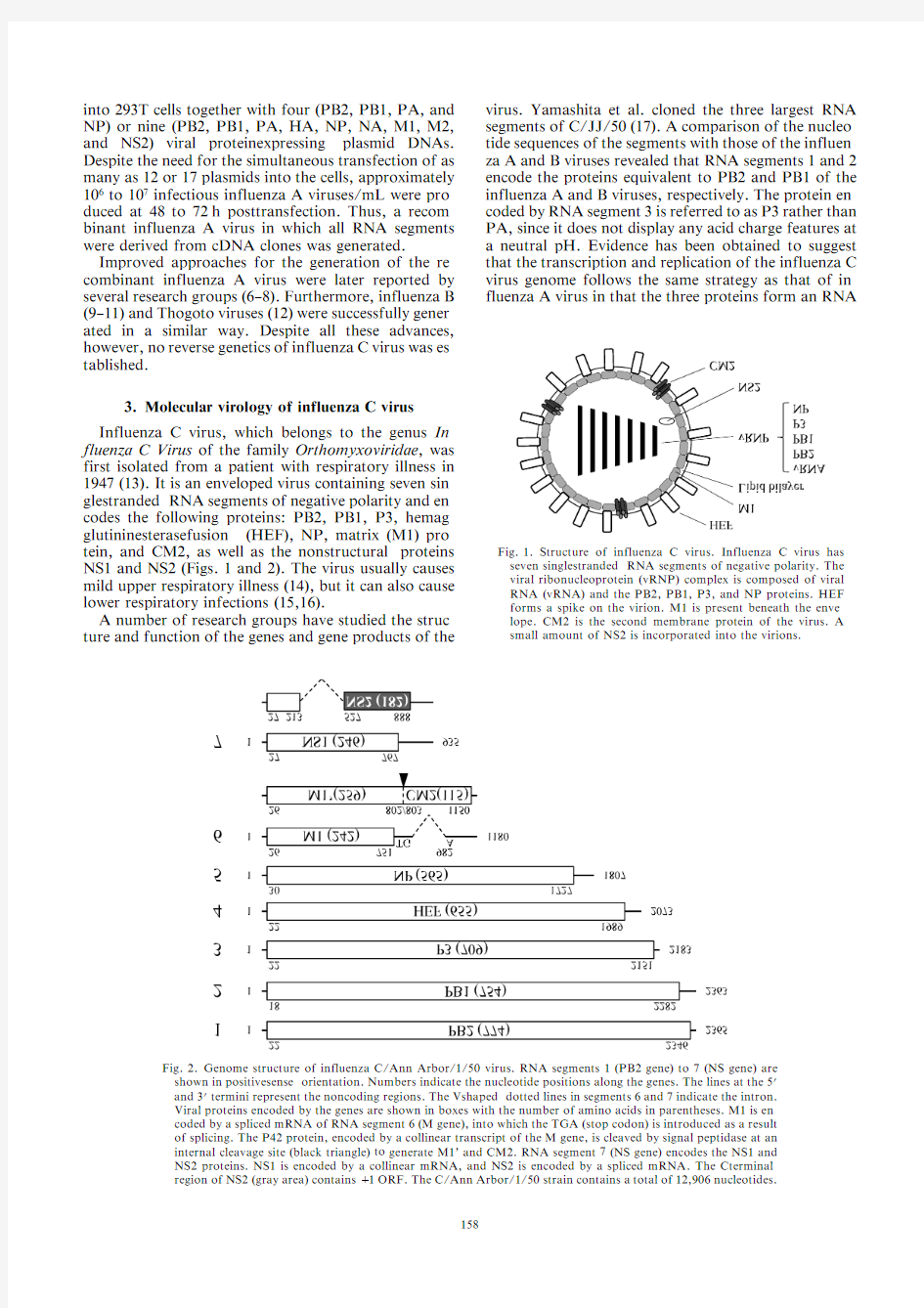
Fig.1.Structure of influenza C virus.Influenza C virus has seven single-stranded RNA segments of negative polarity.The viral ribonucleoprotein (vRNP)complex is composed of viral RNA (vRNA)and the PB2,PB1,P3,and NP proteins.HEF forms a spike on the virion.M1is present beneath the enve-lope.CM2is the second membrane protein of the virus.A small amount of NS2is incorporated into the
virions.
Fig.2.Genome structure of influenza C/Ann Arbor/1/50virus.RNA segments 1(PB2gene)to 7(NS gene)are shown in positive-sense orientation.Numbers indicate the nucleotide positions along the genes.The lines at the 5?and 3?termini represent the noncoding regions.The V-shaped dotted lines in segments 6and 7indicate the intron.Viral proteins encoded by the genes are shown in boxes with the number of amino acids in parentheses.M1is en-coded by a spliced mRNA of RNA segment 6(M gene),into which the TGA (stop codon)is introduced as a result of splicing.The P42protein,encoded by a collinear transcript of the M gene,is cleaved by signal peptidase at an internal cleavage site (black triangle)to generate M1'and CM2.RNA segment 7(NS gene)encodes the NS1and NS2proteins.NS1is encoded by a collinear mRNA,and NS2is encoded by a spliced mRNA.The C-terminal region of NS2(gray area)contains {1ORF.The C/Ann Arbor/1/50strain contains a total of 12,906nucleotides.
158
into 293T cells together with four (PB2,PB1,PA,and NP)or nine (PB2,PB1,PA,HA,NP,NA,M1,M2,and NS2)viral protein-expressing plasmid DNAs.Despite the need for the simultaneous transfection of as many as 12or 17plasmids into the cells,approximately 106to 107infectious influenza A viruses/mL were pro-duced at 48to 72h posttransfection.Thus,a recom-binant influenza A virus in which all RNA segments were derived from cDNA clones was generated.
