
16S rDNA-Based Identi?cation of Bacteria from Conjunctival Swabs by PCR and DGGE Fingerprinting Claudia Schabereiter-Gurtner,1Saskia Maca,2Sabine Ro¨lleke,1Karl Nigl,2Julius Lukas,2 Alexander Hirschl,3Werner Lubitz,1and Talin Barisani-Asenbauer2
P URPOSE.Establishment of a new molecular biology technique for the identi?cation of multiple bacteria from the ocular en-vironment,which can be applied supplementarily to cultiva-tion in cases of severe bacterial infections.
M ETHODS.From60human conjunctivae(29with purulent and 31with nonpurulent conjunctivitis),swabs were taken and DNA was extracted.Fragments of200bp,spanning the V3 region of the eubacterial16S rDNA,were ampli?ed by poly-merase chain reaction(PCR)and separated by denaturing gra-dient gel electrophoresis(DGGE).For phylogenetic identi?ca-tion,DGGE bands were excised and directly sequenced,or16S rDNA clone libraries were constructed and clones were screened by DGGE.Sequences were compared with sequences of known bacteria listed in the EMBL database.Furthermore, the results were compared with results obtained from conven-tional cultivation.
R ESULTS.16S rDNA could be ampli?ed from25of29investi-gated swabs taken from purulent conjunctivitis eyes and from 2of31investigated swabs taken from nonpurulent conjuncti-vitis eyes.Sixteen samples showed monomicrobial and11 samples showed polymicrobial infections.The following gen-era(n is number of samples)were detected:Staphylococcus (n?8),Corynebacterium(n?7),Propionibacterium(n?7),Streptococcus(n?6),Bacillus(n?2),Acinetobacter(n?3),Pseudomonas(n?3),Proteus(n?1),and Brevundimo-nas(n?1).Four sequences could not be identi?ed to the genus level.They had highest sequence similarities both to sequences of Pantoea and Enterobacter(n?1),Kingella and Neisseria(n?1),Serratia and Aranicola(n?1),and Leu-conostoc and Weissella(n?2),respectively.Culture was only positive for coagulase-negative staphylococci(n?9),Coryne-bacteria(n?3),Staphylococcus aureus(n?1),Streptococ-cus sp.(n?1),Proteus sp.(n?1),Klebsiella oxytoca(n?1),and Pseudomonas aeruginosa(n?1).In total,45%of the 60analyzed conjunctival swabs were PCR positive,whereas only22%were culture positive.No sample positive by culture gave negative results by PCR.
C ONCLUSIONS.16S rDNA sequence analyses and DGGE?nger-printing are appropriate methods for the detection and identi-?cation of monomicrobial as well as polymicrobial ocular in-fections of bacteria that might not be detected by conventional cultivation.(Invest Ophthalmol Vis Sci.2001;42:1164–1171)B acterial ocular infections are common.Although many
cases show a benign course,some can be associated with sight-threatening ocular complications.Identi?cation of the causative pathogens in these cases is mandatory but often dif?cult because some bacteria have special growth require-ments.Furthermore,sample size from ocular tissues is usually small,leading to unreliable cultivation results.Initiation of proper therapy can then be delayed with possible devastating visual consequences.
Molecular approaches to the identi?cation of bacteria show promising results.The ampli?cation of16S rDNA of any bac-terial species is possible without prior cultivation when broad-range PCR primers targeted to highly conserved regions are applied.The comparison of ampli?ed and sequenced16S rDNA sequences with sequences of known bacteria in16S rDNA databases facilitates a subsequent phylogenetic identi?-cation.In ophthalmology,the16S rDNA-based identi?cation of pathogens is still at its beginning and,except in a few studies, is rarely applied.Hykin et al.1and Therese et al.2used eubac-terial primers and Propionibacterium-speci?c primers to de-tect bacterial DNA in vitreous samples of patients who had endophthalmitis.Lohmann et al.3and Knox et al.4detected and identi?ed bacteria in corneal scrapings and in vitreous samples of patients who had keratitis and endophthalmitis by ampli?cation and subsequent direct sequencing of16S rDNA. These studies allowed the simple detection of eubacterial DNA or the identi?cation of monomicrobial infections,whereas pathogens of polymicrobial infections could not be identi?ed by direct sequencing.
Nevertheless,bacterial infections of the eye are sometimes polymicrobial.In the studies of Ormerod et al.5and Kunimoto et al.,6it was shown that ocular infections such as endoph-thalmitis were polymicrobial in up to32%.Concerning polymi-crobial communities,the direct sequencing of mixed16S rDNA fragments fails,and sequence information can only be obtained through16S rDNA clone libraries.7,8To avoid the sequencing of clones containing identical sequences,clone libraries can be screened by restriction fragment length polymorphism analysis (RFLP)or by denaturing gradient gel electrophoresis(DGGE). By applying RFLP,16S rDNA amplicons are digested with a set of different restriction endonucleases,and DNA fragments are separated in agarose gels,leading to different RFLP pro?les of individual16S rDNA sequences.9,10
DGGE facilitates pro?ling of monomicrobial as well as polymicrobial communities in polyacrylamide gels because of the sequence-speci?c separation of16S rDNA amplicons of same length.11During gel electrophoresis,short16S rDNA amplicons migrate toward increasing denaturing concentra-tions,leading to a partial melting of the DNA helix and to a decrease and subsequent ending of electrophoretic migration. As a consequence,a band pattern is produced in which each band theoretically represents a bacterial taxon.
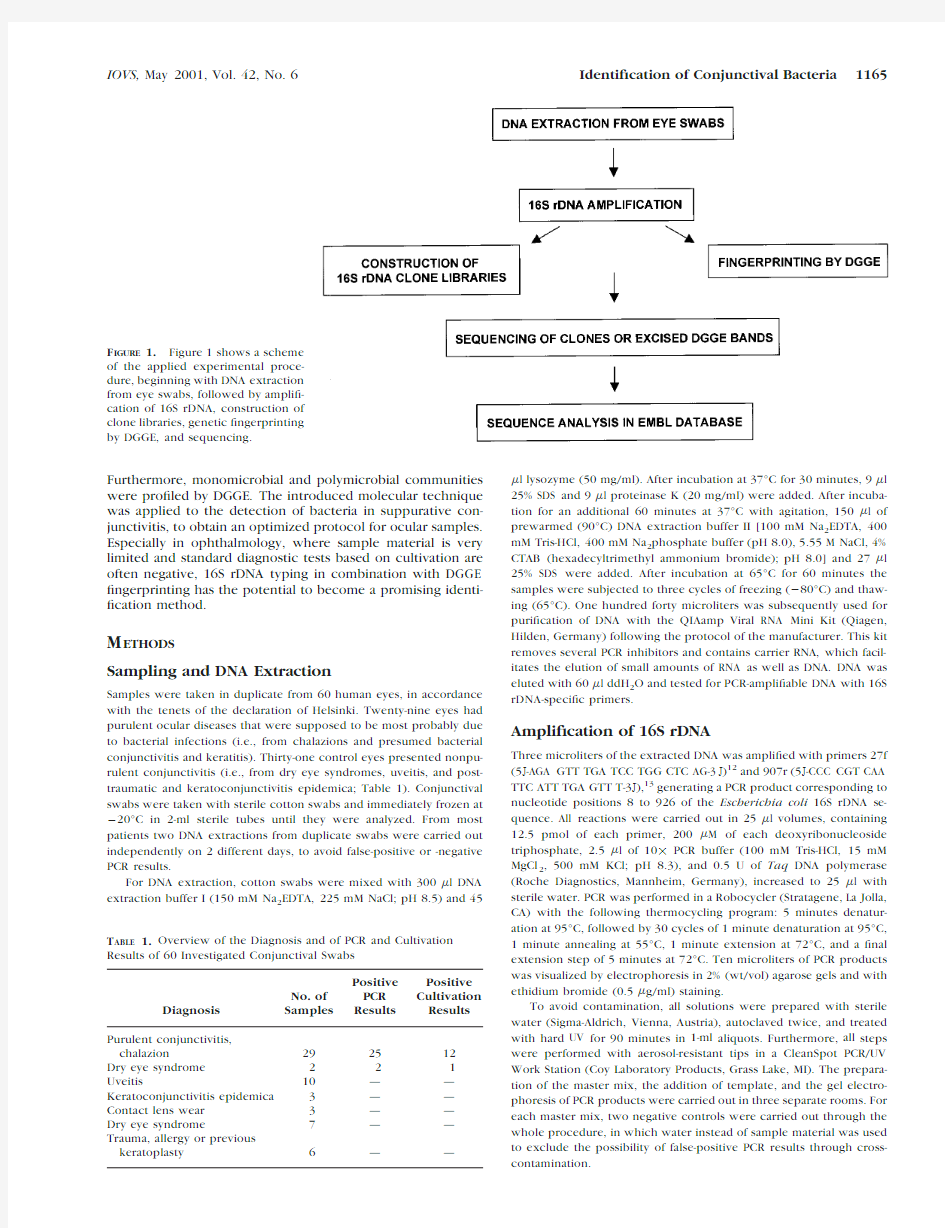
In the present study,a method is proposed that combines 16S rDNA genotyping with DGGE?ngerprinting.Figure1 shows a scheme of the applied experimental procedure.The microbial communities of conjunctival swabs were investi-gated by amplifying,cloning,and sequencing of16S rDNA.
From the1Institute of Microbiology and Genetics and3Depart-
ment of Clinical Microbiology,University of Vienna,Austria;and2De-
partment of Ophthalmology,University of Vienna Medical School,
Vienna,Austria.
Submitted for publication July19,2000;revised November15,
2000;accepted November22,2000.
Commercial relationships policy:N.
The publication costs of this article were defrayed in part by page
charge payment.This article must therefore be marked“advertise-
ment”in accordance with18U.S.C.§1734solely to indicate this fact.
Corresponding author:Talin Barisani-Asenbauer,Department of
Ophthalmology,University of Vienna,Wa¨hringergu¨rtel18–20,1090
Wien,Austria.talin.barisani@aug.akh.magwien.gv.at
Investigative Ophthalmology&Visual Science,May2001,Vol.42,No.6 1164Copyright?Association for Research in Vision and Ophthalmology
Furthermore,monomicrobial and polymicrobial communities were pro?led by DGGE.The introduced molecular technique was applied to the detection of bacteria in suppurative con-junctivitis,to obtain an optimized protocol for ocular samples. Especially in ophthalmology,where sample material is very limited and standard diagnostic tests based on cultivation are often negative,16S rDNA typing in combination with DGGE ?ngerprinting has the potential to become a promising identi-?cation method.
M ETHODS
Sampling and DNA Extraction
Samples were taken in duplicate from60human eyes,in accordance with the tenets of the declaration of Helsinki.Twenty-nine eyes had purulent ocular diseases that were supposed to be most probably due to bacterial infections(i.e.,from chalazions and presumed bacterial conjunctivitis and keratitis).Thirty-one control eyes presented nonpu-rulent conjunctivitis(i.e.,from dry eye syndromes,uveitis,and post-traumatic and keratoconjunctivitis epidemica;Table1).Conjunctival swabs were taken with sterile cotton swabs and immediately frozen at ?20°C in2-ml sterile tubes until they were analyzed.From most patients two DNA extractions from duplicate swabs were carried out independently on2different days,to avoid false-positive or-negative PCR results.
For DNA extraction,cotton swabs were mixed with300?l DNA
extraction buffer I(150mM Na
2EDTA,225mM NaCl;pH8.5)and45
?l lysozyme(50mg/ml).After incubation at37°C for30minutes,9?l
25%SDS and9?l proteinase K(20mg/ml)were added.After incuba-
tion for an additional60minutes at37°C with agitation,150?l of
prewarmed(90°C)DNA extraction buffer II[100mM Na
2
EDTA,400
mM Tris-HCl,400mM Na
2
phosphate buffer(pH8.0),5.55M NaCl,4%
CTAB(hexadecyltrimethyl ammonium bromide);pH8.0]and27?l
25%SDS were added.After incubation at65°C for60minutes the
samples were subjected to three cycles of freezing(?80°C)and thaw-
ing(65°C).One hundred forty microliters was subsequently used for
puri?cation of DNA with the QIAamp Viral RNA Mini Kit(Qiagen,
Hilden,Germany)following the protocol of the manufacturer.This kit
removes several PCR inhibitors and contains carrier RNA,which facil-
itates the elution of small amounts of RNA as well as DNA.DNA was
eluted with60?l ddH2O and tested for PCR-ampli?able DNA with16S
rDNA-speci?c primers.