Improved approaches for the generation of the re-combinant influenza A virus were later reported by several research groups (6 8).Furthermore,influenza B (9 11)and Thogoto viruses (12)were successfully gener-ated in a similar way.Despite all these advances,however,no reverse genetics of influenza C virus was es-tablished.
3.Molecular virology of influenza C virus Influenza C virus,which belongs to the genus In-fluenza C Virus of the family Orthomyxoviridae ,was first isolated from a patient with respiratory illness in 1947(13).It is an enveloped virus containing seven sin-gle-stranded RNA segments of negative polarity and en-codes the following proteins:PB2,PB1,P3,hemag-glutinin-esterase-fusion (HEF),NP,matrix (M1)pro-tein,and CM2,as well as the non-structural proteins NS1and NS2(Figs.1and 2).The virus usually causes mild upper respiratory illness (14),but it can also cause lower respiratory infections (15,16).
A number of research groups have studied the struc-ture and function of the genes and gene products of the
virus.Yamashita et al.cloned the three largest RNA segments of C/JJ/50(17).A comparison of the nucleo-tide sequences of the segments with those of the influen-za A and B viruses revealed that RNA segments 1and 2encode the proteins equivalent to PB2and PB1of the influenza A and B viruses,respectively.The protein en-coded by RNA segment 3is referred to as P3rather than PA,since it does not display any acid charge features at a neutral pH.Evidence has been obtained to suggest that the transcription and replication of the influenza C virus genome follows the same strategy as that of in-fluenza A virus in that the three proteins form an RNA
polymerase complex(18 20),although the precise role(s)of the respective proteins remains to be elucidat-ed.
RNA segment4encodes the HEF glycoprotein,which forms a spike on the virus envelope(Fig.1).The HEF protein has three biological activities:receptor binding, receptor destroying(acetylesterase),and membrane fusing activities.The receptor of the virus is the9-O-acetyl-N-acetylneuraminic acid(Neu5,9Ac2),and the acetylesterase activity of HEF inactivates the virus receptor by releasing the O-acetyl residues from the C-9 position of Neu5,9Ac2(21 23).The fusion activity of HEF is dependent on the proteolytic cleavage of a precursor(HEF0)into HEF1and HEF2,and it requires activation at a low pH upon internalization of the vi-ral particle through the endosome pathway(24,25). Sugawara et al.produced37monoclonal antibodies (MAbs)against HEF and indicated the presence of nine antigenic sites(A-1to A-5and B-1to B-4)on the mole-cule(26).The amino acid(s)recognized by the MAbs were identified by isolating the escape mutants of the MAbs directed against A-1to A-4(27).In1998,Rosen-thal et al.revealed the three-dimensional structure of HEF and showed that the three biological functions were attributable to distinct domains on the molecule (28).Together with a report by Matsuzaki et al.(27), this demonstrated that the antigenic sites A-1,A-2,and A-4were located on the receptor-binding domain and A-2on the acetylesterase domain,indicating that,like the influenza A virus hemagglutinin(HA),the main an-tigenic region of HEF is located on the globular head of the molecule.
HEF has a number of unique characteristics that are different from those of the influenza A virus HA.There are only three amino acids in the cytoplasmic region of HEF,whereas the corresponding region of HA contains 11to12amino acids.The short cytoplasmic region may or may not affect the transportation of HEF to the cell surface(29 32).The fatty acid attached to the cysteine at residue642of HEF is a stearic acid(33),whereas palmitic acid is mainly attached to the influenza A virus HA(34).Whether these facts are significant to the in-fluenza C virus replication remains to be elucidated. Nakada et al.determined the entire sequence of RNA segment5of C/California/78and suggested that the segment codes for NP(35).Sugawara et al.constructed a panel of MAbs against NP and reported that(i)there are at least two antigenic sites(I and II)on the molecule, (ii)the localization of NP in virus-infected cells is con-sistent with that of the influenza A virus NP,and(iii) NP exhibits molecular maturation during transport to the nucleus in virus-infected cells(36,37).NP was also shown to be essential to the transcription/replication processes of an artificial vRNA flanked by the noncod-ing regions(NCRs)of C/Johannesburg/1/66(19,20). Thus,although the features of the NP protein clarified to date are consistent with those of the influenza A virus NP protein,the functional domain(s)of NP remains to be analyzed.
The RNA segments6and7are bicistronic genes(Fig.
2).The spliced mRNA of the RNA segment6encodes M1(38).M1gives rigidity to the virion and is involved in the budding and morphogenesis processes of the virus (39 42).The unspliced mRNA of the segment encodes
the374-amino-acid protein,P42,which is cleaved by signal peptidase at an internal cleavage site to give M1' and CM2(43,44).The biochemical features of CM2,the second membrane protein of the virus,are closely simi-lar to those of M2(45,46),a proton channel of influenza
A virus.Although CM2expressed in Xenopus laevis oo-
cytes forms an ion channel permeable to Cl|(47)and CM2has the ability to modulate the pH of the exocytic pathway(48),the role of CM2in the virus replication cycle remains unclear.M1',composed of the N-terminal 259amino acids of P42,is degraded shortly after cleavage through the signal located in the C-terminal17-amino acid region of the protein(49).The half-life of P42is approximately30min(50),although a part of P42is transported to the cis-Golgi apparatus(51).The significance of M1'and P42in the viral life cycle also remains to be determined.
Analysis of the NS gene of the C/California/78strain initially showed that the gene contained934nucleotides and was capable of encoding both the286-amino-acid NS1and121-amino-acid NS2proteins(52,53).Based on
a comparison of the nucleotide sequences from a larger
number of influenza C virus isolates,including C/ California/78,the gene was later demonstrated to potentially encode NS1(246amino acids)from un-spliced mRNA and NS2(182amino acids)from spliced mRNA(54).In fact,the246-amino-acid NS1and182-amino-acid NS2proteins were identified in virus-infect-ed cells(54,55).The involvement of NS1in persistent in-fection(55)and viral mRNA splicing(56)was also reported.Like the influenza A virus NS2/NEP,the in-fluenza C virus NS2possesses nuclear export activity and is incorporated into virions(57,58).
Crescenzo-Chaigne et al.investigated the functions of the NCR of an influenza C virus RNA segment(19,20).
In their study,the model-type RNA flanked by NCR of the C/Johannesburg/1/66strain NS gene was expressed in COS 1cells from the Pol I plasmid-based system, together with the PB2,PB1,P3,and NP proteins,and the RNA template was shown to be transcribed and replicated.Thus,the involvement of the NCR of the in-fluenza C viral genome in its replication and transcrip-tion was demonstrated.
4.Epidemiology of influenza C virus
The epidemiology of influenza C virus has also been extensively investigated.In the1980s,on the basis of the results obtained from a limited number of influenza C virus isolates,the virus was considered to be antigenical-ly stable,divided into several antigenic groups,and evolved more slowly than influenza A virus(59 61).
Moriuchi et al.developed a tissue culture method for primary virus isolation that is convenient for routine work with a large number of clinical specimens(15,62), and they then initiated surveillance for influenza C virus infection in Sendai and Yamagata Cities in Japan.The established system has enabled us to systematically ana-lyze influenza C virus epidemiology and thereby find that(i)influenza C virus is divided into six antigenic and genetic groups(Taylor/1233/47-,Aichi/1/81-,Sao Paulo/378/82-,Kanagawa/1/76-,Yamagata/26/81-, and Mississippi/80-related lineages)(Fig.3)(63,64),(ii) the evolutionary rate of the virus is lower than that of
159
Fig.3.Phylogenetic tree of influenza C virus HEF genes.The region from nucleotides 64to 1,989of the HEF gene was analyzed.Horizontal distances are proportional to the minimum number of nucleotide differences needed to join the sequences.Numbers above the branches are the bootstrap probabilities (z )of each branch.A total of 74strains were divided into six lineages as indicated on the right of the figure (adapted from reference 64with permis-sion;kindly provided by Yoko Matsuzaki).
160
influenza A virus (63,65);e.g.,the rate of the HEF gene is 0.49~10|3nucleotides per site per year,which is only one-ninth of that of the influenza A virus HA gene,(iii)viruses belonging to different genetic and antigenic groups cocirculate within a limited geographical area during a given period,and reassortment events between different viruses frequently occur (64,66,67),and (iv)viruses containing a proper combination of genetic elements through reassortment events may have an
advantage in spreading among the human population (54,64,68,69).
There is evidence that influenza C virus has the poten-tial to infect animals,as the presence of specific anti-bodies was demonstrated in the serum of pigs (70,71)and dogs (72 74).Furthermore,a number of influenza C viruses were isolated from abattoir pigs in Beijing,China,and pig-to-pig transmission of the virus on ex-perimental infection has been demonstrated (75).Hu-
Fig.4.Reverse genetics of influenza C virus.Seven Pol I plas-mids for the expression of vRNAs were transfected into 293T cells together with viral protein-expressing plasmids for PB2,PB1,P3,HEF,NP,M1,CM2,NS1,and NS2.Recombinant influenza C viruses produced from the transfected 293T cells were then titrated and propagated in the amniotic cavity of em-bryonated chicken eggs.
161
man influenza C virus isolates were found to be geneti-cally and antigenically related to pig isolates,suggesting that interspecies transmission between humans and pigs has occurred in nature (76).Ohwada et al.showed that experimental infection resulted in clinical symptoms and viral replication in dogs (77).The potential role of animals as a reservoir for human influenza C remains to be elucidated,although influenza C viruses seem to be maintained among the human population.
5.Generation of influenza C virus-like particles and a recombinant influenza C virus For a number of negative-sense RNA viruses,the es-tablishment of a reverse genetics has been preceded by the construction of a mini-replicon system.Therefore,we attempted to establish an influenza C VLP genera-tion system,as the packaging of the artificial genome into influenza C virions and gene transfer to susceptible cells by VLPs had not yet been reported at that time.The cDNA of the green fluorescent protein (GFP)gene flanked by the NCR sequences of RNA segment 5(NP gene)of C/Ann Arbor/1/50was cloned into a pHH21vector in an anti-sense orientation between the Pol I promoter and the terminator sequences so that the artificial RNA (GFP-vRNA)was expressed under the control of the human Pol I promoter.The resulting plasmid DNA,pPolI/NP-AA.GFP(|),was transfect-ed into 293T cells,a human embryonic kidney cell line constitutively expressing simian virus 40large T antigen,together with viral protein-expressing plasmids for PB2,PB1,P3,HEF,NP,M1,CM2,NS1,and NS2,and in-cubated for up to https://www.doczj.com/doc/2a5145719.html,ing electron microscopy,we confirmed that the supernatant of the 293T cells con-tained influenza C VLPs,and,by the quantification of GFP-positive HMV-II cells infected with the VLPs and helper virus (C/Ann Arbor/1/50),we determined that the number of VLPs generated was approximately 106/mL (41).
The findings by several groups suggest that influenza C virus proteins function more efficiently at 339C than at 379C.Nagele and Meier-Ewert showed that the maxi-mum RNA polymerase activity of influenza C virus was detected at 339C (18).We also observed that,in the Pol I-based system,a higher amount of luciferase was ex-pressed at 339C than at 379C (data not shown).There-fore,our experiment was carried out at 339C,not 379C,including incubation of the transfected 293T cells and infection of the HMV-II cells with the VLPs (41).
``Cap snatching''is a unique feature of the influenza A virus transcription initiation process,and the roles of the three RNA polymerase subunits involved in that process have been extensively analyzed (78 83).In our study,in determining the sequence of the 3?end of the NP gene,we found that 12nucleotides were added to the 5?end of the NP gene mRNA (unpublished results).Similar results were obtained for the other six gene seg-ments (data not shown,see below).These findings pro-vide evidence that the cap snatching machinery by in-fluenza C virus polymerases is in fact activated in a simi-lar manner as for influenza A virus,although the roles of the respective polymerase subunits in the cap snatch-ing process remain to be clarified.