Ampli?cation of16S rDNA
Three microliters of the extracted DNA was ampli?ed with primers27f
(5?-AGA GTT TGA TCC TGG CTC AG-3?)12and907r(5?-CCC CGT CAA
TTC ATT TGA GTT T-3?),13generating a PCR product corresponding to
nucleotide positions8to926of the Escherichia coli16S rDNA se-
quence.All reactions were carried out in25?l volumes,containing
12.5pmol of each primer,200?M of each deoxyribonucleoside
triphosphate,2.5?l of10?PCR buffer(100mM Tris-HCl,15mM
MgCl
2
,500mM KCl;pH8.3),and0.5U of Taq DNA polymerase
(Roche Diagnostics,Mannheim,Germany),increased to25?l with
sterile water.PCR was performed in a Robocycler(Stratagene,La Jolla,
CA)with the following thermocycling program:5minutes denatur-
ation at95°C,followed by30cycles of1minute denaturation at95°C,
1minute annealing at55°C,1minute extension at72°C,and a?nal
extension step of5minutes at72°C.Ten microliters of PCR products
was visualized by electrophoresis in2%(wt/vol)agarose gels and with
ethidium bromide(0.5?g/ml)staining.
To avoid contamination,all solutions were prepared with sterile
water(Sigma-Aldrich,Vienna,Austria),autoclaved twice,and treated
with hard UV for90minutes in1-ml aliquots.Furthermore,all steps
were performed with aerosol-resistant tips in a CleanSpot PCR/UV
Work Station(Coy Laboratory Products,Grass Lake,MI).The prepara-
tion of the master mix,the addition of template,and the gel electro-
phoresis of PCR products were carried out in three separate rooms.For
each master mix,two negative controls were carried out through the
whole procedure,in which water instead of sample material was used
to exclude the possibility of false-positive PCR results through cross-
contamination.
F IGURE1.Figure1shows a scheme
of the applied experimental proce-
dure,beginning with DNA extraction
from eye swabs,followed by ampli?-
cation of16S rDNA,construction of
clone libraries,genetic?ngerprinting
by DGGE,and sequencing.
T ABLE1.Overview of the Diagnosis and of PCR and Cultivation Results of60Investigated Conjunctival Swabs
Diagnosis
No.of
Samples
Positive
PCR
Results
Positive
Cultivation
Results
Purulent conjunctivitis,
chalazion292512
Dry eye syndrome221
Uveitis10——
Keratoconjunctivitis epidemica3——
Contact lens wear3——
Dry eye syndrome7——
Trauma,allergy or previous
keratoplasty6——
IOVS,May2001,Vol.42,No.6Identi?cation of Conjunctival Bacteria1165
DGGE Analysis
For the genetic?ngerprinting of bacterial16S rDNA from individual eye swabs,nested PCR reactions were carried out.16S rDNA fragments corresponding to nucleotide positions341to534in the E.coli se-quence were ampli?ed with the forward primer341fGC,to which at its5?end a40-base GC-clamp was added(341f:5?-CCT ACG GGA GGC AGC AG-3?;GC-clamp:5?-CGC CCG CCG CGC GCG GCG GGC GGG GCG GGG GCA CGG GGG G-3?)11and the reverse primer518r(5?-ATT ACC GCG GCT GCT GG-3?)11in100?l volumes with4?l of PCR product of the?rst ampli?cation as template DNA.Cycling conditions were as described above.The presence of PCR products was con-?rmed by analyzing10?l of product by electrophoresis in2%(wt/vol) agarose gels and staining with ethidium bromide before DGGE analysis. Ninety microliters of PCR products was precipitated with96%EtOH, resuspended in15?l ddH2O,and separated by DGGE.Gel electro-phoresis was performed as described elsewhere in a linear denaturant gradient from25%to60%in a D GENE-System(Bio-Rad,Munich, Germany).11After completion of electrophoresis,gels were stained in an ethidium bromide solution and documented with a UVP documen-tation system.
16S rDNA Sequencing
Sequence identi?cation was carried out only with samples showing a PCR product after the?rst30ampli?cation cycles.From some samples sequence information was obtained by excising and direct sequencing of reampli?ed200-bp DGGE bands as described elsewhere.14For the direct sequencing of excised DGGE bands,the extracted DNA was reampli?ed with primer341f,including an additional T3sequence at its5?end(T3:5?-AAT TAA CCC TCA CTA AAG-3?)and primer518r.To identify microorganisms on the basis of longer16S rDNA fragments, from most samples900-bp16S rDNA clone libraries were constructed by cloning5?l of PCR product ampli?ed with primers27f and907r. Cloning was performed with the pGEM-T Vector System(Promega, Mannheim,Germany),following the protocol of the manufacturer.The ligation products were subsequently transformed into E.coli XLI-Blue, which allows blue-white screening.15To screen for positive clones, clone inserts were ampli?ed with the vector-speci?c primers SP6 (5?-ATT TAG GTG ACA CTA TAG AAT AC-3?)and T7(5?-TAA TAC GAC TCA CTA TAG GG-3?).Screening for different clones was carried out by comparing inserts reampli?ed with primers341fGC and518r in DGGE.In a second screening the different clones were compared with the DGGE?ngerprint of the corresponding eye swab.Inserts of clones producing PCR products that matched identical positions in the DGGE ?ngerprint of the eye swab were sequenced.
For sequencing of clone inserts,fragments were ampli?ed with primers SP6and T7.One hundred microliters of PCR products was puri?ed with a QIAquick PCR Puri?cation Kit(Qiagen)and sequenced with a LI-COR DNA Sequencer Long Read4200.16Sequencing reac-tions were carried out by cycle sequencing with the SequiTherm system(EPICENTRE,Madison,WI)with2pmol?uorescently labeled primers and5U SequiTherm thermostable DNA polymerase.Clone inserts were partially sequenced(200or500bp)with primers341f or 518r.Reampli?ed DGGE bands were sequenced with primer T3. Phylogenetic Analyses
The obtained sequences were compared with sequences of known bacteria listed in the EMBL nucleotide sequence database.The FASTA search option for the EMBL database was used to search for close evolutionary relatives.17
Nucleotide Sequence Accession Numbers
The sequences obtained in this study have been assigned in the EMBL database under the accession numbers AJ405008–AJ405055. Cultivation
All swabs were cultured by routine in a standard diagnostic labor. Bacterial culture was performed with the aim to detect common causes of conjunctivitis such as Haemophilus in?uenzae,Staphylo-coccus aureus,Streptococcus pneumoniae as well as causes of infec-tions in immunocompromised patients(members of the family Enter-obacteriaceae and Pseudomonas aeruginosa).Anaerobic cultures and cultures for Neisseria gonorrhoeae and enrichment cultures, which are only performed if unusual organisms are suspected,were not carried out.
Brie?y,swabs were placed into a transport medium(Transswab; Medical Wire&Equipment Co.Ltd.,Corsham Wilts,United Kingdom), and after arrival in the laboratory were plated onto columbia agar?5% sheep blood,chocolate agar?isovitalex?bacitracin and McConkey agar(Becton Dickinson Microbiology Systems,Sparks,MD).The media were incubated in5%to7%CO
2
at35°C for up to48hours.Bacteria were identi?ed using standard microbiologic procedures.
R ESULTS
Ampli?cation of16S rDNA and
DGGE Fingerprinting
Generally,PCR was done twice with template DNA obtained from each of the duplicate extractions per sample.Samples producing reproducible PCR products four times were re-garded as PCR positive.In cases of negative PCR results in all ampli?cations,the samples were regarded as PCR negative. Five samples showed very faint bands in agarose gels in only one of the duplicate DNA extractions.Therefore,they were ampli?ed twice more under the same conditions.Because these very faint bands were not reproducible,these samples were also regarded as PCR negative.In total,27of60investi-gated eye swabs showed reproducible PCR products after30 ampli?cation cycles(Table1).Two of the PCR-positive sam-ples(A4and A9)belonged to the eyes with nonpurulent conjunctivitis and were initially not supposed to show bacte-rial colonization(Table2).Four of the PCR-negative samples were initially supposed to be PCR positive according to the symptoms(purulent conjunctivitis).Thirty-three of the60in-vestigated samples were regarded as PCR negative.
DGGE analysis and sequence identi?cation were carried out only with the27PCR-positive samples.DGGE?ngerprints derived from the conjunctival swabs taken in duplicate per patient showed the same patterns(data not shown).Table1 gives an overview of the conjunctival swabs from which16S rDNA could be ampli?ed successfully and furthermore an over-view of the PCR-negative conjunctival swabs.
Sequence Analysis of Individual DGGE Bands
Ten sequences were obtained by the direct sequencing of excised DGGE bands.Thirty-eight sequences were obtained by sequencing of cloned16S rDNA inserts.During the screening no clones could be detected that were not present in the original DGGE band patterns.From samples A6and A9no clones or excised DGGE bands were sequenced(Table2). From the excised and reampli?ed DGGE bands sequence in-formation between106and173bp was obtained,some of which had a high number of ambiguous bases,because of the sometimes bad quality of DNA.From clone inserts,between 199and550bp were sequenced.Table2shows the results of comparative sequence analyses obtained from the EMBL data-base.Sequence homologies to sequences of known bacteria in the EMBL database ranged between90%and100%.Most se-quences had similarity values between98%and99.8%.In total, 16samples showed monomicrobial and11samples showed polymicrobial infections.Of the11samples showing polymi-crobial infections,7samples had infections with two different genera,3samples with three different genera,and1sample with four different genera.Table3gives an overview of how often the identi?ed genera were detected in the27PCR-
1166Schabereiter-Gurtner et al.IOVS,May2001,Vol.42,No.6
positive samples.The most frequently detected genera were Staphylococcus and Corynebacterium,followed by Propi-onibacterium and Streptococcus .The remaining sequences were af?liated with the genera Bacillus,Acinetobacter,Bre-vundimonas,Pseudomonas,and Proteus .For most sequences af?liated with Streptococcus,Staphylococcus,and Corynebac-terium,the sequence similarities obtained from the EMBL database were the same for different species within the same
T ABLE 2.Overview of the Diagnosis,Cultivation,and Sequence Analysis Results of the 27PCR-positive Conjunctival Swabs
Sample Diagnosis
Sequence Name*
Phylogenetic Af?liation
%?
Cultivation
A1Purulent conjunctivitis A1–B1(T3/121/0)Staphylococcus spp.100.0Coagulase-negative staphylococci A2Purulent conjunctivitis and chalazion A2–K1(341f/200/0)Staphylococcus caprae 100.0Coagulase-negative staphylococci A2–K2(518r/455/0)Uncultured Corynebacterium 98.0A3Chalazion A3–K1(341f/503/0)
Corynebacterium accolens/fastidiosum 99.8Coagulase-negative staphylococci A3–K2(341f/497/0)Uncultured Corynebacterium sp.99.6Corynebacterium amycolatum/asperum 99.4
A4
Dry eye syndrome
A4–K1(341f/502/2)Corynebacterium macginleyi 99.0Coagulase-negative staphylococci,Klebsiella oxytoca,Corynebacteria
A4–K2(341f/506/1)Corynebacterium sp.99.8Corynebacterium amycolatum/asperum/variabilis
94.5A5Purulent conjunctivitis A5–K1(341f/503/0)Uncultured Corynebacterium sp.99.6Coagulase-negative staphylococci A6Purulent conjunctivitis A6–B1,not sequenced Has the same position in DGGE as Corynebacterium sp.
Corynebacteria
A7Purulent conjunctivitis A7–K1(518r/453/0)Corynebacterium asperum 99.8Staphylococcus aureus A8Chalazion A8–B1(T3/141/0)Corynebacterium bovis 98.6Corynebacteria A8–B2(T3/151/0)Propionibacterium acnes 99.3A9Dry eye syndrome A9–B1,not sequenced Has the same position in DGGE as Propionibacterium acnes
—
A10Purulent conjunctivitis and keratitis
A10–B1(T3/151/7)Propionibacterium acnes 98.1—
A11Purulent conjunctivitis A11–K1(341f/550/0)
Acinetobacter johnsonii 99.8—A11–K2(341f/449/0)Pseudomonas putida 99.8A11–B2(T3/149/9)Streptococcus sp.91.3A12Purulent conjunctivitis A12–B1(T3/172/0)Acinetobacter lwof?i
99.4—A13
Purulent conjunctivitis
A13–K1(341f/200/1)Acinetobacter johnsonii/lwof?i 98.0—A13–K2(341f/200/0)
Acinetobacter sp.