In the influenza C VLP generation experiment,NS1
expression in the transfected 293T cells was found to be required for efficient gene transfer to HMV-II cells (84).On the other hand,even without NS1expression,in-fluenza A VLPs were successfully generated and found capable of efficiently transmitting the synthetic reporter RNA to susceptible cells (85).This discrepancy may sug-gest that the influenza C virus NS1has a unique fun-ction(s)in the virus replication cycle that differs from those reported for influenza A virus.
To establish the reverse genetics of influenza C virus,we constructed Pol I plasmid DNAs for the seven RNA segments of C/Ann Arbor/1/50,a prototype strain of influenza C virus.The resulting Pol I plasmids were then transfected into 293T cells,together with four (PB2,PB1,P3,and NP)or nine (PB2,PB1,P3,HEF,NP,M1,CM2,NS1,and NS2)viral protein-expressing plasmids (Fig.4).The resultant supernatant was deter-mined to contain 101to 103EID 50/mL of infectious recombinant viruses (42),the titer of which was much lower than that of influenza A virus (106to 107PFU/mL)(see below).
Crescenzo-Chaigne and van der Werf reported the reverse genetics of influenza C virus using a similar ap-proach by which,at 10days posttransfection,they ob-tained 104PFU/mL of the recombinant virus (86).This finding suggests that,as in our observation,a limited number of infectious recombinants existed in the super-natant of the transfected 293T cells.Thus helper virus
Fig.5.Hypothesized virion generation mechanism for influenza C and A viruses.293T cells transfected with plasmid DNAs for reverse genetics are divided into seven (influenza C virus)or eight (influenza A virus)groups according to the number of vRNAs expressed in each cell.The length of the white (influen-za C virus)and black (influenza A virus)arrows indicates the generation efficiencies of the viruses from the respective cells.In the case of influenza C virus,293T cells expressing one RNA segment are the most efficient in the production of virions.In contrast,for influenza A virus,cells expressing one set (eight segments)of vRNA are the most efficient in the production of infectious virions.
162
independent-reverse genetics has currently become available for application to influenza C virology,there-by affording the opportunity to resolve a number of questions regarding the virus.At the moment,however,we must admit that the current system has a disadvan-tage in that recombinants with severe growth defect(s)may not be obtained due to the low generation efficien-cy,a problem that needs to be overcome.
6.Analysis of the structure-function relationship of the M1protein Nishimura et al.reported that cord-like structures (CLSs),composed of numerous filamentous particles in the process of budding,were found to extrude from C/Yamagata/1/88-infected HMV-II cells (39).Each of these particles was covered with a layer of surface projections and aggregated with their long axes.Further analysis of a series of reassortant viruses between C/Yamagata/1/88and C/Taylor/1233/47,the latter of which is a unique strain incapable of forming CLS,showed that reassortants with the M gene from C/Tay-lor/1233/47could not form CLS on infected cells (40).Upon establishment of the influenza C-VLP genera-tion system,we identified CLSs extruding from the VLP-generating 293T cells.By expressing a series of M1mutants in the 293T cells together with the other plas-mids required for VLP generation,we demonstrated that the M1protein is a determinant for CLS formation and residue 24of M1(Ala or Thr)is responsible for CLS formation as well as VLP morphology (filamen-tous or spherical)(41).Furthermore,the generation of an infectious recombinant virus with an M1mutation revealed that residue 24of the M1protein (Ala or Thr)also affects CLS formation on the infected cells and virion morphology (filamentous or spherical).Mem-brane flotation analysis of recombinant virus-infected cells revealed that the wild-type M1protein (possessing Ala at residue 24)showed higher affinity to the plasma membrane than mutant M1(possessing Thr at residue 24),suggesting that an amino acid on M1affects the virion morphology through the membrane affinity of M1to the plasma membrane (42).Thus,using the re-verse-genetics system of influenza C virus,we could demonstrate the structure-function relationship of the M1protein in the context of viral replication.
https://www.doczj.com/doc/2a5145719.html,parison of the generation efficiencies of influenza C and A viruses Initially,we expected that the adoption of a similar approach would allow the recombinant influenza C virus to be generated as efficiently as influenza A virus (106to 107PFL/mL),since (i)the number of RNA seg-ments in influenza C virus is seven,which is smaller by one segment than that in influenza A virus,and (ii)the number of influenza C VLPs generated using a similar system was approximately 106/mL,which is 100-fold higher than that of influenza A VLPs (104/mL)(41,87).In fact,the supernatant of the plasmid-transfected 293T cells showed 4HAU/mL,a titer corresponding to 105PFU/mL of the egg-grown influenza C virus (data not shown).However,the supernatant contained only 101to 103EID 50/mL of infectious viruses (42).The discrepan-
cy between the HA and infectious virus titers could be explained by the presence of large and limited numbers of noninfectious and infectious virus particles,respec-tively,in the supernatant of the plasmid-transfected 293T cells.
Neumann et al.reported that in the plasmid-driven reverse-genetics system,one in 102.8to 103.3293T cells produce the recombinant infectious influenza A virus (5).This means that approximately one in 1,000cells ex-presses one set (eight segments)of vRNA,leading to the generation of infectious influenza A viruses,and that the remaining 999in 1,000cells express less than seven vRNA segments.This suggestion seems to apply in the case of influenza C virus reverse genetics:one in 1,000cells expresses one set (seven segments)of vRNA,and the remaining 999cells express fewer than six RNA seg-ments.Therefore,the above-mentioned discrepancy be-tween the HA and infectious virus titers in the super-natant suggests that the remaining 999cells efficiently produce noninfectious particles.
The hypothesis that the 293T cells expressing less than six RNA segments efficiently produce influenza C parti-cles is supported by the previous observations that (i)interaction of HEF with M1may be sufficient to lead to budding at the cell surface (39,our unpublished result)and (ii)nucleocapsids may not be required to initiate the budding process of the virus (40,88).In other words,in the case of influenza C virion formation,coexpression of M1and HEF in a transfected cell,even if the ratio of M1to HEF in the cell is different from that in virus-in-fected cells,may readily lead to particle formation regardless of the presence or absence of nucleocapsids.Furthermore,it is possible that inefficient interaction of M1with nucleocapsid (39)facilitates the production of noninfectious particles.Alternatively,as a driving force for virion formation,the influenza C virus M1protein may function more efficiently than the influenza A virus M1protein.
There is a significant difference between the VLP generation efficiencies of influenza C (106/mL)and A (104/mL)viruses (41,87).A comparison of the number of VLPs with that of infectious virus particles between influenza A and C viruses suggests that in the case of in-fluenza C virus generation,cells expressing only one
RNA segment may be the most efficient in the produc-tion of virus particles.Based on these findings,we formed a hypothesis explaining the generation of in-fluenza C virus,which is shown in Fig.5.In the case of influenza A virus,293T cells expressing one set(eight segments)of vRNA produce infectious virions most ef-ficiently.This is consistent with the results reported by Fujii et al.,in which cells expressing seven and six seg-ments are less efficient in the production of influenza A viruses(89).In contrast,for influenza C virus,293T cells expressing one vRNA segment may be the most ef-ficient in the production of virions,whereas cells ex-pressing one set(seven segments)of vRNA may be the least efficient,resulting in the presence of a large num-ber of noninfectious virions in the supernatant.
8.Prospects for research on influenza C virology
A number of influenza C virus proteins are consi-dered to be likely targets for future reverse genetics ana-lyses.CM2is the second membrane protein of the virus, and its characteristics have been extensively studied (45 48,90).A recombinant influenza C virus lacking CM2would help elucidate the role of CM2in the virus replication cycle.
NS gene products are also likely candidates for analy-sis.As mentioned above,the influenza C virus NS2pos-sesses a nuclear export activity,and the nuclear export signal(NES)is composed of two separate leucine-rich domains on the NS2protein(57).The role of the NES in virus replication,including the role of the individual amino acids in the NES,remains to be elucidated.We have recently reported that NS1is involved in viral pre-mRNA splicing(56),although the functional domain on the NS1protein has not yet been mapped.
Our research group and others have identified the amino acid(s)on the HEF glycoprotein responsible for receptor binding and esterase activity(21,22,27). Furthermore,analysis of transfected cells expressing HEF revealed the role of the individual glycosylation sites of HEF in its proper conformation,biological ac-tivities and transport(91).These findings should be fur-ther studied in the context of virus replication using recombinant viruses.
Phylogenetic analysis showed that the34NS genes analyzed were split into two distinct groups,A and B, and suggested that influenza C viruses that acquired group B NS genes through reassortment events domi-nantly replaced viruses with group A NS genes,forming a stable lineage(54).This finding leads to the hypothesis that the NS gene and/or NS gene product(s)may pro-vide influenza C virus with a determinant for an epidemiological advantage.A comparison of the growth kinetics of recombinants harboring A or B NS genes would clarify the significance of the gene.
In addition,influenza C virus could be engineered to generate expression vectors,and recombinant influenza C viruses containing a foreign antigen have potential ap-plications as live vaccines.
9.Conclusion
An influenza C virus reverse-genetics system was es-tablished and will be useful in addressing a number of
questions regarding influenza C molecular virology and epidemiology,particularly those associated with the type-specificity requirements of influenza viruses.
Acknowledgments We thank Drs.Y.Kawaoka and H.Goto for providing plasmid DNAs and for their helpful comments.We also thank Drs.K.Sugawara,Y.Matsuzaki and E.Takashita for their ex-cellent advice in establishing the reverse genetics of influenza C virus.
REFERENCES
1.Luytjes,W.,Krystal,M.,Enami,M.,et al.(1989):Amplifica-
tion,expression,and packaging of a foreign gene by influenza vi-
rus.Cell,59,1107 1113.
2.Enami,M.,Luytjes,W.,Krystal,M.,et al.(1990):Introduction
of site-specific mutations into the genome of influenza virus.
https://www.doczj.com/doc/2a5145719.html,A,87,3802 3805.
3.Pleschka,S.,Jaskunas,S.R.,Engelhardt,O.G.,et al.(1996):A
plasmid-based reverse genetics system for influenza A virus.J.
Virol.,70,4188 4192.
4.Zobel,A.,Neumann,G.and Hobom,G.(1993):RNA polymer-
ase-I catalyzed transcription of insert viral cDNA.Nuc.Acid
Res.,21,3607 3614.
5.Neumann,G.,Watanabe,T.,Ito,H.,et al.(1999):Generation of
influenza A viruses entirely from cloned cDNAs.Proc.Natl.
https://www.doczj.com/doc/2a5145719.html,A,96,9345 9350.
6.Fodor,E.,Devenish,L.,Engelhardt,O.G.,et al.(1999):Rescue
of influenza A virus from recombinant DNA.J.Virol.,73,
9679 9682.
7.Hoffmann,E.,Neumann,G.,Kawaoka,Y.,et al.(2000):A
DNA transfection system for generation of influenza A virus
from eight https://www.doczj.com/doc/2a5145719.html,A,97,6108 6113.
8.Neumann,G.,Fujii,K.,Kino,Y.,et al.(2005):An improved
reverse genetics system for influenza A virus generation and its
implications for vaccine https://www.doczj.com/doc/2a5145719.html,A,
102,16825 16829.
9.Hoffmann,E.,Mahmood,K.,Yang,C.F.,et al.(2002):Rescue
of influenza B virus from eight plasmids.Proc.Natl.Acd.Sci.
USA,99,11411 11416.