99.5Acinetobacter johnsonii/lwof?i 99.0A13–K3(341f/200/0)Brevundimonas vesicularis 99.0A14Purulent conjunctivitis A14–B1(T3/139/0)Staphylococcus pasteuri 99.3—A14—B3(T3/106/8)Propionibacterium acnes 91.5A15Purulent conjunctivitis A15–K1(341f/200/0)Staphylococcus capitis 100.0—A15–K2(341f/200/3)Propionibacterium acnes 98.5
A16
Purulent conjunctivitis A16–K1(518r/481/0)Streptococcus salivarius 99.2Coagulase-negative staphylococci A17Purulent conjunctivitis A17–K1(341f/200/1)Staphylococcus pasteuri 98.0—A17–K2(341f/200/1)Staphylococcus pasteuri
99.0A17–K3(341f/200/0)Streptococcus bovis/waiu/suis 96.5
A18Purulent conjunctivitis and keratitis
A18–K1(341f/498/0)
Propionibacterium acnes
99.4Coagulase-negative staphylococci A18–K2(341f/537/4)Streptococcus mitis/oralis 98.1A19
Purulent conjuntivitis and keratitis
A19–K1(341f/475/0)?Kingella kingae/Neisseria sp.90.3—
A19–K2(341f/199/0)Staphylococcus capititis
99.5A19–K3(341f/200/0)Streptococcus bovis/waiu/suis
97.5A19–K4
(341f/518/0)
Serratia proteamaculans/Aranicola proteolyticus
100.0A20
Chalazion and purulent conjunctivitis A20–B1(T3/150/1)Staphylococcus capitis/cohnii/saprophyticus/epidermis
99.3—
A20–K1(341f/536/0)?Enterobacter agglomerans/Pantoea
ananas
99.3
A20–K2(341f/536/0)?Enterobacter agglomerans
99.4Pantoea annans
99.3A20–K3(341f/531/1)Propionibacterium acnes
99.5
A21
Purulent conjunctivitis A21–K1(341f/519/0)Weissella confusa/kandleri
97.1Streptococci,Coagulase-negative staphylococci A21–K2(341f/200/2)Streptococcus mitis/pneumoniae/
oralis/infantis
96.5A21–K3(341f/200/0)Staphylococcus capitis/cohnii/
saprophyticus/epidermis
100.0
A22Purulent conjunctivitis A22–K1(341f/543/0)Leuconostoc mesenteroides
94.7—A22–K2(341f/518/0)?Leuconostoc paramesenteroides
96.7Weissella sp.
96.5
A23Purulent conjunctivitis A23–K1(T3/200/0)Proteus mirabilis 99.5Proteus sp.
A24Purulent conjunctivitis A24–K1(341f/530/1)Pseudomonas aeruginosa 99.6Pseudomonas aeruginosa
A25Purulent conjunctivitis A25–K1(341f/532/1)Pseudomonas sp.
98.9Coagulase-negative staphylococci A25–K2(341f/533/1)Pseudomonas sp.
99.3Pseudomas migulae/putida
98.9A26Purulent conjunctivitis and keratitis
A26–K1(341f/146/0)Bacillus subtilis 97.3—A27
Purulent conjunctivitis A27–B1(T3/173/1)Bacillus clarkii 94.3
—*Sequences numbered with –B were obtained by direct sequencing of excised DGGE bands;sequences numbered with –K were obtained by shotgun cloning of 900-bp 16S rDNA PCR products (sequencing primer/number of sequenced bases/number of ambiguous bases).
?Percent similarity of partial 16S rRNA coding sequences to sequences of their closest bacterial relatives available in the EMBL nucleotide sequence database.
?A phylogenetic identi?cation to the genus level was not possible.
IOVS,May 2001,Vol.42,No.6Identi?cation of Conjunctival Bacteria 1167
genus,which prevented an identi?cation to the species level.For six sequences no identi?cation to the genus level could be obtained because of similar sequence similarity values to the different genera Leuconostoc and Weissella (A22-K1and A22-K2),Pantoea and Enterobacter (A20-K1and A20-K2),Kingella and Neisseria (A19-K1),and to Serratia and Aranicola (A19-K4),respectively.
Comparison of DGGE Fingerprints
To compare the DGGE positions of all sequences,the se-quenced clones and DGGE bands were reampli?ed and ana-lyzed parallel in DGGE.Figure 2shows the DGGE patterns of the reampli?ed clones and excised DGGE bands of all 27PCR-positive samples.DGGE ?ngerprints of 12samples con-tained one band,?ngerprints of 9samples contained two bands,?ngerprints of 5samples contained three bands,and ?ngerprints of 2samples contained four bands.Table 3gives an overview of how many different positions in DGGE could be found among sequences af?liated with the same genus.Five different positions in DGGE were found among 9Staphylococ-cus -af?liated sequences.Three different positions in DGGE were found among 6Streptococcus -af?liated sequences.Nine different positions in DGGE were found among 10Corynebac-
terium -af?liated sequences.Two different positions in DGGE were found among 3Lactobacillaceae-af?liated sequences.Only different positions in DGGE were found among 4Acin-etobacter-,4Pseudomonas-,2Bacillus-,and 2Enterobacter/Pantoea -af?liated sequences.All sequences af?liated with Pro-pionibacterium had the same position in DGGE.
Comparison of Cultivation and PCR
No sample positive by culture gave negative results by PCR,whereas 55%of the PCR-positive results were culture negative.Culture was only positive for coagulase-negative staphylococci (n ?9),Corynebacterium spp.(n ?3),S.aureus (n ?1),Streptococcus sp.(n ?1),Proteus sp.(n ?1),Klebsiella oxytoca (n ?1),and P.aeruginosa (n ?1;Table 3).In seven cases (samples A3,A4,A5,A7,A16,A18,and A25)coagulase-negative staphylococci and in one case (sample A4)K.oxytoca were detected only by cultivation but not by PCR in apparently polymicrobial infections (Tables 2and 3).
D ISCUSSION
In the present study,the conjunctival ?ora of 60eyes was investigated by molecular means,including PCR,DGGE,and
T ABLE 3.Overview of the Abundance of Bacterial Genera and Number of Their Different Positions in DGGE
Identi?ed Genera
Phylogenetic Af?liation
PCR-Positive Swabs*
DGGE?Culture-Positive
Swabs*
Gram-positive,aerobic Corynebacterium Actinomycetales/Corynebacteriaceae 79/103Staphylococcus Bacillus/Staphylococcus group
84/910Streptococcus Bacillus/Clostridium group,Streptococcaceae 63/61Bacillus
Bacillus/Clostridium group,Bacillaceae 22/2—Gram-positive,obligate anaerobic Propionibacterium
Actinomycetales/Propionibacteriaceae 71/7—Gram-positive,faculative anaerobic Leuconostoc/Weissella Bacillus/Clostridium group,Lactobacillaceae
22/3—Gram-negative,aerobic Acinetobacter ?-Proteobacteria,Moraxellaceae
34/4—Pseudomonas ?-Proteobacteria,Pseudomonas group 34/41Brevundimonas ?-Proteobacteria,Caulobacter group 11/1—Kingella/Neisseria ?-Proteobacteria,Neisseriaceae
11/1—Pantoea/Enterobacter ?-Proteobacteria,Enterobacteriaceae 11/1—Proteus
?-Proteobacteria,Enterobacteriaceae 11/11Serratia/Aranicola ?-Proteobacteria,Enterobacteriaceae 11/1—Klebsiella oxytoca
?-Proteobacteria,Enterobacteriaceae
—
—
1
*Number of eye swabs in which the corresponding genera were detected.?Number of different positions in DGGE/number of investigated
sequences.
F IGURE 2.Detail of the ethidium bromide–stained 16S rDNA DGGE ?ngerprints of reampli?ed clones and excised DGGE bands derived from 27eye swabs,from which 16S rDNA fragments could be ampli?ed successfully after 30ampli?cation cycles.DGGE bands A1–B1,A6–B1,A11–B2,and A19–K4were almost not visible in the ?gure and their positions in DGGE were therefore marked with https://www.doczj.com/doc/0a4108382.html,nes and DGGE bands are numbered according to the nomenclature of samples listed in Table https://www.doczj.com/doc/0a4108382.html,nes M represent the standard reference pattern.The following reference species were used:Borrelia sp.(I),Pasteurella canis (II),Listeria monocytogenes (III),Actinobacillus pleuropneumonie (IV),Staphylococcus aureus (V),Bordetella bronchiseptica (VI),Escherichia coli (VII),Pasteurella hemolytica (VIII),Rhodococcus sp.(IX)and Frankia sp.(X).
1168Schabereiter-Gurtner et al.IOVS,May 2001,Vol.42,No.6
16S rDNA sequence analyses.Working with conjunctival swabs,only very little bacterial material is available.The am-pli?cation of16S rDNA is therefore a useful method for the investigation of such samples.However,the extreme sensitiv-ity of nucleic acid ampli?cation techniques enhances the pos-sibility of detection of clinically irrelevant or contaminating target sequences.If used improperly,the ampli?cation of16S rDNA might give misleading results,especially with clinical samples that contain relatively few pathogenic microorgan-isms.We have observed that the exceeding of35ampli?cation-cycles led to ampli?cation of contaminating DNA in some of the reagent-only controls,despite precautions such as the use of aerosol-resistant tips and the spatial separation of DNA extraction,PCR,and the gel electrophoresis of PCR products. As ampli?cation was therefore restricted to30cycles,the normal conjunctival?ora could not be detected under the given laboratory circumstances,because of the low number of bacteria present on the normal conjunctiva.
In total,eubacterial DNA could be ampli?ed from27of60 investigated eye swabs.PCR-positive results were obtained from25of29specimens taken from purulent conjunctivitis and2samples taken from eyes with dry eye syndrome.Four samples derived from eyes with purulent conjunctivitis were unexpectedly PCR negative.Reasons for these negative PCR results were in two cases fungal infections and in two cases herpes simplex virus infection.Negative PCR results from control eyes had been expected.In cases of intraocular disease vitreous?uid and aqueous humor have to be investigated, because conjunctival swabs do not contain the pathogenic organism.Control eyes with contact lens wear history,dry eye syndrome,allergy,and trauma had been suspected to have no bacterial infection.
Comparison of sequences with sequences listed in the EMBL database revealed that most of them had sequence sim-ilarities of98%to100%to sequences of known genera.As the investigated sequences were only parts(between121and536 bp)of the approximately1600-bp-long16S rDNA,a clear phylogenetic af?liation was obtained only to the genus and not to the species level.Generally,a reliable phylogenetic identi-?cation based on partial16S rDNA analysis is often only pos-sible to the genus level.18,19For some sequences a clear iden-ti?cation to the genus level was not possible.Two clones had identical similarities to sequences of the genera Enterobacter and Pantoea,both members of the Enterobacteriaceae and proposed to be reclassi?ed together in the genus Pantoea.20 Three clones had similarities identical to sequences of the genera Leuconostoc and Weissella,both members of the Lac-tobacillaceae and proposed to be reclassi?ed together in the genus Weissella.21One clone had sequence similarities identi-cal to sequences of the genera Serratia and Aranicola,both members of the Enterobacteriaceae.Another clone had a90% similarity value to sequences of the genera Kingella and Neis-seria,both members of the Neisseriaceae.The low sequence similarity value to known bacteria reveals that this sequence represents a new genus related to the Neisseriaceae.The problem of inadequate phylogenetic identi?cation could be improved by cloning and sequencing of the entire16S rDNA. Furthermore,because the number of16S rDNA sequences in public databases is increasing day by day,a better identi?cation of so far unknown bacteria may be possible in the future.
The obtained results are mostly in accordance with results of other studies based on cultivation.Most bacteria frequently observed in this study are expected pathogenic organisms causing infections in human eyes and lids.Different species of Bacillus,Proteus,Pseudomonas,Serratia,and especially of Corynebacterium,Staphylococcus,and Streptococcus have been found to be part of the normal conjunctival?ora as well as to play roles as pathogens in different ocular diseases.22–25Propionibacterium acnes has been detected to be part of the normal anaerobic conjunctival?ora as well as to be one of the causative agents of late-onset endophthalmitis23,26,27and cor-neal ulceration.28Concerning the other detected bacteria,little is known from the literature on a possible pathogenic charac-ter in human eyes.However,in neonates and infants,these bacteria have been associated with more or less severe non-eye diseases.29To our knowledge,Acinetobacter spp.and Enter-obacter agglomerans/Pantoea ananas are not very com-monly detected in the human eye.E.agglomerans and Acin-etobacter lwof?i are rarely isolated from eyes that have endophthalmitis.25,30Pantoea may cause fever,shaking chills, sepsis,and osteomyelitis.Pantoea was previously observed in six conjunctival swabs of patients who had conjunctivitis, unfortunately without knowledge of the clinical course,31and was involved in endophthalmitis after foreign body penetra-tion.32The detected organism with the highest sequence sim-ilarities to the genera Leuconostoc and Weissella has not yet been found in eyes.Members of the genus Leuconostoc are facultatively anaerobic,catalase-negative,Gram-positive cocci and exhibit an intrinsic resistance to vancomycin.Neonates may be colonized during delivery by Leuconostoc inhabiting the maternal genital tract.Leuconostoc was also encountered in cerebrospinal?uid,peritoneal dialysate?uid and wounds but is supposed to have very little virulence for healthy hu-mans.33Kingella kingae is a small Gram-negative rod and may be involved in suppurative arthritis,osteomyelitis,spondylo-diskitis,endocarditis,transient bacteremia,meningitis,pulmo-nary infections,dactylitis,and subglottic and epiglottic infec-tions.K.kingae,also known to cause eyelid abscesses and endophthalmitis,was observed in corneal ulceration.34–36De-tection by conventional culture is dif?cult and cultures should be examined once per week for a total of3weeks.However, only5%of Kingella infections will be detected by this proce-dure.Brevundimonas is a pseudomonad,rarely encountered in human infection.Brevundimonas vesicularis may be a virulent organism involved in central nervous system infections and bacteremia,including nosocomial infections.37 In this study,the presence of polymicrobial infections of coagulase-negative staphylococci and K.oxytoca with Coryne-bacterium spp.,Streptococcus spp.,Pseudomonas spp.or Propionibacterium spp.raised the problem of detecting the coagulase-negative staphylococci and K.oxytoca by PCR.The presence of coagulase-negative staphylococci might have been overestimated by selective cultivation as a result of cultivation-dependent population shifts.Because of the preferential am-pli?cation of the more abundant template DNA of those bac-teria that could not be detected by cultivation,rare bacteria might have failed to be ampli?ed in a suf?cient amount.Con-cerning polymicrobial infections,on the one hand the template DNA of rare bacteria can be outcompeted in the ampli?cation process by template DNAs of bacteria that are present in greater numbers11,38,39;on the other hand selective cultivation can lead to an overestimation of a certain organisms.Both cases lead to discrepancies between PCR and culture.
The combination of cloning and genetic?ngerprinting by DGGE allows the identi?cation of polymicrobial infections. Although the excising and direct sequencing of DGGE bands seems to be a more rapid method,cloning leads to longer and a higher quality of sequence information,which facilitates a more reliable phylogenetic identi?cation.The construction of DGGE markers,containing16S rDNA fragments of bacteria relevant for ocular diseases,would facilitate and accelerate the interpretation of DGGE?ngerprints.In this study,it is notice-able that especially among the genera Corynebacterium,Acin-etobacter,and Pseudomonas sequences could be distin-guished by DGGE.
IOVS,May2001,Vol.42,No.6Identi?cation of Conjunctival Bacteria1169
Although being an elegant method for the investigation of especially polymicrobial infections,DGGE has been used so far only in a few studies for bacterial identi?cation in clinical specimens.40–42The broad-range nature of the method allows the detection of rare,unexpected,or fastidious pathogens. Especially in ophthalmology,where only little sample material is available from the outset and cultivation results are often negative,the sensitive broad-range ampli?cation of16S rDNA in combination with DGGE could become a promising detec-tion and identi?cation method.In the present study,no sample positive by culture gave negative results by PCR,whereas55% of the PCR-positive results were culture negative.This indi-cates that PCR is a more reliable and signi?cantly more sensi-tive method for the detection of bacterial infections than cul-tivation.Similar observations were made by Lohmann et al.,43 whose PCR results in vitreous samples were positive in92%, whereas the cultivation results were positive in only24%. Culturing failed to detect Propionibacterium acnes and Acti-nomyces israelii,which are known to be fastidious and slowly growing bacteria.Therese et al.2investigated vitreous samples, in which44%of culture-negative samples showed positive PCR results.Okhravi et al.44investigated37aqueous and vitreous samples with suspected infection,of which100%were PCR positive and54%were culture negative.Ley et al.41detected bacterial DNA of Pseudomonas spp.,Acinetobacter spp.,Esch-erichia spp.,Moraxella spp.,Staphylococcus spp.,and Bacil-lus spp.in20blood samples that were culture negative.In addition,several other studies in medical microbiology re-vealed that the PCR technique is more sensitive in the detec-tion of bacteria than cultivation.3,7,8,45
The introduced molecular technique was applied to the detection of bacteria in suppurative conjunctivitis,to establish the technique for ocular samples.Results of the present pilot study reveal that16S rDNA genotyping in combination with DGGE is more sensitive than conventional cultivation.Al-though the introduced method is not completely free of all biases,it can be used supplementarily or as an alternative to cultivation,especially in infections caused by bacteria with unusual growth requirements,for patients who have been unsuccessfully treated with antibiotics or who suffer from sight-threatening or chronical bacterial infections that cannot be cultured.
References
1.Hykin PG,Tobal K,McIntyre G,Matheson MM,Towler HMA,
Lightman SL.The diagnosis of delayed post-operative endoph-thalmitis by polymerase chain reaction of bacterial DNA in vitreous samples.J Med Microbiol.1994;40:408–415.
2.Therese KL,Anand AR,Madhavan HN.Polymerase chain reaction
in the diagnosis of bacterial endophthalmitis.Br J Ophthalmol.
1998;82:1078–1082.
3.Lohmann CP,Linde H-J,Reischl U.Die Schnelldiagnostik einer
infektio¨sen Endophthalmitis mittels Polymerase-Kettenreaktion (PCR):Eine Erga¨nzung zu den konventionellen mikrobiologischen Diagnostikverfahren.Klin Monatsbl Augenheilkd.1997;11:22–27.
4.Knox CM,Cevellos V,Dean D.16S ribosomal DNA typing for
identi?cation of pathogens in patients with bacterial keratitis.
J Clin Microbiol.1998;36:3492–3496.
5.Ormerod LD,Paton BG,Haaf J,Topping TM,Baker AS.Anaerobic
bacterial endophthalmitis.Ophthalmology.1987;84:799–808. 6.Kunimoto DY,Das T,Sharma S,et al.Microbiologic spectrum and
susceptibility of isolates:part II.Posttraumatic endophthalmitis.
Endophthalmitis Research Group.Am J Ophthalmol.1999;128: 242–244.
7.Javala J,Ma¨ntymaa M-L,Ekblad U,et al.Bacterial16S rDNA poly-
merase chain reaction in the detection of intra-amniotic infection.
Br J Obstet Gynaecol.1996;103:664–669.
8.Hitti J,Riley DE,Krohn MA,et al.Broad-spectrum bacterial rDNA
polymerase chain reaction assay for detecting amniotic detection
of?uid infection among women in premature labor.Clin Infect Dis.1997;24:1228–1232.
9.Dymock D,Weightman AJ,Scully C,Wade WG.Molecular analysis
of micro?ora associated with dentalveolar abscesses.J Clin Micro-biol.1996;34:537–542.
10.Suau A,Bonnet R,Sutren M,et al.Direct analysis of genes encod-
ing16S rRNA from complex communities reveals many novel molecular species within the human gut.J Clin Microbiol.1999;
65:4799–4807.
11.Muyzer G,De Waal EC,Uitterlinden AG.Pro?ling of complex
microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-ampli?ed genes coding for 16S rRNA.Appl Environ Microbiol.1993;59:695–700.
https://www.doczj.com/doc/0a4108382.html,ne DJ,Pace B,Olson GJ,Stahl DA,Sogin ML,Pace NR.Rapid
determination of16S ribosomal RNA sequences for phylogenetic analyses.Proc Natl Acad Sci USA.1985;82:6955–6959.
13.Teske A,Wawer C,Muyzer G,Ramsing NB.Distribution of sulfate-
reducing bacteria in a strati?ed fjord(Mariager Fjord,Denmark)as evaluated by most-probable-number counts and DGGE of PCR-ampli?ed ribosomal DNA fragments.Appl Environ Microbiol.
1996;62:1405–1415.
14.Ro¨lleke S,Muyzer G,Wawer C,Wanner G,Lubitz W.Identi?cation
of bacteria in a biodegraded wall painting by denaturing gradient gel electrophoresis of PCR-ampli?ed gene fragments coding for 16S rRNA.Appl Environ Microbiol.1996;62:2059–2065.
15.Sambrook J,Fritsch EF,Maniatis T.Molecular Cloning:A Labora-
tory Manual,2nd ed.Cold Spring Harbor,NY:Cold Spring Harbor Laboratory;1989.
16.Middendorf LR,Bruce JC,Bruce RC,et al.Continuous,on-line DNA
sequencing using a versatile infrared laser scanner/electrophoresis apparatus.Electrophoresis.1992;13:487–494.
17.Pearson WR.Rapid and sensitive sequence comparison with FAST
and FASTA.Methods Enzymol.1994;183:63–98.
18.Vandamme P,Pot B,Gillis M,De Vos P,Kersters K,Swings J.
Polyphasic taxonomy,a consensus approach to bacterial system-atics.Microbiol Rev.1996;60:407–438.
19.Fox GE,Wisotzkey JD,Jurtsuk P.How close is close:16S rRNA
sequence identity may not be suf?cient to guarantee species iden-tity.Int J Syst Bacteriol.1992;42:166–170.
20.Gavini F,Mergaert J,Beji A,Mielcarek C,et al.Transfer of Enter-
obacter agglomerans(Beijerinck1888,Ewing and Fife to Pantoea gen.nov.as Pantoea agglomerans comb.nov.and description of Pantoea dispersa sp.nov.Int J Syst Bacteriol.1989;1972:39:337–345.
21.Collins MD,Samelis J,Metazopoulos J,Wallbanks S.Taxonomic
studies of some leuconostoc-like organisms from fermented sau-sages-description of a new genus Weissella for the Leuconostoc paramesenteroides group of species.J Appl Bacteriol.1993;75: 595–603.
22.Mao LK,Flynn HW,Miller D,P?ugfelder SC.Endophthalmitis
caused by streptococcal species.Arch Ophthalmol.1992;110: 789–801.
23.Thiel H-J,Schuhmacher U.U¨ber die Standort?ora der menschli-
chen Bindehaut:Untersuchungen von135Personen unterschiedli-chen Alters.Klin Monatsbl Augenheilkd.1994;205:348–357. 24.Grasbon T,de Kaspar HM,Klau?V.Koagulasenegative Staphy-
lokokken auf der blanden und chronisch entzu¨ndeten Bindehaut.
Ophthalmologe.1995;92:793–801.
25.Crawford PM,Conway MD,Peyman GA.Trauma-induced Acineto-
bacter lwof?i endophthalmitis with multi-organism recurrence: strategies with intravitreal treatment.Eye.1997;11:863–864. 26.Keyser BJ,Maguire JI,Halperin LS.Propionibacterium acnes en-
dophthalmitis after Staphylococcus epidermis endophthalmitis.
Am J Ophthalmol.1993;116:505–506.
27.De Queiro′z Campos MS,de Queiro′z Campos L,Rehder JRCL,Lee
MB,O’Brien T,McDonnell PJ.Anaerobic?ora of the conjunctival sac in patients with AIDS and with anophthalmia compared with normal eyes.Acta Ophthalmol(Copenh).1994;72:241–245. 28.Jones DB.Decision-making in the management of microbial kera-
titis.Ophthalmology.1981;88:814–820.
29.Feigin RD,Chery JD,eds.Textbook of Pediatric Infectious Dis-
eases,4th ed.Philadelphia:WB Saunders;1998.
1170Schabereiter-Gurtner et al.IOVS,May2001,Vol.42,No.6
30.Zeiter JH,Koch DD,Parke DW2d,Font RL.Endogenous endoph-
thalmitis with lenticular abscess caused by Enterobacter agglom-erans(Erwinia species).Ophthalmic Surg.1989;20:9–12.