10.Jackson,D.,Cadman,A.,Zurcher,T.,et al.(2002):A reverse
genetics approach for recovery of recombinant influenza B
viruses entirely from cDNA.J.Virol.,76,11744 11747.
11.Hatta,M.and Kawaoka,Y.(2003):The NB protein of influenza
B virus is not necessary for virus replication in vitro.J.Virol.,77,
6050 6054.
12.Wagner,E.,Engelhardt,O.G.,Gruber,S.,et al.(2001):Rescue
of recombinant Thogoto virus from cloned cDNA.J.Virol.,75,
9282 9286.
13.Taylor,R.M.(1949):Studies on survival of influenza virus be-
tween epidemics and antigenic variants of the virus.Am.J.Pub-
lic Health,39,171 178.
14.Katagiri,S.,Ohizumi,A.and Homma,M.(1983):An outbreak
of type-C influenza in a childrens home.J.Infect.Dis.,148,
51 56.
15.Moriuchi,H.,Katsushima,N.,Nishimura,H.,et al.(1991):
Community-acquired influenza-C virus-infection in children.J.
Pediatr.,118,235 238.
16.Matsuzaki,Y.,Katsushima,N.,Nagai,Y.,et al.(2006):Clinical
features of influenza C virus infection in children.J.Infect.Dis.,
193,1229 1235.
17.Yamashita,M.,Krystal,M.and Palese,P.(1989):Comparison
of the3large polymerase proteins of influenza A,B,and C
viruses.Virology,171,458 466.
18.Nagele,A.and Meier-Ewert,H.(1984):Influenza-C-virion-asso-
ciated RNA-dependent RNA-polymerase activity.Biosci.Rep.,4,
703 706.
19.Crescenzo-Chaigne,B.,Naffakh,N.and van der Werf,S.(1999):
Comparative analysis of the ability of the polymerase complexes
of influenza viruses type A,B and C to assemble into functional
RNPs that allow expression and replication of heterotypic model
RNA templates in vivo.Virology,265,342 353.
20.Crescenzo-Chaigne,B.and van der Werf,S.(2001):Nucleotides
at the extremities of the viral RNA of influenza C virus are in-
volved in type-specific interactions with the polymerase complex.
J.Gen.Virol.,82,1075 1083.
21.Herrler,G.,Multhaup,G.,Beyreuther,K.,et al.(1988a):Serine
163
71of the glycoprotein HEF is located at the active site of the acetylesterase of influenza C virus.Arch.Virol.,102,269 274.
22.Herrler,G.,Durkop,I.,Becht,H.,et al.(1988b):The
glycoprotein of influenza C virus is the hemagglutinin,esterase and fusion factor.J.Gen.Virol.,69,839 846.
23.Schauer,R.,Reuter,G.,Stoll,S.,et al.(1988):Isolation and
characterization of sialate9(4)-O-acetylesterase from influenza C virus.Biol.Chemist.Hoppe-Seyler.,369,1121 1130.
24.Kitame,F.,Sugawara,K.,Ohwada,K.,et al.(1982):Proteolytic
activation of hemolysis and fusion by influenza C virus.Arch.
Virol.,73,357 361.
25.Ohuchi,M.,Ohuchi,R.,Mifune,K.,et al.(1982):Demonstra-
tion of hemolytic and fusion activities of influenza C virus.J.
Virol.,42,1076 1079.
26.Sugawara,K.,Nishimura,H.,Hongo,S.,et al.(1993):Construc-
tion of an antigenic map of the haemagglutinin-esterase protein of influenza C virus.J.Gen.Virol.,74,1661 1666.
27.Matsuzaki,M.,Sugawara,K.,Adachi,K.,et al.(1992):Location
of neutralizing epitopes on the hemagglutinin-esterase protein of influenza C virus.Virology,189,79 87.
28.Rosenthal,P.B.,Zhang,X.D.,Formanowski,F.,et al.(1998):
Structure of the haemagglutinin-esterase-fusion glycoprotein of influenza C virus.Nature,396,92 96.
29.Szepanski,S.,Veit,M.,Pleschka,S.,et al.(1994):Post-transla-
tional folding of the influenza C virus glycoprotein HEF:defec-tive processing in cells expressing the cloned gene.J.Gen.Virol., 75,1023 1030.
30.Muraki,Y.,Hongo,S.,Sugawara,K.,et al.(1999):Location of a
linear epitope recognized by monoclonal antibody S16on the hemagglutinin-esterase glycoprotein of influenza C virus.Virus Res.,61,53 61.
31.Oeffner,F.,Klenk,H.D.and Herrler,G.(1999):The cytoplasmic
tail of the influenza C virus glycoprotein HEF negatively affects transport to the cell surface.J.Gen.Virol.,80,363 369.
32.Pekosz,A.and Lamb,R.A.(1999):Cell surface expression of
biologically active influenza C virus HEF glycoprotein expressed from cDNA.J.Virol.,73,8808 8812.
33.Veit,M.,Herrler,G.,Schmidt,M.F.,et al.(1990):The hemag-
glutinating glycoproteins of influenza B and C viruses are acylat-ed with different fatty acids.Virology,177,807 811.
34.Kordyukova,L.V.,Serebryakova,M.V.,Baratova,L.A.,et al.
(2008):S acylation of the hemagglutinin of influenza viruses: mass spectrometry reveals site-specific attachment of stearic acid to a transmembrane cysteine.J.Virol.,82,9288 9292.
35.Nakada,S.,Creager,R.S.,Krystal,M.,et al.(1984):Complete
nucleotide sequence of the influenza C/California/78virus nucleoprotein gene.Virus Res.,1,433 441.
36.Sugawara,K.,Nishimura,H.,Hongo,S.,et al.(1991):Antigenic
characterization of the nucleoprotein and matrix protein of in-fluenza C virus with monoclonal-antibodies.J.Gen.Virol.,72, 103 109.