31.Bottone E,Schneiderson SS.Erwinia species:an emerging human
pathogen.Am J Clin Pathol.1972;57:400–405.
32.Oesterle CS,Kronenberg HA,Peyman GA.Endophthalmitis caused
by an Erwinia species.Arch Ophthalmol.1977;95:824–825. 33.Facklam R,Elliott JA.Identi?cation,classi?cation and clinical rel-
evance of catalase-negative,Gram-positive cocci,excluding the Streptococci and Enterococci.Clin Microbiol Rev.1995;8:479–495.
34.Henriksen SD.Corroding bacteria from the respiratory tract.Acta
Pathol Microbiol Scand.1969;75:85–90.
35.Carden SM,Colville DJ,Gonis G,et al.Kingella kingae endoph-
thalmitis in an infant.Aust NZ L J Ophthalmol.1991;19:217–220.
36.Goutzmanis JJ,Gonis G,Gilbert GL.Kingella kingae infection in
children:ten cases and a review of the literature.Pediatr Infect Dis J.1990;10:677–683.
37.Gilad J,Borer A,Peled N,et al.Hospital-acquired Brevundimonas
vesicularis septicaemia following open heart surgery:case report and literature review.Scand J Infect Dis.2000;32:90–91.
38.Murray AE,Hollibaugh JT,Orrego C.Phylogenetic compositions of
bacterioplankton from two Californian estuaries compared by de-
naturing gradient gel electrophoresis of16S rDNA fragments.Appl Environ Microbiol.1996;62:2676–2680.
39.Polz MF,Cavanaugh CM.Bias in template-to-product ratios in
multitemplate PCR.Appl Environ Microbiol.1998;64:3724–3730.
https://www.doczj.com/doc/0a4108382.html,lar MR,Linton CJ,Cade A,Glancy D,Hall M,Jalal H.Application
of16S rRNA gene PCR to study bowel?ora of preterm infants with and without necrotizing enterocolitis.J Clin Microbiol.1996;34: 2506–2510.
41.Ley BE,Linton CJ,Bennett DMC,Jalal H,Foot ABM,Millar MR.
Detection of bacteraemia in patients with fever and neutropenia using16S rRNA gene ampli?cation by polymerase chain reaction.
Eur J Clin Microbiol Infect Dis.1998;17:247–253.
42.Zoetendal EG,Akkermans ADL,de Vos W.Temperature gradient
gel electrophoresis analysis of16S rRNA from human fecal samples reveals stable and host-speci?c communities of active bacteria.
Appl Environ Microbiol.1998;64:3854–3859.
43.Lohmann CP,Linde H-J,Reischl U.Improved detection of micro-
organisms by polymerase chain reaction in delayed endophthalmi-tis after cararact surgery.Ophthalmology.2000;107:1047–1052.
44.Okhravi N,Adamson P,Carroll N,et al.PCR-based evidence of
bacterial involvement in eyes with suspected intraocular infection.
Invest Ophthalmol Vis Sci.2000;41:3474–3479.
45.Rantakokko-Javala K,Nikkari S,Javala J,et al.Direct ampli?cation
of rRNA genes in diagnosis of bacterial infections.J Clin Microbiol.
2000;38:32–39.
IOVS,May2001,Vol.42,No.6Identi?cation of Conjunctival Bacteria1171
智能制造技术路线图 摘要: 新一代信息通信技术产业、高档数控机床和机器人、航空航天装备、海洋工程装备及高技术船舶、先进轨道交通装备、节能与新能源汽车、电力装备、农业装备、新材料、生物医药及高性能医疗器械十大重点领域进入《技术路线图》,意味着互联网和传统工业的融合将是发展的制高点,智能制造将是中国制造未来的主攻方向。 日前,国家制造强国建设战略咨询委员会在京正式发布《〈中国制造2025〉重 点领域技术路线图(2015版)》。新一代信息通信技术产业、高档数控机床和 机器人、航空航天装备、海洋工程装备及高技术船舶、先进轨道交通装备、节能与新能源汽车、电力装备、农业装备、新材料、生物医药及高性能医疗器械十大重点领域进入《技术路线图》。 引领发展方向 2010年以来我国制造业增加值连续五年超过美国成为制造大国,一些优势领域 已达到或接近世界先进水平。然而,我国制造业大而不强,创新能力、整体素质和竞争力与发达国家相比仍有明显差距。加快实现从制造大国向制造强国的转变,已成为新时期我国经济社会发展的重大战略任务。 为了推进这一历史性的转变,国务院组织编制并于今年5月19日正式发布《中国制造2025》,对我国制造业转型升级和跨越发展做了整体部署,提出了我国 制造业由大变强“三步走”战略目标,明确了建设制造强国的战略任务和重点,是我国实施制造强国战略的第一个十年行动纲领。 制造业覆盖面很广,为了确保我国十年后能够迈入制造强国行列,必须坚持整 体推进、重点突破的发展原则。受国家制造强国建设战略咨询委员会委托,中国工程院围绕《中国制造2025》确定的新一代信息通信技术产业、高档数控机床 和机器人、航空航天装备、海洋工程装备及高技术船舶、先进轨道交通装备、节能与新能源汽车、电力装备、农业装备、新材料、生物医药及高性能医疗器械等十大重点领域未来十年的发展趋势、发展重点和目标等进行了研究,提出了十大
技术路线图(Technology Roadmap) 什么是技术路线图 技术路线图是指应用简洁的图形、表格、文字等形式描述技术变化的步骤或技术相关环节之间的逻辑关系。它能够帮助使用者明确该领域的发展方向和实现目标所需的关键技术,理清产品和技术之间的关系。它包括最终的结果和制定的过程。技术路线图具有高度概括、高度综合和前瞻性的基本特征。 技术路线图是一种结构化的规划方法,我们可以从三个方面归纳:它作为一个过程,可以综合各种利益相关者的观点,并将其统一到预期目标上来。同时,作为一种产品,纵向上它有力地将目标、资源及市场有机结合起来,并明确它们之间的关系和属性,横向上它可以将过去、现在和未来统一起来,既描述现状,又预测未来;作为一种方法,它可以广泛应用于技术规划管理、行业未来预测、国家宏观管理等方面。 技术路线图的缘起 技术路线图最早出现在美国汽车行业,汽车企业为降低成本要求供应商提供他们产品的技术路线图。20世纪70年代后期和80年代早期,摩托罗拉和康宁公司先后采用了绘制技术路线图的管理方法对产品开发任务进行规划。摩托罗拉主要用于技术进化和技术定位,康宁公司主要用于公司的和商业单位战略。继摩托罗拉和康宁公司之后,许多国际大公司,如微软、三星、朗讯公司,洛克-马丁公司和飞利普公司等都广泛应用这项管理技术。2000年英国对制造业企业的一项调查显示,大约有10%的公司承认使用了技术路线图方法,而且其中80%以上用了不止一次(C.J.Farrukh, R.Phaal, 2001)[1]。不仅如此,许多国家政府、产业团体和科研单位也开始利用这种方法来对其所属部门的技术进行规划和管理。 技术路线图真正的奠基人是摩托罗拉公司当时的CEO—Robert Galvin。当时,Robert Galvin在全公司范围内发动了一场绘制技术路线图的行动,主要目的是鼓励业务经理适当地关注技术未来并为他们提供一个预测未来过程的工具。这个工具为设计和研发工程师与做市场调研和营销的同事之间提供了交流的渠道,建立了各部门之间识别重要技术、传达重要技术的机制,使得技术为未来的产品开发和应用服务。 摩托罗拉的经验引起了全球企业高层管理者的注意。20世纪90年代后,企业对于技术路线图的兴趣空前高涨,技术路线图被迅速应用到各个领域,而技术路线图作为一种工具和方法也在不断发展、完善。目前,技术路线图已经是公认的技术经营和研究开发管理的基本工具之一。 [编辑]
技术路线 一般是指研究的准备,启动,进行,再重复,取得成果的过程。 多见于理工科和软科学。 技术路线是指申请者对要达到研究目标准备采取的技术手段、具体步骤及解决关键性问题的方法等在内的研究途径.合理的技术路线可保证顺利的实现既定目标.技术路线的合理性并不是技术路线的复杂性. 技术路线是指进行研究的具体程序的操作步骤,应尽可能详尽.每一步骤的关键点要阐述清楚并具有可操作性.如有可能,可以使用流程图或示意图加以说明,以达到一目了然的效果. 在开题报告中,可以先写技术路线,再写研究方法,再略述可行性分析。论文研究方法 (在一个课题研究过程中,根据不同的研究目的和要求,往往会用到两种以上方法。在开题报告中说明两~三个即可) 调查法 调查法是科学研究中最常用的方法之一。它是有目的、有计划、有系统地搜集有关研究对象现实状况或历史状况的材料的方法。调查方法是科学研究中常用的基本研究方法,它综合运用历史法、观察法等方法以及谈话、问卷、个案研究、测验等科学方式,对教育现象进行有计划的、周密的和系统的了解,并对调查搜集到的大量资料进行分析、综合、比较、归纳,从而为人们提供规律性的知识。 调查法中最常用的是问卷调查法,它是以书面提出问题的方式搜集资料的一种研究方法,即调查者就调查项目编制成表式,分发或邮寄给有关人员,请示填写答案,然后回收整理、统计和研究。 观察法 观察法是指研究者根据一定的研究目的、研究提纲或观察表,用自己的感官和辅助工具去直接观察被研究对象,从而获得资料的一种方法。科学的观察具有目的性和计划性、系统性和可重复性。在科学实验和调查研究中,观察法具有如下几个方面的作用:①扩大人们的感性认识。②启发人们的思维。③导致新的发现。 实验法 实验法是通过主支变革、控制研究对象来发现与确认事物间的因果联系的一种科研方法。其主要特点是:第一、主动变革性。观察与调查都是在不干预研
研究方法和技术路线 调查法 调查法是科学研究中最常用的方法之一。它是有目的、有计划、有系统地搜集有关研究对象现实状况或历史状况的材料的方法。调查方法是科学研究中常用的基本研究方法,它综合运用历史法、观察法等方法以及谈话、问卷、个案研究、测验等科学方式,对教育现象进行有计划的、周密的和系统的了解,并对调查搜集到的大量资料进行分析、综合、比较、归纳,从而为人们提供规律性的知识。 调查法中最常用的是问卷调查法,它是以书面提出问题的方式搜集资料的一种研究方法,即调查者就调查项目编制成表式,分发或邮寄给有关人员,请示填写答案,然后回收整理、统计和研究。 观察法 观察法是指研究者根据一定的研究目的、研究提纲或观察表,用自己的感官和辅助工具去直接观察被研究对象,从而获得资料的一种方法。科学的观察具有目的性和计划性、系统性和可重复性。在科学实验和调查研究中,观察法具有如下几个方面的作用:①扩大人们的感性认识。②启发人们的思维。③导致新的发现。 实验法 实验法是通过主支变革、控制研究对象来发现与确认事物间的因果联系的一种科研方法。其主要特点是:第一、主动变革性。观察与调查都是在不干预研究对象的前提下去认识研究对象,发现其中的问题。而实验却要求主动操纵实验条件,人为地改变对象的存在方式、变化过程,使它服从于科学认识的需要。第二、控制性。科学实验要求根据研究的需要,借助各种方法技术,减少或消除各种可能影响科学的无关因素的干扰,在简化、纯化的状态下认识研究对象。第三,因果性。实验以发现、确认事物之间的因果联系的有效工具和必要途径。 文献研究法 文献研究法是根据一定的研究目的或课题,通过调查文献来获得资料,从而全面地、正确地了解掌握所要研究问题的一种方法。文献研究法被子广泛用于各种学科研究中。其作用有:①能了解有关问题的历史和现状,帮助确定研究课题。②能形成关于研究对象的一般印象,有助于观察和访问。③能得到现实资料的比较资料。④有助于了解事物的全貌。 实证研究法 实证研究法是科学实践研究的一种特殊形式。其依据现有的科学理论和实践的需要,提出设计,利用科学仪器和设备,在自然条件下,通过有目的有步骤地操纵,根据观察、记录、测定与此相伴随的现象的变化来确定条件与现象之间的因果关系的活动。主要目的在于说明各种自变量与某一个因变量的关系。 定量分析法 在科学研究中,通过定量分析法可以使人们对研究对象的认识进一步精确化,以便更加科学地揭示规律,把握本质,理清关系,预测事物的发展趋势。
科研项目技术路线 一、"技术路线"的解释 1、技术路线是指申请者对要达到研究目标准备采取的技术手段、具体步骤及解决关键性问题的方法等在内的研究途径。技术路线在叙述研究过程的基础上,采用流程图的方法来说明,具有一目了然的效果。技术路线强调以研发项目为主线,完成项目研究内容的流程、顺序、各项研究内容间的内在联系和步骤。合理的技术路线可保证顺利的实现既定目标,技术路线的合理性并不是技术路线的复杂性。 2、技术路线是指进行研究的具体操作步骤,应尽可能详尽.每一步骤的关键点要阐述清楚并具有可操作性.如有可能,可以使用流程图或示意图加以说明,以达到一目了然的效果 二、技术路线编写格式(包括研究路线流程图和生产工艺流程图) (一)、研究路线流程图即产品开发流程图 1、做成树形图,按照研究内容流程来写,一般包括研究对象、方法、拟解决的问题,相互之间关系。 2、做成结构示意图:根据研究项目的子内容、研究顺序、相互关系,方法、解决问题做成结构示意图。 (二)、产品生产工艺流程图 三、示例
1、某产品开发流程图
2、研制途径流程示意图 3、工艺流程 本项目的工艺流程简单,生产成本低廉,概要如下:工艺流程图:
4、关键技术 ①经过抗菌肽氨基酸残基的突变体改造,获得了一个由原来抗革兰氏阳性菌又抗革兰氏阴性菌的抗菌肽改造成了只抗革兰氏阴性菌而对革兰氏阳性菌没有抑制作用的新型抗菌肽,在国际上首次研制成功了用动物肠道益生菌的嗜醋乳酸菌携带抗革兰氏阴性菌的抗菌肽基因工程菌,此基因工程菌能在动物胃肠道中分泌表达抗革兰氏阴性菌的抗菌肽,对其它革兰氏阳性菌无效,只要在动物饲料中添加这种活菌制剂,便可防治病原性革兰氏阴性菌的感染。 ②为了产品质量稳定性,我们需要确定一吨以上发酵罐的工艺条件,包括最佳发酵液配方、发酵过程中pH调节方法、温度控制、最适宜的发酵时间和搅拌形式等。 其它解释 1、什么是技术特征?技术方案一般由若干技术特征组成。技术特征可以大概分为三类:1、例如产品技术方案的技术特征可以是零件、部件、材料、器具、设备、装置的形状、结构、成分、尺寸等等; 2、方法技术方案的技术特征可以是工艺、步骤、过程,所涉及的时间、温度、压力以及所采用的设备和工具等等。 2、技术路线:是指对要达到研究目标准备采取的技术手段、具体步骤及解决关键性问题的方法等在内的研究途径。技术路线在叙述研究过程的基础上,可采用流程图的方法来说明,具有一目了然的效果。技术路线强调以科学问题为主线,完成项目研究内容的流程、顺序、各项研究内容间的内在联系和步骤。 3、研究方案包括有关方法、技术路线、实验手段、关键技术等。实
毕业论文的研究方法或技术路线 研究思路、研究方法、技术路线和实施步骤 1、研究什么?——怎样确定研究课题 一切科学研究始于问题——问题即课题;教学即研究(掌握方法很重要,否则就不是研究);进步与成果即成长。 教育科研课题主要来源于两大方面: A.实践来源——客观存在的或潜在的教育实际问题,教育教学实践本身存在的问题。 教育教学与其外部的矛盾(教师与家长、教师与学校、学校与社会、教育与社会发展)。 B.理论来源——现有教育理论所揭示的问题以及理论体系中的空白和矛盾点(例如《关于“信息技术与课程整合”的冷思考》一文产生的过程) 2、怎样进行研究课题的论证? 我们既然已选定了一个课题,我们就必须对这个课题的所有情况进行全面的了解。了解这个课题目前在国外、国内的研究情况,包括研究已取得的成果和存在的问题,了解这一课题所属的理论体系等等。对课题的全面了解,可以使我们在研究过程中少走弯路,确立研究的主攻方向,这就是我们常说的:“知己知彼,百战百胜”。 怎样对一个课题进行论证呢?论证一个课题主要是弄清如下几个问题: A.所要研究的问题是什么性质和类型的问题? B.要研究的问题具有什么现实意义?它的理论价值(即在理论上预计有哪些突破?) C.要研究的问题目前已有哪些研究成果?研究的方向是什么? D.要研究的问题所应具备的条件分析。 E.课题研究的策略和步骤如何? F.课题研究的成果及其表现形式有哪些? 3、教育课题研究的基本方法有: ⑴ 观察法⑵ 调查法⑶ 测验法⑷ 行动研究法⑸ 文献法⑹ 经验总结法⑺ 个案研究法⑻ 案例研究法 ⑼ 实验法(在一个课题研究过程中,根据不同的研究目的和要求,往往会用到两种以上方法) 3.1 观察法:为了了解事实真相,从而发现某种现象的本质和规律。 观察法的步骤:观察法的实施分为以下三个步骤,步骤之一就是进行观察研究的设计,此步骤可分为如下几个方面: 3.1.1 作大略调查和试探性观察。 这一步工作的目的不在于搜集材料,而在于掌握基本情况,以便能正确地计划整个观察过程。例如:要观察某一教师的教学工作,便应当预先到学校大致了解这位教师的工作情况,学生的情况,有关的环境和条件等等。这可以通过跟教
调查报告的研究方案及技术路线 一、研究背景及意义分析 1、研究背景 随着我国蔬菜生产力的提高,蔬菜产业成为农民增收的重要产业,蔬菜供应也由短缺到供需平衡,并出现结构性过剩,大大提高了人们的生活水平。但我国蔬菜质量并未同步跟进,由于农药、化肥等农业投入品的过度不合理使用,使得蔬菜中农药等有害物质残留超标问题突出。蔬菜质量安全问题不仅影响着蔬菜产业的持续发展,影响农民增收,也影响着大众消费安全,影响社会和谐定。基于此,本课题拟对呈贡区蔬菜生产过程中农药的使用情况做出调查,以了解当前呈贡区蔬菜生产中存在的问题,并提出相应对策及建议。 2、研究意义 蔬菜质量安全工作,是一项涉及生态、环境、资源、经济、人口、社会等问题的系统工程,因此,选择“呈贡县蔬菜质量安全控制的研究”,发展无公害蔬菜产销事业,顺应了当前的国际、国内形势,是农业、农村经济发展到现阶段的客观需要,对于推进农业结构调整,全面提高蔬菜产品竞争力,切实增加农民收入具有较强的现实意义。 本研究试图通过调查呈贡县蔬菜生产过程中存在的问题,分析造成蔬菜质量安全问题的原因及对农民增收的影响,找出主要因素,重点从蔬菜生产过程中的投入品使用、管理等生产源头方面探索有效的蔬菜质量安全控制措施,以为推动呈贡区蔬菜无害化生产、提升蔬菜产品质量、保障消费安全、增强呈贡蔬菜的市场竞争力、实现农业增效和农民增收提供有益的借鉴。 二调查方案设计、研究内容及技术路线 1、研究内容 本论文的研究内容主要包括以下几部分: (1)、呈贡县蔬菜质量存在的问题及原因。通过对呈贡县农药、肥料等农业投入品经营使用情况、违禁农药销售使用情况、农药残留超标情况等的调查,分析存在问题的主要因素。 (2)、呈贡县蔬菜质量安全管理现状及存在问题。调查分析呈贡县蔬菜质量安全管理现行体制、采取的措施、制度、蔬菜标准化生产管理情况,目前管理取得的成效及存在的漏洞。 (3)、进一步保证和加强呈贡县蔬菜安全生产的对策建议。通过前文对呈贡县蔬菜安全生产中存在的问题及原因的分析,对如何保证蔬菜的安全生产给出针对性的对策建议。 2、研究方法 本研究立足呈贡市实际,研究方法主要有: (1)、实地调研法 ①对呈贡县蔬菜生产全过程进行实地调查,采用问卷法和访谈法获得第一手资料,了解农户农药、化肥使用的品种、数量、安全间隔期、使用次数等情况,了解当前蔬菜主要种植品种。 (2)、定性分析和定量分析相结合。 在实地调查的基础上,对呈贡县蔬菜质量状况及蔬菜质量安全控制现状进行
磷酸铁锂的生产工艺与技术路线选择锂离子电池作为一种高性能的二次绿色电池,具有高电压、高能量密度(包括体积能量、质量比能量)、低的自放电率、宽的使用温度范围、长的循环寿命、环保、无记忆效应以及可以大电流充放电等优点。锂离子电池性能的改善,很大程度上决定于电极材料性能的改善,尤其是正极材料。目前研究最广泛的正极材料有LiCoO2、LiNiO2以及LiMn2O4等,但由于钴有毒且资源有限,镍酸锂制备困难,锰酸锂的循环性能和高温性能差等因素,制约了它们的应用和发展。因此,开发新型高能廉价的正极材料对锂离子电池的发展至关重要。 1997年,Padhi等报道了具有橄榄石结构的磷酸铁锂(LiFePO4)能够可逆地嵌脱锂,且具有比容量高、循环性能好、电化学性能稳定、价格低廉等特点,是首选的新一代绿色正极材料,特别是作为动力锂离子电池材料。磷酸铁锂的发现引起了国内外电化学界不少研究人员的关注,近几年,随着锂电池的越来越广的应用,对LiFePO4的研究越来越多。 2.1 磷酸铁锂的结构和性能 磷酸铁锂(LiFePO4)具有橄榄石结构,为稍微扭曲的六方密堆积,其空间群是P mnb型,晶型结构如图2.1所示。 图2.1 磷酸铁锂的空间结构图 LiFePO4由FeO6八面体和PO4四面体构成空间骨架,P占据四面体位置,而Fe和Li则填充在八面体空隙中,其中Fe占据共角的八面体位置,Li则占据共边的八面体位置。晶格一个FeO6八面体与两个FeO6八面体和一个PO4四面
体共边,而PO4四面体则与一个FeO6八面体和两个LiO6八面体共边。由于近乎六方堆积的氧原子的紧密排列,使得锂离子只能在二维平面上进行脱嵌,也因此具有了相对较高的理论密度(3.6g/cm3)。在此结构中,Fe2+/Fe3+相对金属锂的电压为3.4V,材料的理论比容量为170mA·h/g。在材料中形成较强的P-O-M 共价键,极大地稳定了材料的晶体结构,从而导致材料具有很高的热稳定性。 Wang等对LiFePO4的电化学性能做了详细的分析,图2.2是LiFePO4的循环载荷伏安图,在C-V图中形成两个峰,在阳极扫描时Li+从Li x FePO4结构中脱出,在3.52V形成氧化峰;当在4.0~3.0扫描时Li+嵌入到Li x FePO4结构中,相应的在3.32V形成还原峰;C-V曲线中的氧化还原峰表明在L iFePO4电极上发生着可逆的锂离子嵌脱反应。 图2.2 磷酸铁锂的循环载荷伏安图 2.2 磷酸铁锂的制备方法及研究 LiFePO4正极材料的性能在一定程度上取决于材料的形态、颗粒的尺寸以及原子排列,因此制备方法尤为重要。目前主要有固相法和液相法,其中固相法包括高温固相反应法、碳热还原法、微波合成法和脉冲激光沉积法;液相法包括溶胶·凝胶法、水热合成法、沉淀法以及溶剂热合成法等。 2.2.1 固相法 2.2.1.1 高温固相反应法… 2.2.1.2 碳热还原法 碳热还原法也是固相法中的一种,是比较容易工业化的合成方法,以廉价的