37.Sugawara,K.,Muraki,Y.,Takashita,E.,et al.(2006):Confor-
mational maturation of the nucleoprotein synthesized in influen-za C virus-infected cells.Virus Res.,122,45 52.
38.Yamashita,M.,Krystal,M.and Palese,P.(1988):Evidence that
the matrix protein of influenza C virus is coded for by a spliced mRNA.J.Virol.,62,3348 3355.
39.Nishimura,H.,Hara,M.,Sugawara,K.,et al.(1990):Charac-
terization of the cord-like structures emerging from the surface of influenza C virus-infected cells.Virology,179,179 188.
40.Nishimura,H.,Hongo,S.,Sugawara,K.,et al.(1994):The abil-
ity of influenza C virus to generate cord-like structures is in-fluenced by the gene coding for M-protein.Virology,200, 140 147.
41.Muraki,Y.,Washioka,H.,Sugawara,K.,et al.(2004):Identifi-
cation of an amino acid residue on influenza C virus M1protein responsible for formation of the cord-like structures of the virus.
J.Gen.Virol.,85,1885 1893.
42.Muraki,Y.,Murata,T.,Takashita,E.,et al.(2007):A mutation
on influenza C virus M1protein affects virion morphology by al-tering the membrane affinity of the protein.J.Virol.,81, 8766 8773.
43.Pekosz,A.and Lamb,R.A.(1998):Influenza C virus CM2in-
tegral membrane glycoprotein is produced from a polypeptide precursor by cleavage of an internal signal sequence.Proc.Natl.
https://www.doczj.com/doc/2a5145719.html,A,95,13233 13238.
44.Hongo,S.,Sugawara,K.,Muraki,Y.,et al.(1999):Influenza C
virus CM2protein is produced from a374-amino-acid protein
(P42)by signal peptidase cleavage.J.Virol.,73,46 50.
45.Hongo,S.,Sugawara,K.,Muraki,Y.,et al.(1997):Characteriza-
tion of a second protein(CM2)encoded by RNA segment6of in-
fluenza C virus.J.Virol.,71,2786 2792.
46.Pekosz,A.and Lamb,R.A.(1997):The CM2protein of influen-
za C virus is an oligomeric integral membrane glycoprotein struc-
turally analogous to influenza A virus M2and influenza B virus
NB proteins.Virology,237,439 451.
47.Hongo,S.,Ishii,K.,Mori,K.,et al.(2004):Detection of ion
channel activity in Xenopus laevis oocytes expressing influenza C
virus CM2protein.Arch.Virol.,149,35 50.
48.Betakova,T.and Hay,A.J.(2007):Evidence that the CM2pro-
tein of influenza C virus can modify the pH of the exocytic path-
way of transfected cells.J.Gen.Virol.,88,2291 2296.
49.Pekosz,A.and Lamb,R.A.(2000):Identification of a membrane
targeting and degradation signal in the p42protein of influenza C
virus.J.Virol.,74,10480 10488.
50.Hongo,S.,Gao,P.,Sugawara,K.,et al.(1998):Identification of
a374amino acid protein encoded by RNA segment6of influenza
C virus.J.Gen.Virol.,79,2207 2213.
51.Li,Z.N.,Muraki,Y.,Takashita,E.,et al.(2004):Biochemical
properties of the P42protein encoded by RNA segment6of in-
fluenza C virus.Arch.Virol.,149,275 287.
52.Nakada,S.,Graves,P.N.,Desselberger,U.,et al.(1985):In-
fluenza C virus RNA7codes for a nonstructural protein.J.
Virol.,56,221 226.
53.Nakada,S.,Graves,P.N.and Palese,P.(1986):The influenza C
virus NS gene:evidence for a spliced mRNA and a second NS
gene product(NS2protein).Virus Res.,4,263 273.
54.Alamgir,A.S.M.,Matsuzaki,Y.,Hongo,S.,et al.(2000):
Phylogenetic analysis of influenza C virus nonstructural(NS)
protein genes and identification of the NS2protein.J.Gen.
Virol.,81,1933 1940.
55.Marschall,M.,Helten,A.,Hechtfischer,A.,et al.(1999):The
ORF,regulated synthesis,and persistence-specific variation of in-
fluenza C viral NS1protein.Virology,253,208 218.
56.Muraki,Y.,Furukawa,T.,Kohno,Y.,et al.(2010):Influenza C
virus NS1protein up-regulates the splicing of viral mRNAs.J.
Virol.,84,1957 1966.
57.Paragas,J.,Talon,J.,O'Neill,R.E.,et al.(2001):Influenza B
and C virus NEP(NS2)proteins possess nuclear export activities.
J.Virol.,75,7375 7383.
58.Kohno,Y.,Muraki,Y.,Matsuzaki,Y.,et al.(2009):Intracellular
localization of influenza C virus NS2protein(NEP)in infected
cells and its incorporation into virions.Arch.Virol.,154,
235 243.
59.Chakraverty,P.(1978):Antigenic relationship between influenza
C viruses.Arch.Virol.,58,341 348.
60.Buonagurio,D.A.,Nakada,S.,Desselberger,U.,et al.(1985):
Noncumulative sequence changes in the hemagglutinin genes of
influenza C virus isolates.Virology,146,221 232.
61.Buonagurio,D.A.,Nakada,S.,Fitch,W.M.,et al.(1986):
Epidemiology of influenza C virus in man:multiple evolutionary
lineages and low rate of change.Virology,153,12 21.
62.Nishimura,H.,Sugawara,K.,Kitame,F.,et al.(1989):A hu-
man-melanoma cell-line highly susceptible to influenza C virus.J.
Gen.Virol.,70,1653 1661.
63.Muraki,Y.,Hongo,S.,Sugawara,K.,et al.(1996):Evolution of
the haemagglutinin-esterase gene of influenza C virus.J.Gen.
Virol.,77,673 679.
64.Matsuzaki,Y.,Mizuta,K.,Sugawara,K.,et al.(2003):Frequent
reassortment among influenza C viruses.J.Virol.,77,871 881.