四、研究方法及路线 (一)研究方法 1.文献资料法:利用图书馆、档案馆及互联网等广泛查相关的文献资料,加以分析 与研究。 2.文本分析法。以《文化产业振兴规划》、《文化科技创新工程纲要》、《本市“十二五”文化发展规划》、《本市促进文化大发展大繁荣的实施意见》、《关于打造“文化五城”建设文化强市的意见》等权威文本为研究对象,通过分析研究法律整体,深刻理解精神实质,分析其中的条 文关于文化科技融合发展的相关规定及发展文化产业的具体要求。 3.实地调查法。为更好地了解本市文化科技融合发展的真实现状,在三区各选择3 个能体现文化科技融合发展的主体公园、游乐园、旅游景区、产业园区及科技馆等进行现场 观察和询问,并做好记录。 4.访谈法。计划选择20 名专家学者、知名企业家及科技、文化、旅游及宣传等相关 部门的政府工作人员,针对本市文化科技融合发展的相关问题分别进行半小时左右的访谈, 并根据情况,召开 2 到 3 次的小型座谈会。 5.案例分析法。对国内外省市及企业文化科技融合发展成功的典型案例进行持续追 踪调查,进行剖析,深入研究,总结经验。 6.比较研究法。比较研究美、日、德等发达国家文化科技融合发展的做法与特点, 总结成功经验,得出启示,以供借鉴。 7.统计分析法。统计 2002-2008 年本市居民家庭人均文化消费支出数额及其占消费支 出的比重,并与发达城市对比,分析本市居民文化消费水平和质量提升的发展空间。 8.分析归纳法。研究分析查阅的文献资料,归纳总结其研究内容并合理分类;根据 比较研究及案例分析的结果,总结归纳国内外文化科技融合发展中好的做法和经验。 (二)技术路线 1.研究的总体思路 首先,根据本市 2013 软科学研究计划申报指南,结合研究条件和自身研究优势,确 定文化科技融合发展战略研究这一选题。其次,查阅大量相关文献和权威政策文本,了解国内 外研究现状,奠定课题研究的理论基础。再次,选取文化科技融合发展方面有成功经验的美国、 日本和德国,从理念、体制机制、路径、模式及政策等方面进行比较研究,总结他们的经验和 做法,得出一些对我国有益的启示;同时,选取北京海淀、上海张江、深圳华强文化科技集团、 美国的电影、日本的桑蚕与丝绸以及德国柏林的创意经济产业发展等六个代表 性的案例做分析研究,总结其成功的发展经验,为本市文化科技融合发展战略构想提供参考。 在此基础上,从动力机制、组织机构、管理制度、监督机制、激励机制及人才培养机制等方 面探索文化科技融合发展的长效体制机制。然后,运用访谈法、文献资料法、文本分析法等 研究本市文化科技融合发展的现状及存在的问题。最后,结合比较研究、案例研究得出的总 结与启示,针对具体问题,提出本市文化科技融合发展的战略构想。 2.技术路线图 (见下页)
磷酸铁锂的生产工艺与技术路线选择 锂离子电池作为一种高性能的二次绿色电池,具有高电压、高能量密度(包括体积能量、质量比能量)、低的自放电率、宽的使用温度范围、长的循环寿命、环保、无记忆效应以及可以大电流充放电等优点。锂离子电池性能的改善,很大程度上决定于电极材料性能的改善,尤其是正极材料。目前研究最广泛的正极材料有LiCoO2、LiNiO2以及LiMn2O4等,但由于钴有毒且资源有限,镍酸锂制备困难,锰酸锂的循环性能和高温性能差等因素,制约了它们的应用和发展。因此,开发新型高能廉价的正极材料对锂离子电池的发展至关重要。 1997年,Padhi等报道了具有橄榄石结构的磷酸铁锂(LiFePO4)能够可逆地嵌脱锂,且具有比容量高、循环性能好、电化学性能稳定、价格低廉等特点,是首选的新一代绿色正极材料,特别是作为动力锂离子电池材料。磷酸铁锂的发现引起了国内外电化学界不少研究人员的关注,近几年,随着锂电池的越来越广的应用,对LiFePO4的研究越来越多。 2.1 磷酸铁锂的结构和性能 磷酸铁锂(LiFePO4)具有橄榄石结构,为稍微扭曲的六方密堆积,其空间群是P mnb型,晶型结构如图2.1所示。 图2.1 磷酸铁锂的空间结构图 LiFePO4由FeO6八面体和PO4四面体构成空间骨架,P占据四面体位置,而Fe和Li则填充在八面体空隙中,其中Fe占据共角的八面体位置,Li则占据共边的八面体位置。晶格一个FeO6八面体与两个FeO6八面体和一个PO4四面体共边,而PO4四面体则与一个FeO6八面体和两个LiO6八面体共边。由于近乎六方堆积的氧原子的紧密排列,使得锂离子只能在二维平面上进行脱嵌,也因此具有了相对较高的理论密度(3.6g/cm3)。在此结构中,Fe2+/Fe3+相对金属锂的电压为3.4V,材料的理论比容量为170mA·h/g。在材料中形成较强的P-O-M共价键,极大地稳定了材料的晶体结构,从而导致材料具有很高的热稳定性。
研究思路、研究方法、技术路线和实施步骤 2009年04月04日星期六 12:35 1、研究什么?——怎样确定研究课题 一切科学研究始于问题——问题即课题;教学即研究(掌握方法很重要,否则就不是研究);进步与成果即成长。 教育科研课题主要来源于两大方面: A.实践来源——客观存在的或潜在的教育实际问题,教育教学实践本身存在的问题。 教育教学与其外部的矛盾(教师与家长、教师与学校、学校与社会、教育与社会发展)。 B.理论来源——现有教育理论所揭示的问题以及理论体系中的空白和矛盾点(例如《关于“信息技术与课程整合”的冷思考》一文产生的过程) 2、怎样进行研究课题的论证? 我们既然已选定了一个课题,我们就必须对这个课题的所有情况进行全面的了解。了解这个课题目前在国外、国内的研究情况,包括研究已取得的成果和存在的问题,了解这一课题所属的理论体系等等。对课题的全面了解,可以使我们在研究过程中少走弯路,确立研究的主攻方向,这就是我们常说的:“知己知彼,百战百胜”。 怎样对一个课题进行论证呢?论证一个课题主要是弄清如下几个问题: A.所要研究的问题是什么性质和类型的问题? B.要研究的问题具有什么现实意义?它的理论价值(即在理论上预计有哪些突破?) C.要研究的问题目前已有哪些研究成果?研究的方向是什么? D.要研究的问题所应具备的条件分析。 E.课题研究的策略和步骤如何? F.课题研究的成果及其表现形式有哪些? 3、教育课题研究的基本方法有: ⑴ 观察法⑵ 调查法⑶ 测验法⑷ 行动研究法⑸ 文献法⑹ 经验总结法⑺ 个案研究法⑻ 案例研究法 ⑼ 实验法(在一个课题研究过程中,根据不同的研究目的和要求,往往会用到两种以上方法) 3.1 观察法:为了了解事实真相,从而发现某种现象的本质和规律。 观察法的步骤:观察法的实施分为以下三个步骤,步骤之一就是进行观察研究的设计,此步骤可分为如下几个方面: 3.1.1 作大略调查和试探性观察。 这一步工作的目的不在于搜集材料,而在于掌握基本情况,以便能正确地计划整个观察过程。例如:要观察某一教师的教学工作,便应当预先到学校大致了解这位教师的工作情况,学生的情况,有关的环境和条件等等。这可以
目前国内外研究者尚未就如何提高电子农务平台的服务能力进行全面的研究,本项目的研究抓住项目的推广和实践这二个重点,使人才培养目标和实践的具体内容、实践方法与要求有机统一,“学”与“做”相统一。 项目的研究方法主要采用问卷调查法,文献法、个案研究法等多种研究方法,同时注重我国电子农务的背景分析,注意吸收社会学、经济学、哲学、农业学等相关学科的最新研究成果,并注重对大众群体的调研,找到符合农业户的改革发展道路,构建和谐的社会新农村与专业相挂钩的新模式。 采用问卷调查法主要是通过对买家电子农务、卖家电子农务、第三方电子农务进行调研,了解目前农业市场的缺陷与不足。利用网络平台,实地考察等多种方法对农户进行回访,收集实践性的反馈意见:着重加强对新平台的推广,让农户朋友了解新的农业模式,不断完善实践技能的针对性。 运用文献法,就是利用期刊、网络及图书资料把国内外电子农业改革的相关理论与实践研究成果进行搜集和积累,促使研究能够超越国内外已有的研究,使研究具有前沿性与全球视野,找到最适合农户朋友们的农业模式。 采用个案研究法,利用个别典型案例启发并引导教师与学生项目实践,并通过学生在实际工作中的实践水平、动手能力与项目完成程度来检验实践,本项目的研究要源于实践,高于实践,服务于实践,学习与实践完全统一。 技术路线: 查找相关文献——>找出问题存在的方向——>设计问卷——>探测性调研——>修改问卷——>征询专家——>问卷确定——>收集市场信息——>分类整理、加工、分析市场信息——>形成调研报告——>完成 进度安排: 第一阶段:(2012年4月-2012年6月) 全面搜索相关书籍、文献资料、网络资料数据; 学习相关科研知识、科研技能、提高科研基本素质;
技术路线图 技术路线图是指应用简洁的图形、表格、文字等形式描述技术变化的步骤或技术相关环节之间的逻辑关系。它能够帮助使用者明确该领域的发展方向和实现目标所需的关键技术,理清产品和技术之间的关系。它包括最终的结果和制定的过程。技术路线图具有高度概括、高度综合和前瞻性的基本特征。 技术路线图是一种结构化的规划方法,我们可以从三个方面归纳:它作为一个过程,可以综合各种利益相关者的观点,并将其统一到预期目标上来。同时,作为一种产品,纵向上它有力地将目标、资源及市场有机结合起来,并明确它们之间的关系和属性,横向上它可以将过去、现在和未来统一起来,既描述现状,又预测未来;作为一种方法,它可以广泛应用于技术规划管理、行业未来预测、国家宏观管理等方面。 [编辑] 技术路线图的缘起 技术路线图最早出现在美国汽车行业,汽车企业为降低成本要求供应商提供他们产品的技术路线图。20世纪70年代后期和80年代早期,摩托罗拉和康宁公司先后采用了绘制技术路线图的管理方法对产品开发任务进行规划。摩托罗拉主要用于技术进化和技术定位,康宁公司主要用于公司的和商业单位战略。继摩托罗拉和康宁公司之后,许多国际大公司,如微软、三星、朗讯公司,洛克-马丁公司和飞利普公司等都广泛应用这项管理技术。2000年英国对制造业企业的一项调查显示,大约有10%的公司承认使用了技术路线图方法,而且其中80%以上用了不止一次(C.J.Farrukh, R.Phaal, 2001)[1]。不仅如此,许多国家政府、产业团体和科研单位也开始利用这种方法来对其所属部门的技术进行规划和管理。 技术路线图真正的奠基人是摩托罗拉公司当时的CEO—Robert Galvin。当时,Robert Galvin 在全公司范围内发动了一场绘制技术路线图的行动,主要目的是鼓励业务经理适当地关注技术未来并为他们提供一个预测未来过程的工具。这个工具为设计和研发工程师与做市场调研和营销的同事之间提供了交流的渠道,建立了各部门之间识别重要技术、传达重要技术的机制,使得技术为未来的产品开发和应用服务。 摩托罗拉的经验引起了全球企业高层管理者的注意。20世纪90年代后,企业对于技术路线图的兴趣空前高涨,技术路线图被迅速应用到各个领域,而技术路线图作为一种工具和方法也在不断发展、完善。目前,技术路线图已经是公认的技术经营和研究开发管理的基本工具之一。 [编辑] 技术路线图的研究现状[2]
技术路线的写法及示例-标准化文件发布号:(9456-EUATWK-MWUB-WUNN-INNUL-DDQTY-KII