65.Tada,Y.,Hongo,S.,Muraki,Y.,et al.(1997):Evolutionary
analysis of influenza C virus M genes.Virus Genes,15,53 59.
66.Matsuzaki,Y.,Muraki,Y.,Sugawara,K.,et al.(1994):Cocircu-
lation of two distinct groups of influenza C virus in Yamagata
City,Japan.Virology,202,796 802.
67.Peng,G.,Hongo,S.,Muraki,Y.,et al.(1994):Genetic reassort-
ment of influenza C viruses in man.J.Gen.Virol.,75,
3619 3622.
68.Peng,G.,Hongo,S.,Kimura,H.,et al.(1996):Frequent occur-
rence of genetic reassortment between influenza C virus strains in
nature.J.Gen.Virol.,77,1489 1492.
69.Matsuzaki,Y.,Mizuta,K.,Kimura,H.,et al.(2000):Characteri-
zation of antigenically unique influenza C virus strains isolated in
Yamagata and Sendai Cities,Japan,during1992 1993.J.Gen.
Virol.,81,1447 1452.
70.Yamaoka,M.,Hotta,H.,Itoh,M.,et al.(1991):Prevalence of
164
antibody to influenza C virus among pigs in Hyogo Prefecture, Japan.J.Gen.Virol.,72,711 714.
71.Brown,I.H.,Harris,P.A.and Alexander,D.J.(1995):Serologi-
cal studies of influenza viruses in pigs in Great Britain1991 2.
Epidemiol.Infect.,114,511 520.
72.Ohwada,K.,Kitame,F.,Sugawara,K.,et al.(1987):Distribution
of the antibody to influenza C virus in dogs and pigs in Yamagata Prefecture,Japan.Microbiol.Immunol.,31,1173 1180.
73.Manuguerra,J.C.and Hannoun,C.(1992):Natural infection of
dogs by influenza C virus.Res.Virol.,143,199 204.
74.Manuguerra,J.C.,Hannoun,C.,Simon,F.,et al.(1993):Natu-
ral infection of dogs by influenza C virus:a serological survey in Spain.New Microbiol.,16,367 371.
75.Guo,Y.J.,Jin,F.G.,Wang,P.,et al.(1983):Isolation of in-
fluenza C virus from pigs and experimental infection of pigs with influenza C virus.J.Gen.Virol.,64,177 182.
76.Kimura,H.,Abiko,C.,Peng,G.,et al.(1997):Interspecies trans-
mission of influenza C virus between humans and pigs.Virus Res.,48,71 79.
77.Ohwada,K.,Kitame,F.and Homma,M.(1986):Experimental
infections of dogs with type C influenza virus.Microbiol.Im-munol.,30,451 460.
78.Honda, A.,Mizumoto,K.and Ishihama, A.(1999):Two
separate sequences of PB2subunit constitute the RNA cap-bind-ing site of influenza virus RNA polymerase.Gene Cell,4, 475 485.
79.Li,M.L.,Rao,P.and Krug,R.M.(2001):The active sites of the
influenza cap-dependent endonuclease are on different poly-merase subunits.EMBO J.,20,2078 2086.
80.Fechter,P.,Mingay,L.,Sharps,J.,et al.(2003):Two aromatic
residues in the PB2subunit of influenza A RNA polymerase are crucial for cap binding.J.Biol.Chem.,278,20381 20388. 81.Guilligay,D.,Tarendeau,F.,Resa-Infante,P.,et al.(2008):The
structural basis for cap binding by influenza virus polymerase subunit PB2.Nat.Struct.Biol.,15,500 506.
82.Dias,A.,Bouvier,D.,Crepin,T.,et al.(2009):The cap-snatch-
ing endonuclease of influenza virus polymerase resides in the PA
subunit.Nature,458,914 918.
83.Yuan,P.,Bartlam,M.,Lou,Z.,et al.(2009):Crystal structure of
an avian influenza polymerase PA N reveals an endonuclease ac-
tive site.Nature,458,909 913.
84.Muraki,Y.,Sugawara,K.,Takashita,E.,et al.(2005):Evidence
for the involvement of M1protein in the formation of cord-like
structures of influenza C virus.p.90 93.In Y.Kawaoka(ed.),
Options for the Control of Influenza V.Elsevier,Amsterdam.
85.Mena,I.,Vivo,A.,Perez,E.,et al.(1996):Rescue of a synthetic
chloramphenicol acetyltransferase RNA into influenza virus-like
particles obtained from recombinant plasmids.J.Virol.,70,
5016 5024.
86.Crescenzo-Chaigne,B.and van der Werf,S.(2007):Rescue of in-
fluenza C virus from recombinant DNA.J.Virol.,81,
11282 11289.
87.Neumann,G.,Watanabe,T.and Kawaoka,Y.(2000):Plasmid-
driven formation of influenza virus-like particles.J.Virol.,74,
547 551.
88.Herrler,G.,Nagele,A.,Meier-Ewert,H.,et al.(1981):Isolation
and structural analysis of influenza C virion glycoproteins.Virol-
ogy113,439 451.
89.Fujii,Y.,Goto,H.,Watanabe,T.,et al.(2003):Selective incor-
poration of influenza virus RNA segments into virions.Proc.
https://www.doczj.com/doc/2a5145719.html,A,100,2002 2007.
90.Li,Z.N.,Hongo,S.,Sugawara,K.,et al.(2001):The sites for fat-
ty acylation,phosphorylation and intermolecular disulphide bond
formation of influenza C virus CM2protein.J.Gen.Virol.,82,
1085 1093.
91.Sugahara,K.,Hongo,S.,Sugawara,K.,et al.(2001):Role of in-
dividual oligosaccharide chains in antigenic properties,intracellu-
lar transport,and biological activities of influenza C virus hemag-
glutinin-esterase protein.Virology,285,153 164.
165