技术路线的写法及示例 技术路线一般是指研究的准备,启动,进行,再重复,取得成果的过程。 多见于理工科和软科学。 技术路线是指申请者对要达到研究目标准备采取的技术手段、具体步骤及解决关键性问题的方法等在内的研究途径。合理的技术路线可保证顺利的实现既定目标。技术路线的合理性并不是技术路线的复杂性。 技术路线是指进行研究的具体程序的操作步骤,应尽可能详尽.每一步骤的关键点要阐述清楚并具有可操作性。如有可能,可以使用流程图或示意图加以说明,以达到一目了然的效果。 1、研究背景 研究背景即提出问题,阐述研究该课题的原因。研究背景包括理论背景和现实需要。还要综述国内外关于同类课题研究的现状:①人家在研究什么、研究到什么程度?②找出你想研究而别人还没有做的问题。③他人已做过,你认为做得不够(或有缺陷),提出完善的想法或措施。④别人已做过,你重做实验来验证。 2、目的意义 目的意义是指通过该课题研究将解决什么问题(或得到什么结论),而这一问题的解决(或结论的得出)有什么意义。有时将研究背景和目的意义合二为一。 3、成员分工 成员分工应是指课题组成员在研究过程中所担负的具体职责,要人人有事干、个个担责任。组长负责协调、组织。 4、实施计划
实施计划是课题方案的核心部分,它主要包括研究内容、研究方法和时间安排等。研究内容是指可操作的东西,一般包括几个层次:⑴研究方向。⑵子课题(数目和标题)。⑶与研究方案有关的内容,即要通过什么、达到什么等等。研究方法要写明是文献研究还是实验、调查研究若是调查研究是普调还是抽查如果是实验研究,要注明有无对照实验和重复实验。实施计划要详细写出每个阶段的时间安排、地点、任务和目标、由谁负责。若外出调查,要列出调查者、调查对象、调查内容、交通工具、调查工具等。如果是实验研究,要写出实验内容、实验地点、器材。实施计划越具体,则越容易操作。 5、可行性论证 可行性论证是指课题研究所需的条件,即研究所需的信息资料、实验器材、研究经费、学生的知识水平和技能及教师的指导能力。另外,还应提出该课题目前已做了哪些工作,还存在哪些困难和问题,在哪些方面需要得到学校和老师帮助等等。 6、预期成果及其表现形式 预期成果一般是论文或调查(实验)报告等形式。成果表达方式是通过文字、图片、实物和多媒体等形式来表现。 这部分要写课题的实施方案,也就是你计划通过什么样的方法来实现你的课题的研究任务,换而言之,需要您给出一个比较可行的(理论上即可)设计方案来。 研究思路、研究方法、技术路线和实施步骤 1、研究什么— —怎样确定研究课题
SNP开发/验证的研究方法和技术路线 1分子标记: 分子标记,我想这部分是我们分子标记组最核心的任务。现在,我们没有任何可用的标记检测我们的定位材料。即使想要验证已经定位的QTLs,我们也需要相对应的区间的分子标记,尤其是SNP标记。 1.1 全基因组SNP—Affymetrix芯片: 一套完整的全基因组的SNP芯片,相对于Douglas体系,其操作简单,高通量。可以直接对定位群体进行初定位的扫描或是对育种材料的背景进行分析。在国家玉米改良中心,有一套3k的Illumina芯片,就是用来对玉米材料进行高通量检测,基因型检测结果通常可以用来QTLs初定位,育种材料的群体划分与纯度鉴定以及低密度的关联分析等。在此,我建议我们应该开发一套番茄基因型检测的芯片。 目前,只是查找到Illumina芯片有一套全基因SNP信息,包含7,720条探针。而Affymetrix公司目前并没有相应的产品。但是通过跟Affymetrix公司了解,可以利用Illumina芯片已有的结果进行开发。 番茄目前测序结果显示其全基因组大小为~760Mb,而玉米为~2,500Mb,但是他们包括的基因数目~30,000个,整体情况相近。另外,番茄作为自交植物,其LD的衰减值应该更大,有效的历史重组会更少,遗传多样性低。因此,综合考虑,我建议我们可以开发~3k芯片,应该可以满足大多数研究材料、育种材料的基因型检测需求。虽然目前下一代测序技术蓬勃发展,但是对于用于基因型检测来讲,其数据分析与成本相对于芯片都要更复杂和更高。总之,我们番茄处于刚刚发展阶段,我认为就基因型检测方面,芯片有其很高的应用价值。即使像玉米,这样测序技术发展很多年的材料,芯片技术也在应用。 1.2全基因组SNP—Douglas: 当用Affymetrix芯片检测鉴定完番茄基因型并完成基因型分析之后,1)对于优良的QTLs或是基因,我们可以直接选择覆盖整个区间的分子标记运行Douglas系统进行分子标记辅助育种,2)对于需要进一步验证的QTLs,我们也
国内外技术路线图方法一、技术路线图的绘制方法目前技术路线图的绘制方法一共有三种即基于专家的方法ExpertBased Approach、基于工作组的方法workshopbased Approach和基于计算机的方法Computerbased Approach当然还有一种方法就是将三种方法结合起来。基于专家的方法就是将一组或几组专家召集到一起确定技术路线图中的节点以及节点之间的属性。例如美国半导体协会的技术路线图绘制就先后有600多名科学家参与其中。该方法实际上就是一种结构化的“头脑风暴法”它依据专家头脑中的知识和经验主观的判断出技术路线图中各种节点或连接的定量与定性属性。基于工作组的方法指由来自不同部门的人员组成团队分成几个Workshop利用参与者的知识和经验确定技术路线图节点和连接的属性。如行业路线图就由行业政府学术界研究人员及其他利益相关者参加共同绘制。基于计算机的方法就是通过对科学、技术、工程和最终产品的大型文本数据库进行计算机分析这些数据库可能包括学术论文、研究报告、信件等等。利用各种常用的方法如引文分析确定技术、工程以及产品的研究领域同时估计并量化他们之间的相关性而且他们与其他领域之间的关系也可以确定并量化出来。与上述两种方法相比基于计算机的方法更加客观。摩托罗拉是最早制定技术路线图的公司。威尔亚德和麦克莱利研究了该公司早期的技术路线图认为摩托罗拉制定的技术路线
图的内容主要包括对摩托罗拉企业的描述、企业技术预测、技术路线矩阵、产品质量和技术进步、技术资源分配、专利组合、产品描述和现状报告及概略图、少数派意见等8个方面并且列出了8个与上述8个方面相关的图形。巴克尔D.Baker和史密斯H.smith认为制作技术路线图共有两种思路:1、从上往下正推法t叩一down。2、从下往上倒推法bottom 一uP。美国圣蒂亚sandia国家实验室总结了一套一般性技术路线图的制作方法其步骤包括三个阶段:1、前期准备。2、路线图的开发。3、随后的行动。二、国外技术路线图的绘制过程2.1 行业技术路线图绘制模板澳大利亚工业部曾对国际上不同的技术路线图绘制方法作了总结提出了一个技术路线图绘制模板具体步骤如图。1确定技术路线图的需求和收益这可以由政府和行业联合确定当然也可以由行业独自完成。2确定路线图的参与者和领导领导的作用是领导技术路线图的绘制并支持参与者的工作。3确定技术路线图绘制的资源首先基于技术路线图的范围、使用目的、时间规划和技术细节确定资源需求。接着路线图的领导者与政府一起落实资金来源。资金来源于政府的相关计划以及相关行业。4确立技术路线图的绘制过程就技术路线图绘制的方法达成一致甄选专家顾问推动技术路线图的绘制并设定愿景1确定需求和收益是否需要绘制技术路线图它会带来什么好处2确定路线图的参与者和领导行业内谁将领导技术
研究方法和研究技术路线
1论文研究方法 调查法 调查法是科学研究中最常用的方法之一。它是有目的、有计划、有系统地搜集有关研究对象现实状况或历史状况的材料的方法。调查方法是科学研究中常用的基本研究方法,它综合运用历史法、观察法等方法以及谈话、问卷、个案研究、测验等科学方式,对教育现象进行有计划的、周密的和系统的了解,并对调查搜集到的大量资料进行分析、综合、比较、归纳,从而为人们提供规律性的知识。 调查法中最常用的是问卷调查法,它是以书面提出问题的方式搜集资料的一种研究方法,即调查者就调查项目编制成表式,分发或邮寄给有关人员,请示填写答案,然后回收整理、统计和研究。 观察法 观察法是指研究者根据一定的研究目的、研究提纲或观察表,用自己的感官和辅助工具去直接观察被研究对象,从而获得资料的一种方法。科学的观察具有目的性和计划性、系统性和可重复性。在科学实验和调查研究中,观察法具有如下几个方面的作用:①扩大人们的感性认识。②启发人们的思维。③导致新的发现。 实验法 实验法是通过主支变革、控制研究对象来发现与确认事物间的因果联系的一种科研方法。其主要特点是:第一、主动变革性。观察与调查都是在不干预研究对象的前提下去认识研究对象,发现其中的问题。而实验却要求主动操纵实验条件,人为地改变对象的存在方式、变化过程,使它服从于科学认识的需要。第二、控制性。科学实验要求根据研究的需要,借助各种方法技术,减少或消除各种可能影响科学的无关因素的干扰,在简化、纯化的状态下认识研究对象。第三,因果性。实验以发现、确认事物之间的因果联系的有效工具和必要途径。 文献研究法 文献研究法是根据一定的研究目的或课题,通过调查文献来获得资料,从而全面地、正确地了解掌握所要研究问题的一种方法。文献研究法被子广泛用于各种学科研究中。其作用有:①能了解有关问题的历史和现状,帮助确定研究课题。②能形成关于研究对象的一般印象,有助于观察和访问。③能得到现实资料的比较资料。④有助于了解事物的全貌。 实证研究法 实证研究法是科学实践研究的一种特殊形式。其依据现有的科学理论和实践的需要,提出设计,利用科学仪器和设备,在自然条件下,通过有目的有步骤地操纵,根据观察、记录、测定与此相伴随的现象的变化来确定条件与现象之间的因果关系的活动。主要目的在于说明各种自变量与某一个因变量的关系。 实证研究又分为:定性研究和定量研究。
“技术路线” 因为要申报全国规划办的国家级规划课题,于是需要讨论表格中给“研究思路、研究方法、技术路线和操作步骤”一栏。这四个概念相对独立,却又紧密联系,因此填写困难,特别是对于基层学校和一线教师来说,更是不知道如何下手。于是,我上网搜索了一种解释和一个案例,发至网上,以供参考。 经过研究,我觉得,这四个概念表现的四个方面应该综合起来表述,也就是说把“研究思路、研究方法、技术路线和操作步骤”放在一起写,也就是说,“技术路线”也需要通过“研究思路”“研究方法”“操作步骤”才能表现,也只有这样才能重复和交叉。 什么是“技术路线”: 技术路线一般是指研究的准备,启动,进行,再重复,取得成果的过程。多见于理工科和软科学。 技术路线是指申请者对要达到研究目标准备采取的技术手段、具体步骤及解决关键性问题的方法等在内的研究途径。 合理的技术路线可保证顺利的实现既定目标。技术路线的合理性并不是技术路线的复杂性。技术路线是指进行研究的具体程序的操作步骤,应尽可能详尽.每一步骤的关键点要阐述清楚并具有可操作性。 如有可能,可以使用流程图或示意图加以说明,以达到一目了然的效果。 “技术路线”的案例: 本课题(基础教育中学习评价的研究)的研究思路、研究方法、技术路线和实施步骤建议 本课题的研究从哲学的层面来说,将采取由具体到抽象、由抽象到具体的思维方法开展研究。具体的方法还有: ⑴文献研究法
查阅国内外有关文献,了解和掌握本课题相关的研究动向,供借鉴。 ⑵调查研究法 我们将主要运用调查法研究目前基础教育中学习评价实施的现状、效果、功能及其成因,掌握第一手材料,为课题研究提供充足的事实依据。 ⑶经验总结法 运用科学的方法,对相关的教学实践经验进行分析概括,较全面、深入系统地揭示经验的实质,使之上升到理性高度,找到可以运用和遵循的规律性的东西。 ⑷行动研究法 运用行动研究法研究新课程条件下学习评价目标的确立、内容设置、状态评估、成果评价、评价者的地位。使学习评价的总体设计符合科学性、开放性、探究性、人本化的原则。为此将采取:a.个案研究法我们将主要运用个案研究法研究被评价主体的差异性,对个别学生、实验班级、某一年级、某一类群体等典型的个案进行深入全面的调查和剖析,力图揭示其中的某些规律和本质。b.问卷调查法通过网络平台,根据研究的目的,设置必要的提问方法,要求被调查者回答,以此来掌握学生对评价的认知状况,并及时反馈评价设计的效度。这种方法是广为采用的一种研究方法。我们将采用的问卷法,其回答形式主要有自由记述法、选择法、分类法、等级法等。C.观察研究法在自然的状态下,有目的、有计划地观察学生在学习过程性评价中个体行为变化的外部表现,以了解其发展情况。根据观察的目的和任务的不同,可以进行长期观察,也可以进行短期观察;可以有选择地进行重点观察,也可以进行全面观察。必须注意的是观察法应在自然状态下进行,不能使被观察者觉察自己是观察的对象。要善于做好观察记录,以便事后进行整理分析。d.谈话研究法研究者初步拟定一定的问题同被评价主体进行谈话,以了解其自我对评价的看法,了解其在被评价过程中的心理和观念的变化。e.建立网络个人评价档案袋 f.其他可供借鉴的研究方法。